Abstract
One hundred twenty six doubled-haploid (DH) rice lines were evaluated in nine diverse Asian environments to reveal the genetic basis of genotype × environment interactions (GEI) for plant height (PH) and heading date (HD). A subset of lines was also evaluated in four water-limited environments, where the environmental basis of G × E could be more precisely defined. Responses to the environments were resolved into individual QTL × environment interactions using replicated phenotyping and the mixed linear-model approach. A total of 37 main-effect QTLs and 29 epistatic QTLs were identified. On average, these QTLs were detectable in 56% of the environments. When detected in multiple environments, the main effects of most QTLs were consistent in direction but varied considerably in magnitude across environments. Some QTLs had opposite effects in different environments, particularly in water-limited environments, indicating that they responded to the environments differently. Inconsistent QTL detection across environments was due primarily to non- or weak-expression of the QTL, and in part to significant QTL × environment interaction effects in the opposite direction to QTL main effects, and to pronounced epistasis. QTL × environment interactions were trait- and gene-specific. The greater GEI for HD than for PH in rice were reflected by more environment-specific QTLs, greater frequency and magnitude of QTL × environment interaction effects, and more pronounced epistasis for HD than for PH. Our results demonstrated that QTL × environment interaction is an important property of many QTLs, even for highly heritable traits such as height and maturity. Information about QTL × environment interaction is essential if marker-assisted selection is to be applied to the manipulation of quantitative traits.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Most plant traits are quantitative in nature and are influenced by many genes or quantitative trait loci (QTLs). Quantitative traits are also influenced by the environment and tend to show varied degrees of genotype × environment interactions (GEIs). GEIs occur when two or more genotypes perform differently in different environments, and are thus described as differential genotypic sensitivities to environments (Falconer 1981). Plants, particularly self-pollinated plants, tend to show a high level of GEIs that allow better adaptation to their changing environments and the maintenance of genetic variation in populations (Jain and Marshall 1967). In plant breeding, GEIs must be considered to identify superior and stable genotypes when breeding materials are tested in different environments. Because of their importance in plant breeding and evolution, GEIs of quantitative traits have been the subject of extensive investigations (cf. Baker 1988; Cooper and Hammer 1996). Classical studies on GEIs using segregating plant populations have been few, but they have yielded valuable information regarding the importance of GEIs for quantitative traits (Mather and Jinks 1982).
DNA markers and high-density genetic maps of major crops developed since the late 1980s have facilitated efforts to understand the genetic basis of quantitative traits through QTL mapping. Main-effect QTLs (M-QTLs) affecting a wide range of agronomic traits in many plant species have been reported (cf. Paterson 1995; Georges 1997; Stuber 1997). Despite the technical difficulties, QTL × environment interaction has been revealed by inconsistent detection and variable effects of M-QTLs across environments in tomato (Paterson et al. 1991), maize (Bubeck et al. 1993; Veldboom and Lee 1996; Austin and Lee 1998; Crossa et al. 1999; Jiang et al. 1999), barley (Hayes et al. 1993), and rice (Lu et al. 1996; Zhuang et al. 1997). In soybean, QTLs were inconsistent across environments for plant height and lodging resistance, but consistent for maturity, indicating that QTL × environment interaction is trait dependent (Lee et al. 1996).
In most previous studies, QTL × environment interaction was inferred by comparing QTLs detected in different environments. This inference about the presence of QTL × environment interaction has two shortcomings. First, individual QTL × environment interaction effects were not properly quantified, largely because of a lack of appropriate analytical methodology. Second, because only a single threshold was used in most QTL mapping studies, it remains unknown whether inconsistent QTL detection was due to the type-II error arising from the use of single thresholds or to true differential trait expression across environments. Using composite interval mapping, Tinker et al. (1996) were able to detect considerable QTL × environment interaction for seven agronomic traits in two barley crosses, even though many of the detected QTLs were highly consistent across environments. Yan et al. (1999) reported significant QTL × environment interaction associated with common QTLs for plant-type traits in a rice doubled-haploid (DH) population in two different environments. Unfortunately, none of these studies was able to dissect GEIs in the presence of epistasis, which underlies complex phenotypes (Yu et al. 1997; Li et al. 2001, Luo et al. 2001). Thus, many important questions regarding the genetic aspects of GEIs remain largely unanswered. These include the following: Which QTLs are more environment-specific? How frequently do QTLs interact with environments? If present, how important are QTL × environment interaction effects as compared to QTL main effects?
This manuscript describes a large study of QTL × environment interaction in a diverse range of environments for the genetic control of two highly heritable traits, plant height (PH) and heading date (HD), in a well-known doubled-haploid (DH) rice population. The primary objective of this work is to quantify M-QTLs and epistasis with regard to their interactions with environments.
Materials and methods
The experimental population and field trials
The mapping population used in this study comprised 135 DH lines derived from the cross between an indica variety, IR64, and an upland japonica variety, Azucena, as described previously (Huang et al. 1994). Two phenotyping experiments were conducted. Experiment 1 was conducted during 1994 and 1995 in nine diverse environments including seven locations in four Asian countries (Philippines, China, India and Thailand) and two different growing seasons at two of the locations. The geography of the environments covered a wide range of latitudes and longitudes from 13.5° to 31.5° N and from 76° to 121.5° E (Table 1). The same set of DH lines and the parents were evaluated in each of the nine environments in a randomized complete block design with two or three replications. A spacing of 30×20 cm between rows and between plants within a row was used at all locations except E2 (30×25 cm) and E7 (30×15 cm); plot size was 3 to 6 rows, with 15 plants per row. All experiments were established by transplanting and maintained as lowland fields with standing water present for most of the season. The management of the field experiments was in accordance with local standard practices. Twelve traits, including plant height (PH), heading date (HD), and grain yield and its components, were measured on five representative plants in each plot. PH (in cm) was measured from the soil surface to the tip of the tallest panicle of each plant at maturity. HD was recorded as days from the time of sowing to that of the first panicle flowering in 50% of the plants in each plot. Because nine DH lines were segregating for the measured traits, apparently because of outcrossing, they were not included in subsequent data analyses. Experiment 2 was conducted with the parents plus a subset of 82 DH lines during the dry seasons of 1998 and 1999 in aerobic soil conditions under two water levels. The lines were selected to reduce the range of heading dates. These experiments were established by direct sowing at a seed rate of 80 kg ha−1. Entries were replicated twice in the control section and twice in the stress section. In 1998, plots were 3-m long on beds and 0.9-m apart. Three rows were sown on each bed and the spacing between rows within a plot was 18 cm. The area received sprinkler irrigation twice each week until 30 days after planting, after which furrow irrigation was applied twice each week. This level of irrigation resulted in a fully aerobic soil profile where soil moisture status remained near field capacity. In the stress section, water was withheld from 52 to 64 days after planting and again from 70 to 84 days after planting. In 1999, plots were 3 m by 1 m, with 25 cm between rows. The crop was established using sprinkler irrigation, and then changed to drip irrigation three times a week. This type of water management applied water equivalent to about 1.4-times the potential evapotranspiration, and the soil remained aerobic. In the stress treatment, water was withheld for 14 days, beginning, on average, 12 days before HD in that plot. After the stress treatment, irrigation three-times per week was resumed. Data on PH and HD were collected as described in experiment 1.
Linkage map construction and data analyses
A total of 178 markers, including 147 RFLPs, 8 isozymes, 11 RAPDs and 12 cloned genes, was used to construct a complete linkage map for the DH population, as described previously (Huang et al. 1997). This map covers all 12 rice chromosomes with a total genome size of 2,003.4 cM and an average distance of 12.4 cM between adjacent markers. Analysis of variance was performed to evaluate differences among the DH lines and between the parents, among the environments, and the GEIs for the measured traits using the SAS PROC GLM (SAS Institute 1996). Correlation between the two traits in each of the environments and between lines for the same traits across the environments was determined using the SAS PROC CORR (SAS Institute 1996).
Mixed linear models for QTL analyses
According to classical quantitative genetics theory (Falconer 1981), the phenotypic value of a DH line (y hk ) in a specific environment can be described by the following genetic model:
where y hk is the phenotypic effect of the kth DH line in the hth environment, μ is the population mean, G k ~ (0, σ 2 G ) is the genotypic effect of the kth DH line in the hth environment, E h ~ (0, σ 2 E ) is the effect of hth environment, GEI hk ~ (0, σ2 GE ) is the GEI effect between the kth genotype and the hth environment, and ε hk ~ (0, σ2 e ) is the residual. The genotypic effects (G k ), environmental effects (E h ) and GEI effects (GEI hk ) can be predicted by the adjusted unbiased prediction method, in which y k(G)=μ+G k and y hk(GE)=μ+E h +GEI hk (Zhu and Weir 1996; Yan et al. 1998). The mean phenotypic values, y hk , of individual DH lines and their GEI effects (GEI hk =y hk −y k(G)−E h +μ) were used as input data for identifying M-QTL and epistasis affecting PH and HD using the following mixed linear model and the corresponding computer software, QTLMAPPER v. 1.0 (Wang et al. 1999):
where y k is the phenotypic value of a quantitative trait measured on the kth DH line (k=1, 2,..., n); µ is the population mean; A i and A j are the additive effects (fixed) of two putative QTLs (Q i and Q j ), respectively; AA ij is the additive by additive effect (fixed) between Q i and Q j ; x Aik , x Ajk and x AAijk are coefficients of QTL effects derived according to the observed genotypes of the markers (M i−, M i+ and M j−, M j+) and the test positions (rM i− Q i and rM j− Q j ); \(e_{{M_{f} }} \sim N{\left( {0,\sigma ^{2}_{M} } \right)}\) is the random effect of marker f with indicator coefficient \(\mu _{{M_{{fk}} }} \) (1 for M f M f and −1 for m f m f ); \(e_{{MM_{l} }} \sim N{\left( {0,\sigma ^{2}_{{MM}} } \right)}\) is the random effect of the lth marker interaction (between marker K l and marker L l ) with indicator coefficient \(\mu _{{M_{{lk}} }} \) (1 for M K M K M L M L or m K m K m L m L and −1 for M K M K m L m L or m K m K M L M L ); and \(\varepsilon _{k} \sim N{\left( {0,\sigma ^{2}_{e} } \right)}\) is the random residual effect. The inclusion of \(e_{{M_{f} }} \) and \(e_{{MM_{l} }} \) in the model is intended to absorb the additive and epistatic effects of background QTLs (additional segregating QTLs other than the loci examined) to control the noise caused by them (Wang et al. 1999).
Mapping QTLs and quantifying QTL × environment interaction for data of experiment 1 were carried out in two steps. In the first step, GEI effects (GEI hk ) of individual DH lines in each of the environments were obtained according to model (1) (Zhu and Weir 1996). Because the sampled environments represented a random sample of the major rice-growing environments in Asia, E h and GEI hk in model (1) were considered as random factors. Next, the mean trait values (y hk ) and GEI values (GEI hk ) of individual DH lines from each of the environments were used as input data to identify QTLs contributing to trait variation among the DH lines and QTLs contributing to the GEI variation in each of the environments using model (2) (Wang et al. 1999). Thus, M-QTLs and epistatic QTLs (E-QTLs) identified by model (2) using the mean trait values (y hk ) of individual DH lines as input data were expected to contain confounded QTL effects (A i and AA ij ) and QTL × environment interaction effects (A i E h and AA ij E h ), while those QTLs obtained using the predicted GEI effects (GEI hk ) as input data were expected to be largely due to A i E h and AA ij E h effects. The software, QTLMAPPER v. 1.0 based on model (2) (Wang et al. 1999) first identified significant markers (M-QTLs) and marker pairs (E-QTLs) associated with the mean trait or GEI values using stepwise regression with a threshold of P≤0.005 and P≤0.001, respectively. Then, QTL parameters (locations, effects and test statistics) of all putative M-QTL and E-QTL pairs were estimated using the interval mapping and the restricted maximum-likelihood estimation method with all those markers identified in the first step fixed in the model to control the background genetic variation. The permutation method was used to obtain the empirical thresholds (2.97 for HD, 2.86 for HD-GEI effects, 2.87 for PH, and 2.85 for PH-GEI effects) of the experiment based on 1,000 runs of randomly shuffling the trait values, which were expected to have a genome-wide type-I error of P=0.05 (Churchill and Doerge 1994). Thus, the LOD=2.9 was used for claiming significant M-QTLs in this study. For E-QTLs, a threshold of LOD=3.0 was used. We realized that the use of a single arbitrary threshold in QTL mapping could easily detect a QTL in one environment but not in another. Thus, while all QTLs detected at the selected thresholds are presented, any QTL detected in only 1 or 2 environments was interpreted with caution. Also, to examine the extent of type-II error causing inconsistent QTL detection across the environments, all identified M-QTLs and E-QTLs were re-examined in the environments using the mean trait values and trait GEI values of individual DH lines from specific environments under the minimum threshold of P<0.05. In other words, when a QTL was identified using the mean trait or GEI values in one environment, this QTL was also tested by the data from all other environments, and the test statistics and QTL parameters associated with the QTL are also reported as long as the QTL reached the minimum threshold. For experiment 2, only M-QTLs were examined using the mean trait values of individual DH lines because of the small population size.
Results
Phenotypic variation of the DH lines
Table 2 shows summary statistics of the phenotypic performance of the DH lines and parents for PH and HD in the nine environments. Azucena was much taller than IR64 in all environments, but the difference in height between the parents varied considerably across the environments, ranging from 34.1 cm in E1 to 66.0 cm in E2. On average, the parents had a similar HD (102.4 days for Azucena and 103.2 days for IR64), but significant differences in HD between the parents were detected in all environments except E4. Azucena headed earlier than IR64 in E3, E5 and E7, but later than IR64 in the other environments. In experiment 2, water stress was severe in 1998 and caused reduced height and delayed heading in the parents (26.6 cm and 6 days for Azucena, and 22.6 cm and 11 days for IR64). In 1999, the stress was milder, but still caused 11-days heading delay and 8.9 cm of height reduction for Azucena, and 6-days heading delay and 24.1 cm of height reduction for IR64. Similar to the parents, the DH lines, on average, had 11.8-days delayed heading and 16 cm of reduced height in 1998 and only 1.7-days heading delay and 1.4 cm of height reduction in 1999.
The DH lines showed transgressive segregation for both traits in all environments (Table 2) and showed much greater GEIs than the parents. ANOVA indicated that variances among the genotypes (the parents and DH lines), the nine environments, and GEIs were highly significant for both PH and HD. However, the relative contributions of the variance components to the total variation were different for the two traits. HD was influenced more by the environments and showed a much greater GEI than PH. For PH, the variances among the genotypes, among the environments, and the GEIs accounted for 63%, 16% and 12% of the total phenotypic variation, respectively. For HD, these components explained 18%, 54% and 20% of the total phenotypic variation, respectively. In experiment 2, ANOVA indicated that for PH, the variances among the DH lines (R2=70%), between the years (R2=5%) and between the water levels (R2=8%), and genotype × water × year (R2=4%) were significant except for genotype × water. For HD, only the variances among the DH lines (R2=27%) and between different water levels (R2=56%) were statistically significant.
In experiment 1, the mean height of the DH lines varied greatly across the environments, indicating the large environmental effects on height. The DH lines and parents had a below-average PH in E1, E2, E3 and E4, but were taller in E5, E7, E8 and E9. Similarly, heading tended to be earlier in E1, E2 and E8, whereas heading was delayed for both DH lines and the parents in E5, E7 and E9. There was no obvious correspondence between delayed heading and short daylength in the environments, indicating that the parents and DH lines were not photoperiod-sensitive. The coefficients of variation of the DH population for both traits remained largely consistent across the environments, except that the DH lines showed a much greater variation for HD in E1. There was a low but significant positive correlation between PH and HD in the DH lines (r=0.24, 0.23, 0.17, 0.48, 0.31 and 0.23 in E1, E2, E3, E4, E6 and E9, respectively), but correlation was not significant in E5, E7 and E8. In experiment 2, the trait variation in the DH population remained unchanged across years and water-stress conditions, but PH showed slightly reduced variation and HD exhibited increased variation as compared to experiment 1 (Table 2). Significant positive correlation (r=0.34) between the two traits was detected only in 1998 under stress.
M-QTLs and E-QTLs for PH and their interactions with environments
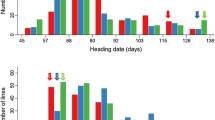
In experiment 1, QTL mapping based on the data of individual environments led to the identification of 17 M-QTLs on all 12 rice chromosomes except chromosomes 6 and 10 (Table 3, Fig. 1). The average number of detectable M-QTLs per environment was 9.8±2.0, ranging from 5 in E6 to 12 in E7. The M-QTLs with the largest effect mapped between RZ730 and RG810 on chromosome 1 in the same region as the semidwarf gene, sd-1. Here, the IR64 allele reduced height by an average of 13.0±3.3 cm (ranging from 8.2 to 18.3 cm). Detected in all 13 environments, this QTL showed significant interactions with E1, E3, E4, E5 and E9, with significant additive × environment (AE) effects ranging from −2.8 to 6.1 cm. Other important M-QTLs for PH included QPh5 detectable in all nine lowland environments; QPh3b, QPh3c, QPh4a, QPh4c and QPh9a detected in eight of the lowland environments; and QPh1 and QPh2a in seven of the environments. Of the M-QTLs mentioned above, we noted that QPh1, QPh3b, QPh5 and QPh9a showed large variation in their effects (in both magnitude and direction). The remaining M-QTLs appeared to be more or less environment-specific. These included QPh8 detected in four of the lowland environments; QPh2b, QPh3a, QPh7, QPh11 and QPh12 in two of the lowland environments; and QPh4b and QPh9b in only one lowland environment. Significant AE effects were detected in 46 cases (30%) for all M-QTLs, except for Ph2b, QPh3c and QPh9b. The IR64 allele at QPh2a, QPh2b, QPh4a, QPh5, QPh7 and QPh12 increased height in lowland environments, while it reduced height at the remaining M-QTLs except for QPh1, QPh3b and QPh11, at which QTL main effects differed in both magnitude and direction across the lowland environments.
Genomic locations of identified main-effect QTLs affecting plant height (PH) and heading date (HD) of the IR64 × Azucena DH population, detected in the nine environments of experiment 1. Ph2, Ph3, etc. were M-QTLs affecting PH detected in the same population reported by Yan et al. (1998)
In experiment 2, 14 of the 17 PH M-QTLs detected in experiment 1 were also identified in one or more cases except for QPh2a, QPh2b and QPh9b (Table 3). Aerobic soil conditions resulted in a reverse in the effect of the IR64 allele at QPh3a, QPh4a and QPh8 compared to the flooded experiment 1. Water stress caused further differences in the detected M-QTLs. Drought reduced the phenotypic effects of sd-1, QPh3c, QPh4a and QPh4b, increased the effects of QPh3b, QPh8 and QPh12, and switched the direction of the QTL effects relative to the well-watered aerobic control and the flooded trials in experiment 1 for both QPh5 and QPh9a.
In addition to the M-QTLs mentioned above, ten E-QTL pairs were identified for PH in experiment 1. Of these, five interactions occurred between an M-QTL and a modifying factor (Table 4). On average, each of the E-QTLs was detectable in 6.7 (74%) of the environments. Of these, two E-QTL pairs were identified in all nine environments, two in eight of the environments, one in seven of the environments, one in six of the environments, one in five of the environments, one in four of the environments and two in three of the environments. Interestingly, the AA effects of the E-QTL were consistent in direction, varying only in magnitude across the environments. Significant AA ij E effects were detected for six of the 11 E-QTL pairs and these AA ij E effects differed greatly in both direction and magnitude across the environments.
M-QTLs and E-QTLs for HD and their interactions with environments
Twenty M-QTLs affecting HD were identified in experiment 1 and they were mapped to all 12 rice chromosomes (Table 5, Fig. 1). The average number of detectable HD M-QTLs per environment was 11.3±2.5, ranging from 7 in E3 to 15 in E7. Of these, QHd8a was the only one detected in all nine environments. However, this QTL showed strong interactions with environments. The IR64 allele at this QTL delayed heading in E3 and E9 but caused early heading in the remaining environments. Significant AE effects were detected between QHd8a and E5, E6 and E9. Other important M-QTLs included QHd2b, QHd3a and QHd7 detected in eight of the environments, QHd1c and QHd3c in seven of the environments, QHd1a and QHd9 in six of the environments, and QHd1b, QHd3b, QHd4 and QHd12 in five of the environments. The remaining M-QTLs appeared to be more environment-specific. These included QHd5a and QHd6b detected in four of the environments, QHd2a, QHd6a and QHd8b in three of the environments, QHd11 in two of the environments, and QHd5b and QHd10 in only one of the environments. Four M-QTLs (QHd1b, QHd2a, QHd6b, QHd8 and QHd9) were unique in that their main effects became detectable only when each one was involved in epistasis with another locus in certain environments (Table 6). Significant additive × environment (AE) effects were detected in 39 cases (22%) for all M-QTLs except QHd5 and QHd11. The phenotypic effects of most M-QTLs varied considerably in magnitude, and five M-QTLs (QHd1a, QHd2b, QHd8, QHd11 and QHd12) showed varied effects in both magnitude and direction across the environments.
In experiment 2, 18 of the 20 M-QTLs identified in experiment 1 were also detected in one or more cases except for QHd5a and QHd11 (Table 5). Drought appeared to induce the expression of QHd1a, QHd2b, QHd3b, QHd4 and QHd10. QHd3c, QHd6a and QHd8b had reduced effects under drought, while QHd1c, QHd5b and QHd8a had increased effects under drought. For QHd3c, the IR64 allele was associated with delayed heading under the high-density, furrow irrigated treatment in 1998. QHd8b also appeared to have a contrasting effect in lowland and aerobic environments.
Table 6 shows 19 E-QTL pairs affecting HD in experiment 1. On average, the number of detectable E-QTL pairs in each of the environments was 8.6±1.9, ranging from 5 in E8 to 11 in E5 and E6. Of these, only one E-QTL pair was identified in all nine environments, one in seven of the environments, one in five of the environments, six in four of the environments, six in three of the environments, and four in two of the environments. Significant AA ij E effects were detected for 12 of the 19 E-QTL pairs and the AA ij E effects of individual E-QTL pairs identified in multiple environments differed greatly in both direction and magnitude across the environments. We noted that 12 of the interactions occurred between an M-QTL and a modifying factor. Detected as a M-QTL in three flooded environments and three aerobic environments, QHd9 (RZ206) interacted simultaneously with four other loci on chromosomes 1, 9, 10 and 11, suggesting its regulatory function in determining HD in rice.
Discussion
In this study, the genetics of two highly heritable traits of rice were dissected into QTL main effects and their interactions with environments. We included epistasis in the linear model so that the interactions of both M-QTLs and E-QTLs with environments could be characterized. It should be pointed out that, according to our models, the QTL effects (Tables 3 and 5) estimated using trait mean values from individual environments were expected to be the QTL phenotypic effects (A+AE or AA+AAE) and those detected by GEI effects were largely attributable to AE or AAE effects. This decomposition allowed us to quantify QTL × environment interaction more accurately. The number of QTLs detected in this study was much more than any previous single-environment QTL mapping studies in rice and other species. Furthermore, we found that most (73%) M-QTLs identified in this study fell within the vicinity of the M-QTLs affecting the same traits identified in the same or different rice mapping populations reported previously (Courtois et al. 1995; Li et al. 1995; Xiao et al. 1995, 1996; Huang et al. 1996; Yano et al. 1997; Zhuang et al. 1997; Yan et al. 1998; Zhou et al. 2001), indicating that these QTLs covered a significant portion of the loci affecting PH and HD in rice at which allelic differences exist. In this study, all QTLs but one (QPh9b) were detected in more than one environment.
The pattern, frequency, magnitude, and causes of QTL × environment interaction
In this study, the identified M-QTLs and E-QTLs were, on average, undetectable in 64% (58% for M-QTLs and 74% for E-QTLs) of the environments for PH and in 51% (57% for M-QTLs and 45% for E-QTLs) of the cases for HD. As the genetic basis of GEIs, QTL × environment interaction presumably arises from differential gene expression in different environments and may conceptually occur in any of the following three cases: (1) a QTL expresses in one environment but not in another, as reflected by inconsistent detection of the QTL across environments; (2) a QTL expresses strongly in one environment but weakly in another, as indicated by the variation in its effects across environments; and (3) a QTL expresses very differently and has opposite effects in different environments. All three cases were observed in this study and the pattern of the observed differential QTL expression in this study appeared to be very complex.
Non-expression of QTLs appeared to be the primary cause of the undetectable QTLs in certain environments. In these cases (37% of M-QTLs and 30% of E-QTLs for PH, and 31% of M-QTLs and 50% of E-QTLs for HD), QTLs were undetectable even under the minimum threshold of P≤0.05. On the other hand, weak QTL expression in certain environments could have resulted in a high probability of type-II error in this study if the identified QTL had not been re-examined under the minimum threshold. In this study, the average number of detectable M-QTLs per environment was 9.8 for PH and 11.3 for HD. However, with the selected threshold of LOD=2.9, this number became only 6.9 for PH and 5.5 for HD. Similarly, with the threshold of LOD=3.0, the average number of E-QTLs detectable per environment was 4.0 for PH and 5.0 for HD, respectively, which became 6.9 for PH and 8.2 for HD under the minimum threshold (Tables 4 and 6). In other words, on average, 4.4 (41.2%) of the M-QTLs and 3.1 (40.4%) of the E-QTLs per environment would have gone undetected because of the type-II error of the selected single thresholds arising from weak QTL expression in certain environments. The third cause of inconsistent QTL detection across environments was the significant QTL × environment interaction effects. In this study, statistically significant AE effects were detected in 26% (30% for PH and 22% for HD) of the QTLs by environment combinations for the M-QTLs, and in 27% of the cases (28% for PH and 26% for HD) for the E-QTLs, as summarized in Table 7. In contrast to the QTL phenotypic effects, which were largely consistent in direction, individual AE and AAE effects varied considerably in both magnitude and direction, and appeared to be responsible for 8.2% (6.8% for M-QTLs and 9.7% for E-QTLs) of the undetected QTLs in certain environments. In these cases, the estimated AE or AAE effects were in the opposite direction to the QTL main effects. The average magnitude of the AE effects was 2.25 cm for PH and 1.42 days for HD, or 42% and 79% of the mean QTL main effects estimated in single environments, respectively. The average magnitude of AAE was 1.89 cm for HD and 1.40 days for HD, or 37% and 81% of the mean QTL epistatic effects estimated in single environments, respectively (Table 7). This was not surprising since AE or AAE effects were part of the QTL main (phenotypic) effects according to model (1).
In seven cases (QPh1, QPh3b, QPh11, QHd1a, QHd2, QHd8 and QHd12), the two alleles at an M-QTL detected in Experiment 1 had opposite effects in different environments. This suggests that the two alleles at these loci responded differently to the environments. Similarly, five HD E-QTL pairs showed opposite AAE effects in different environments. These represented the extreme situation of QTL × environment interaction arising from differential reactions of the identified QTLs to the unspecified environmental factors. Results from experiment 2 suggested that differential responses of the detected QTLs in the sampled environments could be attributable to different cultural conditions and/or abiotic and biotic stresses. For instance, all three types of QTL × environment interaction detected in experiment 1 were also observed in experiment 2, where they could largely be attributed to a specific environmental factors: soil water status and plant density. Opposite effects of QTLs on PH or HD in aerobic or high density conditions compared to lowland fields have important implications for the development of rice cultivars that can be grown using less irrigation water. Sripongpangkul et al. (2000) reported QTL × environment interaction affecting plant height under different levels of submergence where different sets of M-QTLs and E-QTLs affecting internode and leaf elongation were detected, and the expression of most M-QTLs was much stronger under the more stressful condition.
Importance of epistasis
The importance of epistasis in determining quantitative trait variation has been well demonstrated in several recent QTL mapping studies (Doebley et al. 1995; Lark et al. 1995; Li et al. 1997; Yu et el. 1997; Li et al. 2001; Luo et al. 2001). The large number of E-QTLs identified and the involvement of many M-QTLs in epistasis lend strong support to this notion. Our finding that E-QTLs tended to show a greater level of QTL × environment interaction than the M-QTLs was expected because E-QTLs should more likely be influenced by environments than M-QTLs. Furthermore, we noted that six HD M-QTLs (QHd1b, QHd1c, QHd2a, QHd6b, QHd8a and QHd9) would have gone undetected in 19 single-environment cases had their involvement in epistasis been unexamined (Table 6). Strong evidence for the presence of epistatic interactions between and among different rice HD M-QTLs and their differential responses to daylength has been clearly demonstrated using near-isogenic lines (Lin et al. 2000).
Trait- and gene-specificity of QTL × environment interaction
An important finding of this study was that GEIs were trait- and gene (QTL)-specific. As described above, the greater level of GEIs observed for HD in the DH population was reflected by more environment-specific QTLs, greater frequency and magnitude of AE and AAE effects, and more pronounced epistasis for HD than for PH. The level of QTL × environment interaction varied considerably among the M-QTLs or E-QTLs. Some M-QTLs and E-QTLs were detectable in all or most of the environments and/or showed less variation in their effects. Very often, when detectable in multiple environments, the QTL (phenotypic) effects tended to be in the same direction and showed relatively small variation in magnitude across the environments. These non-environment-specific QTLs included 11 M-QTLs (9 for PH and 2 for HD) and 9 E-QTLs (7 for PH and 2 for HD). These QTLs should be particularly useful in marker-aided manipulation of plant height and flowering time in rice. For those QTLs that were more environment-specific and/or involved in epistasis, one should be cautious in applying marker-aided selection.
References
Austin DF, Lee M (1998) Detection of quantitative trait loci for grain yield and yield components in maize across generations in stress and nonstress environments. Crop Sci 38:1296–1308
Baker RJ (1988) Differential response to environmental stress. In: Weir BS, Eisen EJ, Goodman MM, Namkoong G (eds) Proc 2nd Int Conf on Quantitative Genetics. Sinauer, Sunderland, Massachusetts, pp 492–504
Bubeck DM, Goodman MM, Beavis WD, Grant D (1993) Quantitative trait loci controlling resistance to gray leaf spot in maize. Crop Sci 33:838–847
Churchill GA, Doerge RW (1994) Empirical threshold values for quantitative trait mapping. Genetics 138:963–971
Cooper M, Hammer GL (1996) Plant adaptation and crop improvement. CAB International, Oxon, UK
Courtois B, Huang N, Guiderdoni E (1995) RFLP mapping of genes controlling yield components and plant height in an indica × japonica doubled-haploid population. In: Proc Int Rice Research Conf, Los Baños, The Philippines, pp 963–976
Crossa J, Vargas M Van Eeuwijk FA, Jiang C, Edmeades GO, Hoisington D (1999) Interpreting genotype × environment interaction in tropical maize using linked molecular markers and environmental covariables. Theor Appl Genet 99:611–625
Doebley J, Stec A, Gustus C (1995) teosinte branched l and the origin of maize: evidence for epistasis and the evolution of dominance. Genetics 141:333–346
Falconer DS (1981) Introduction to quantitative genetics, 2nd edn. London and New York
Georges M (1997) Case history in animal improvement: mapping complex traits in ruminants. In: Paterson AH (ed) Molecular dissection of complex traits. , CRC Press, Boca Raton, Florida, USA, pp 229–240
Hayes PM, Liu BH, Knapp SJ, Chen F, Jones B (1993) Quantitative trait loci effects and environmental interaction in a sample of North American barley germplasm. Theor Appl Genet 87:392–401
Huang N, McCouch S, Mew T, Parco A, Guiderdoni E (1994) Development of an RFLP map from a doubled-haploid population in rice. Rice Genet Newslett 11:134–137
Huang N, Courtois B, Khush GS, Lin H, Wang G (1996) Association of quantitative trait loci for plant height with major dwarfing genes in rice. Heredity 77:130–137
Huang N, Parco A, Mew T, Magpantay G, McCouch S (1997) RFLP mapping of isozymes, RAPD and QTLs for grain shape and brown plant hopper resistance in a doubled-haploid rice population. Mol Breed 3:105–113
Jain SK, Marshall DR (1967) Population studies in predominantly self-pollinated species. X. Variation in natural populations of Avena fatua and A. barbata. Am Nat 101:19–33
Jiang C, Edmeades GO, Armstead I, Laffite HR, Hayward MD (1999) Genetic analysis of adaptation difference between highland and lowland tropical maize using molecular markers. Theor Appl Genet 99:1106–1119
Lark KG, Chase K, Adler F, Mansur LM, Orf JH (1995) Interactions between quantitative trait loci in soybean in which trait variation at one locus is conditional upon a specific allele at another. Proc Natl Acad Sci USA 92:4656–4660
Lee SH, Bailey MA, Mian MAR, Carter TE, Ashley Jr DA, Hussey RS, Parrott WA, Boerma HR (1996) Molecular markers associated with soybean plant height, lodging, and maturity across locations. Crop Sci 36:728–735
Li ZK, Pinson SRM, Stansel JW, Park WD (1995) Identification of quantitative trait loci (QTLs) for heading date and plant height in cultivated rice (Oryza sativa L.). Theor Appl Genet 91:374–381
Li ZK, Pinson SRM, Park WD, Paterson AH, Stansel JW (1997) Epistasis for three grain yield components in rice (Oryza sativa L.). Genetics 145:453–465
Li ZK, Luo LJ, Mei HW, Shu QY, Wang DL (2001) Overdominant epistatic loci are the primary basis of inbreeding depression and heterosis in rice. I. Biomass and grain yield. Genetics 158:1737–1753
Lin HX, Yamamoto T, Sasaki T, Yano M (2000) Characterization and detection of epistatic interactions of three QTLs, Hd1, Hd2 and Hd3, controlling heading date in rice using nearly isogenic lines. Theor Appl Genet 101:1021–1028
Luo LJ, Li ZK, Mei HW, Shu QY, Tabien R (2001) Overdominant epistatic loci are the primary basis of inbreeding depression and heterosis in rice. II. Grain yield components. Genetics 158:1755–1771
Lu CL, Shen L, Tan Z, Xu Y, He P (1996) Comparative mapping of QTLs for agronomy traits of rice across environments using a doubled-haploid population. Theor Appl Genet 93:1211–1217
Mather K, Jinks JL (1982) Biometrical genetics, Chapman and Hall, London
Paterson AH (1995) Molecular dissection of quantitative traits: progress and prospects. Genome Res 5:321–333
Paterson AH, Damon S, Hewitt JD, Zamir D, Rabinowitch HD (1991) Mendelian factors underlying quantitative traits in tomato: comparison across species, generations, and environments. Genetics 127:181–197
SAS institute Inc (1996) SAS users guide: statistics. SAS Institute, Cary, North Carolina
Sripongpangkul K, Posa GBT, Senadhira DW, Brar DS, Huang N, Khush GS, Li ZK (2000) Genes/QTLs affecting flood tolerance in rice. Theor Appl Genet 101:1074–1081
Stuber CW (1997) Case history in crop improvement: yield heterosis in maize. In: Paterson AH (ed) Molecular dissection of complex traits. CRC Press, Boca Raton, Florida, USA, pp 197–206
Tinker NA, Mather DE, Rossnagel BG, Kasha KJ, Kleiohofs A (1996) Regions of the genome that affect agronomic performance in two barley cultivars. Crop Sci 36:1053–1062
Veldboom LR, Lee M (1996) Genetic mapping of quantitative trait loci in maize in stress and nonstress environments. I. Grain yield and yield components. Crop Sci 36:1310–1319
Wang DL, Zhu J, Li ZK, Paterson AH (1999) Mapping QTLs with epistatic effects and QTLs by environment interaction by mixed linear model approaches. Theor Appl Genet 99:1255–1264
Xiao JH, Li J, Yuan L, Tanksley SD (1995) Dominance is the major genetic basis of heterosis in rice as revealed by QTL analysis using molecular markers. Genetics 140:745–754
Xiao JH, Li J, Yuan L, Tanksley SD (1996) Identification of QTLs affecting traits of agronomic importance in a recombinant inbred population derived from a subspecific rice cross. Theor Appl Genet 92:230–244
Yan JQ, Zhu J, He C, Benmoussa M, Wu P (1998) Molecular dissection of developmental behavior of plant height in rice (Oryza sativa L.). Genetics 150:1257–1265
Yan JQ, Zhu J, He C, Benmoussa M, Wu P (1999) Molecular marker-assisted dissection of genotype × environment interaction for plant type traits in rice (Oryza sativa L.). Crop Sci 39:538–544
Yano M, Marushima Y, Nagamura Y, Kurata N, Minobe Y (1997) Identification of quantitative trait loci controlling heading date in rice using a high-density linkage map. Theor Appl Genet 95:1025–1032
Yu SB, Li JX, Xu CG, Gao YF, Li XH (1997) Importance of epistasis as the genetic basis of heterosis in an elite rice hybrid. Proc Natl Acad Sci USA 94:9226–9231
Zhou Y, Li W, Wu W, Chen Q, Mao D (2001) Genetic dissection of heading time and its components in rice. Theor Appl Genet 102:1236–1242
Zhu J, Weir B (1996) Mixed model approaches for dialelle analysis based on a bio-model. Genet Res Cambridge 68:233–240
Zhuang JY, Lin HX, Lu J, Qian HR, Hittalmani S, Huang N, Zheng KL (1997) Analysis of QTL × environment interaction for yield components and plant height in rice. Theor Appl Genet 95:799–808
Acknowledgements
We are very grateful for valuable comments and suggestions on the manuscript from Dr. Wenzel and two anonymous reviewers. We also thank B. Hardy for his help in editorial work of the MS. Financial support from a BMZ/GTZ grant of the German government and from the Rockefeller Foundation is greatly appreciated.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by G. Wenzel
Rights and permissions
About this article
Cite this article
Li, Z.K., Yu, S.B., Lafitte, H.R. et al. QTL × environment interactions in rice. I. Heading date and plant height. Theor Appl Genet 108, 141–153 (2003). https://doi.org/10.1007/s00122-003-1401-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-003-1401-2