Abstract
In the present study, fifty genotypes were evaluated over 5 years. The analysis of variance revealed significant interaction (P < 0.001) between genotype and year (GYI). The results of different stability statistics illustrate that the Kang’s rank-sum is a good statistic and based on that, Azadi, Roshan, Mahdavi, Marvdasht, and Naz identified as desirable cultivars. Seven environmental factors including maximum temperature, minimum temperature, average temperature, precipitation, relative humidity, daylight hours, and soil temperature over eleven twenty-day periods and sixteen molecular markers associated with vernalization and photoperiod were used as covariates to interpret the interaction. The Partial least square regression biplot with environmental covariates explained 28.23% and with genotypic covariates explained 40.24% of the GYI. The results also showed that genotypes do not respond similarly to environmental variables at different stages of development. Genotypes have been classified into three groups, the first group being more related to environmental factors at the end of the growing season, the second group being more influenced by environmental factors at the beginning of the season, and the third group being genotypes of environmental factors throughout the season, especially the mid-season was affected. Among environmental factors, relative humidity except for period 9 had a special role in GYI in all periods. On the other hand, Ppd.D1D001.KASP, Vrn.B1.B.KASP, Inter1.D.deletion, VRN.A1, Vrn.A1.E7.FT.KASP and Vrn.A1.E4.vern.KASP markers had the most impact on the GYI among the different molecular markers. This information on the causes of the GEI can be useful in future breeding and management programs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Wheat (Triticum aestivum L.) is one of the most important cereals in the world. Studies show that higher-performance lines may not always be stable in all environments. This is due to genotype × environment interactions (GEI) which, together with environmental and genotype effects, are influencing factors in the phenotype of traits (De Leon et al. 2016). Predicting which genotypes have high yield and stability over different years is an important challenge for plant breeders (Monteverde et al. 2019). This is because the weather conditions, including rainfall, especially in arid and semi-arid regions, change unpredictably from year to year. From the farmers' point of view, yield stability over the years is critical to reduce their income fluctuations (Herrera et al. 2020). Also, from a breeding point of view, the patterns of narrow adaptation of genotypes to specific locations should be repetitive over different years. Therefore, it is important to evaluate the genotype over different years.
To accurately predict the performance of genotypes in different environments, researchers need to understand the causes of GEI. Understanding the genotypic responses to each of these factors can help in the interpretation and exploitation of GEI. Some statistical models can use external information directly to study GEI. These models are among the analytical models that, unlike empirical models, understand the morpho-physiological causes of genotype response (Richards 1982; Voltas et al. 2005). Partial least square regression (PLSR) and factorial regression (FR) are statistical models that combine external variables, both environmental and genotypic (such as molecular markers), to study and interpret GEI (Elias et al. 2016). When the number of variables is greater than the observations and there is high collinearity among variables, it is useful to use PLS method (Pacheco et al. 2015). This method is an excellent tool to identify environmental factors affecting grain yield and other wheat traits (Crossa et al. 2010; Kondić-Špika et al. 2019). On the other hand, the PLS model with molecular markers and environmental covariates can predict performance in untested years (Monteverde et al. 2019).
Environmental factors are the most important and influential factors on plant growth. This has become more important with climate change in recent years (Taranto et al. 2018; Mansour et al. 2018). For this reason, it has been suggested to use envirotyping along with phenotyping and genotyping to understand GEI (Xu 2016). In addition, environmental factors do not have the same effects at different stages of plant growth. Zorić et al. (2017) showed that environmental parameters had the most significant impact in particular months. In bread wheat, spike primordia growth stage is more sensitive to environmental factor (Reynolds et al. 2002), or part of the GEI was due to the sensitivity of maize genotypes to the minimum temperature at flowering and the amount of light during grain filling (Malosetti et al. 2013). However, in durum wheat, it was reported that the maximum and mean temperature during the entire crop cycle were the most important environmental factors influencing GEI (Chairi et al. 2020). Another study in durum wheat found that freezing days played critical role in genotype × year interactions under rainfed conditions (Mohammadi 2017). Identification of the responsible environmental factors to the GYI and stability of genotypes will help us to determine the genetic basis of these effects and enhances the predictability of the GEI (Heslot et al. 2014).
GEI controls by many regions of the genome (De Leon et al. 2016). In the other words, GEI can be due to the nonlinear response of the QTL to the environment. Therefore, molecular markers are another important covariates. Functional markers (FMs) are the most valuable markers for crop breeding programs. QTL mapping in environments that are different in terms of factors helps to understand GEI (Xu 2016). Among the markers included in the model for predicting GEI, vernalization and photoperiod markers showed more variability and importance (Heslot et al. 2014). It has been reported that these genes, along with their interaction with environment temperature, determine the potential of wheat yield in different environments (Gororo et al. 2001; Iqbal et al. 2011). So, these genes are generally yield QTLs and affect performance (Whittal et al. 2018; Schmidt et al. 2019; Alipour and Abdi 2020). They affect traits dependently or independently of environmental cues (Arjona et al. 2018). Therefore, it is necessary to identify the genes involved in GYI. Crossa et al. (1999) reported that 30 molecular markers out of 86 markers play a special role in their interaction and maximum temperature which introduced as the most important environmental covariate. The simultaneous use of molecular markers and environmental covariates can provide a clear perspective on GEI (Crossa et al. 1999; Monteverde et al. 2019). The present study was conducted to investigating the yield stability of some Iran wheat cultivars and determine the GEI causes for wheat grain yield using environmental and genotypic covariates.
Materials and methods
Plant material and field evaluation
Fifty wheat cultivars (Table 1) in five cropping years including 2013, 2014, 2015, 2017 and 2018 at the research farm of Tehran University with latitude 50.58 E and latitude 35.56 N and 1112.5 m above sea level were evaluated in a randomized complete block design with two replications. The dimensions of the plots consisted of four lines with a length of 1 m. The distance between the rows was 20 cm and the distance between the plants within rows was 5 cm.
Environmental covariates
Climatic parameters as environmental covariates are presented in Table S1. The environmental factors included maximum temperature, minimum temperature, average temperature, precipitation, relative humidity, daylight hours, and soil temperature, which were divided into 11 periods of 20 days from date 11 November to date 18 June.
Genotypic covariates
Genomic DNA of all 260 investigated samples were extracted using a modified cetyltrimethyl ammonium bromide (CTAB) method (Saghai-Maroof et al. 1984) from five two-week-old seedlings. For each STS reaction, a PCR mix contained 50 ng of genomic DNA, 0.2 mM of each dNTP, 1 × ammonium sulfide PCR buffer, 0.1 µM of forward primer, 0.15 µM of reverse primer, 2.5 mM of Mg2+, 0.05 µM of dye-labeled M13 primer and 1 unit of Taq polymerase. A touchdown program for PCR amplification started at 95 °C for 5 min, followed by five cycles of 45 s at 95 °C, 5 min of annealing at 68 °C which decreased by 2 °C in each subsequent cycle, and 1 min extension at 72 °C. In the subsequent five cycles, the annealing time was reduced to 2 min with a decrease in 2 °C in each subsequent cycle. PCR was continued for an additional 25 cycles of 45 s at 94 °C, 2 min at 50 °C, and 1 min at 72 °C, with a final extension at 72 °C for 5 min. PCR products were detected using an ABI Prism 3730 DNA Analyzer, and the fragment size was scored using GeneMarker version 1.97 (SoftGenetics, LLC). Some of the most closely linked SNPs to vernalization and photoperiod genes have been converted to Kompetitive allele-specific PCR (KASP) assay were also used to screen the investigated samples. KASP assays were performed in a 6-µl reaction volume (3 µl 2X KASP Master Mix, 0.0825 µl KASP primer mix and 3 µl genomic DNA at 25 ng/µl) and data were analyzed in an ABI 7900HT Real-Time PCR System (Life Technology, Grand Island, NY) following the instruction for KASP analysis (http://www.lgcgroup.com). The list of investigated markers is given in Table S2. Only polymorph markers were considered as genotypic covariates.
Statistical analysis
Three different types of statistics were used to evaluate yield stability. Univariate statistics included Wricke’s ecovalance (Wricke 1962), Shukla’s stability variance (Shukla 1972), and Kang’s rank-sum (Kang 1988). Multivariate statistics included AMMI stability value (Purchase 1997), AMMI stability index (Jambhulkar et al. 2014), sums of the absolute value of the IPC scores (Sneller et al. 1997), absolute value of the relative contribution of IPCs to the interaction (Zali et al. 2012) and averages of the squared eigenvector values (Sneller et al. 1997). Finally, statistics based on mixed models included a harmonic mean of genotypic values, the relative performance of genotypic values and harmonic mean of the relative performance of genotypic values (Resende 2007). To investigate the relationship between statistics and yield, the principal component analysis (PCA) was performed based on the rankings of the genotypes.
PLS method was used to understand the effect of covariates. This model uses the cultivar responses (Y) over environments on environmental and genotypic covariables (Z) and is as follows:
where matrix T contains the Z-scores, matrix P contains the Z-loadings, matrix Q contains the Y-loadings, and E and F are the residuals matrix. Therefore:
where \({Y}^{^{\prime}}\) contains genotypic response over environments (the rows of Y), T contains the Z-scores (indexed by environments). W the Z-loadings (or weight, indexed by environmental and genotypic variables), Q the Y-loadings (indexed by genotypes), \(\varsigma\) contains the approximation of PLS to regression coefficient of the responses, \({Y}^{^{\prime}}\) to the explanatory variables in Z.
Therefore, in the PLS biplot, projection of the jth environment (row) of T on the ith genotype (row) of Q approximates the GE; projection of the hth environmental covariables (row) of Q on the ith genotype (row) of Q approximates the regression coefficient of the ith genotype on the hth environmental covariable.
Analysis of variance and AMMI analysis were performed using agricolae R-package (de Mendiburu 2016). Yield stability measures were calculated using ammistability (Ajay et al. 2019) and metan (Olivoto and Lúcio 2020) packages in RStudio 1.0.136. The STABILITYSOFT online program (Pour‐Aboughadareh et al. 2019) was also used to calculate some of the stability statistics. PCA loading plot was drawn with factoextra R-package (Kassambara and Mundt 2017). Finally, GEA-R software (Pacheco et al. 2015) was used for PLSR analysis.
Result
After performing the homogeneity of variance test by Bartlett test and assuring that this assumption was valid, the results of combined analysis of variance showed significant differences among environments, genotypes, and genotype × year interaction. Also, based on AMMI analysis, the two components were significant and explained a total of 70% of GEI changes (Table 2). The box plot showed that the genotypes had the highest yield in the second and first year, respectively (Fig. 1). Also, according to this graph, genetic variance is homogeneous across environments. The average grain yield of the genotypes at all years is presented in Table 3. Genotypes Alvand, Roshan, Naz, and Mahdavi had the highest yield, respectively, while genotype Shahpassand had the lowest performance.
AMMI1 biplot is very important in the simultaneous and straightforward evaluation of performance and stability by placing the average yield of genotypes and environments against the first principal component (PC1). Accordingly, genotypes Azadi, and Shiroodi with high yield and PC1 were close to zero were desirable (Fig. 2a). The AMMI2 biplot provides only information about the stability of genotypes. Genotype Moghan2 was the most stable in all years due to its proximity to the origin of the biplot. On the other hand, the first and second years of the experiment played the most important role in GYI (Fig. 2b).
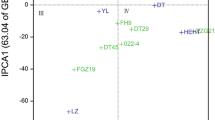
Genotypes Marvdasht, Dez, Inia, and Hamoon had the lowest ecovalence and stability variability, respectively, therefore had high stability. Genotypes Azadi and Roshan ranked first in yield and stability, and they were desirable genotypes in terms of Kang’s rank-sum, but genotype DN11 was undesirable in terms of these two features. Based on the parameters of the AMMI model, the genotypes Moghan2, Azadi, Dez, and Hamoon had high yield stability. According to HMGV, RPGV and HMRPGV statistics, genotypes Alvand, Roshan and Falat had the highest values and the most desirable genotypes in terms of yield, stability and adaptability, while, genotypes Azar, Shahpassand and VEE/NAC were the worst (Table 3). Examination of the relationship between statistics using PCA showed that the HMGV, RPGV and HMRPGV statistics were highly positive correlated with grain yield because the angle between their vectors was less than 90°. Other statistics, except for the Kang’s rank-sum, had a 90° angle with yield, so they had no correlation with it. As expected, Kang’s rank-sum was moderate between the two groups (Fig. 3).
Loading plot obtained from principal component analysis with genotype yield and stability statistics ranks. GY mean grain yield, Wricke Wricke’s ecovalence, Shukla Shukla’s stability variance, KR Kang’s rank-sum, ASV AMMI stability value, ASI AMMI stability index, SIPC sums of the absolute value of the IPC scores, ZA absolute value of the relative contribution of IPCs to the interaction, EV averages of the squared eigenvector values, HMGV harmonic mean of genotypic values, RPGV relative performance of genotypic values, HMRPGV harmonic mean of relative performance of genotypic values
The PLSR biplot with environmental covariates explained 28.23% of the GEI variance (Fig. 4a). Genotypes were divided into three groups based on this biplot. In the first group, genotypes Akbari, Arta, Atrak, Bayat, Biston, Darya, DN11, Hamoon, Karaj1, Karaj3, Kavir, Pishtaz, Roshan, Sabalan, Shiraz, and Zagros were present. This group was associated with year four and the parameters of maximum temperature (mx1 and mx10), minimum temperature (mn9 and mn10), average temperature (at9 and at10), precipitation (pr7 and pr9), relative humidity (rh4, rh5, rh7 and r8), daylight hours (dl6 and dl10) and soil temperature (st9 and st10) had the most influence on them. The second group consisted of genotypes Adl, Azadi, Chamran, Ghods, Mahdavi, Maroon, Marvdasht, Moghan1, Moghan3, Sepahan, Shahi, Shahpassand, VEE/NAC, and Zarrin were scattered between the first and second year environmental vectors. This group had a positive interaction with environmental factors such as maximum temperature (m × 4, m × 8 and m × 9), minimum temperature (mn1, mn2 and mn8), average temperature (at4 and at8), precipitation (pr1 and pr2), relative humidity (rh1, rh2 and rh3), daylight hours (dl7, and dl9) and soil temperature (st1, st2 and st8). Genotypes Alborz, Alvand, Azar, Bam, Darab2, Dez, Falat, Fong, Golestan, Inia, Karaj2, Kaveh, Khazar1, Moghan2, Navid, Naz, Nicknejad, Shiroodi, Siosson, and Sistan belonged to the third group and were associated with the fifth year. This group was influenced by environmental factors of maximum temperature (m × 2, m × 3, m × 5 and m × 7), minimum temperature (mn3, mn5 and mn7), average temperature (at2, at3, at5 and at7), precipitation (pr4, pr6, pr10 and pr11), relative humidity (rh6, rh10 and rh11), daylight hours (dl1, dl2 and dl4) and soil temperature (st3, st5 and st11).
The PLSR biplot with genotypic covariates (molecular markers) explained 40.24% of the GEI variance (Fig. 4b). Unlike the previous biplot, genotypes did not have a clear distribution in this biplot. However, genotypes Golestan, Mahdavi, Moghan2 and Naz were associated with the first year and were influenced by the Ppd.D1D001.KASP marker. Genotypes Khazar1 and Sistan were in the interval between first and second year and the Vrn.B1.B.KASP marker was influenced them. Genotypes Siosson, Pishtaz, Alvand, Sepahan, Moghan2, and Falat were associated with the second year and were influenced by the Inter1.D.deletion marker. Finally, genotypes Arta, Azar, Chamran, Kaveh, and Shahi were located between the second and third year environmental vectors and were associated with the VRN.A1, Vrn.A1.E7.FT.KASP and Vrn.A1.E4.vern.KASP markers.
Discussion
Significant differences among genotypes indicate genetic differences between investigated genotypes. Differences between environments are due to different climatic conditions, as it has been previously reported that climate change from year to year clearly affects wheat production (Macholdt and Honermeier 2019). We found that environmental variables do not have the same effect in different years. In the third year, no specific variable related to GEI was identified, and it had a smaller vector length in the PLS diagram, while the first and fifth years were associated with more variables and showed the longest vector length. In the fifth year, the temperature at the beginning of the growing season and relative humidity at the end of the growing season affected yield. Such a trend was somewhat reversed in the fourth year, with temperatures at the end of the growing season and relative humidity in the middle of the growing season having the remarkable effect. In the first and second years, environmental parameters were also effective almost in the middle of the season. The above, along with other unknown factors, led to fluctuations in the performance of genotypes during these five years. In this regard, Mohammadi (2017) reported that the effect of genotype × year on grain yield and some other traits in durum wheat is significant.
We found that the wheat cultivars introduced in Iran have different yields and stability. Among them, Alvand and Roshan cultivars had the best grain yield. This difference can be examined with different statistics. Our results showed that multivariate and univariate statistics other than Kang’s rank-sum independently of yield lead to a similar ranking of genotypes in terms of yield stability. Similar results have been observed before (Olivoto et al. 2019; Vaezi et al. 2019). These statistics only provide information on yield stability, while yield is more important. Therefore, researchers have used different methods to simultaneously examine genotypes in terms of performance, stability and adaptability. Among them the promising methodologies are HMGV, RPGV and HMRPGV statistics (Rosado et al. 2019). In the present study, selection based on the above three statistics had similar results to selection based on performance. This similarity is also mentioned in the study of Coan et al. (2018). Part of the similarity in our research could be that cultivars have only been evaluated in one place over the years. The Kang’s rank-sum statistic seems to be useful because it pays attention to both yield and stability. Based on that, Azadi, Roshan, Mahdavi, Marvdasht and Naz cultivars were recommended.
The presence of all environmental covariates in the biplot indicates the complexity of the GYI. Environmental factors far from the center of the biplot played a major role in GYI and since they are close together, they are highly correlated. The presence of environmental vectors in different directions indicates different responses of genotypes in different years. The GYI in the first and fifth years appear to be greater and more important than in the other years. This is certainly due to changes in known and unknown environmental factors. In addition, genotypes do not respond similarly to environmental variables at different stages of development. This is related to changes in gene expression (Fusi et al. 2013). Among environmental factors, relative humidity except for period 9 had a special role in GYI in all periods. Genotypes do not respond equally to environmental factors at different stages of development. The genotypes of the first group were mainly influenced by environmental factors at the end of the growing season. In the second group, the main effects of environmental parameters are observed at the beginning of the growing season. The influence of factors on seed germination during this period can be one of the reasons. On the other hand, rainfall at the beginning of the growing season can stimulate the disease. In the third group, environmental variables at different stages of growth, especially in mid-season, had more effect on genotypes. Voltas et al. (2005) showed that spring and winter wheat cultivars respond differently to environmental variables.
A small percentage of the GEI variance was explained by biplot. This result was expected and agrees with the results of other researchers (Crossa et al. 2010; Zorić et al. 2017; Kondić-Špika et al. 2019). As the number of genotypes, environments, and covariates increase, interpretation of the PLS biplot will become difficult and require expertise (Ramburan et al. 2012). But model crop could help us to parameterize and reduce the environment data dimensions from daily weather variables to a few covariates based on the crop growth stages (Heslot et al. 2014). It is not easy to reduce the number of covariates. In particular, only climatic parameters were used here, while soil quality, disease resistance and environmental stress could be other covariates (Heslot et al. 2014).
As with environmental covariates, genotypic covariates that play a role in the GYI have small main effects. The first dimension was well able to separate molecular markers. Approximately 30% of the markers used had a role in the GYI. Similar results have been reported by Crossa et al. (1999). Vernalization genes played a greater role in GYI than in Photoperiod genes. In this regard, Kamran et al. (2014) reported that vernalization genes had a greater effect on phenological stages. However, the Ppd-D1 gene is also of particular importance. The Ppd-D1 marker has been reported to be the most variable marker in a subset of markers with particularly effects across environments (Heslot et al. 2014). As Likhenko et al. (2015) stated, the compounds Ppd-D, Vrn-A1 and Vrn-B1 have a greater effect on traits. Allelic compounds in these genes may provide a better interpretation of GYI. Some of these allelic compounds can reduce the negative effects of climate change (Arjona et al. 2020). A review of the literature shows very few studies in this area. Perhaps one of the reasons for the lack of statistical methods is appropriate.
As a final conclusion, it can be stated that there is a high difference between Iranian wheat cultivars in terms of yield and yield stability during the studied years. Part of this diversity is due to differences in functional markers of Vrn and Ppd. Also, the GYI pattern could almost be interpreted by environmental factors, where relative humidity and temperature-related parameters played the most important role. Although obviously, the causes of the GYI are not limited to the above and there are various factors involved, even unknown ones. However, the same information on the causes of environmental factors responsible for the GYI can be influential in future management plans. In this regard, identifying of molecular markers in the selection process can be used in breeding programs.
References
Ajay BC, Aravind J, Fiyaz RA (2019) Ammistability: R package for ranking genotypes based on stability parameters derived from AMMI model. Indian J Genet Plant Breed 79:460–466
Alipour H, Abdi H (2020) Interactive effects of vernalization and photoperiod loci on phenological traits and grain yield and differentiation of Iranian wheat landraces and cultivars. J Plant Growth Regul. https://doi.org/10.1007/s00344-020-10260-8
Arjona JM, Royo C, Dreisigacker S, Ammar K, Villegas D (2018) Effect of ppd-a1 and ppd-b1 allelic variants on grain number and thousand kernel weight of durum wheat and their impact on final grain yield. Front Plant Sci 9:888
Arjona JM, Royo C, Dreisigacker S, Ammar K, Subirà J, Villegas D (2020) Effect of allele combinations at Ppd-1 loci on durum wheat grain filling at contrasting latitudes. J Agron Crop Sci 206:64–75
Chairi F, Aparicio N, Serret MD, Araus JL (2020) Breeding effects on the genotype× environment interaction for yield of durum wheat grown after the green revolution: the case of Spain. Crop J. https://doi.org/10.1016/j.cj.2020.01.005
Coan MMD, Marchioro VS, Franco FDA, Pinto RJB, Scapim CA, Baldissera JNC (2018) Determination of genotypic stability and adaptability in wheat genotypes using mixed statistical models. J Agric Sci Technol 20:1525–1540
Crossa J, Vargas M, Van Eeuwijk FA, Jiang C, Edmeades GO, Hoisington D (1999) Interpreting genotype × environment interaction in tropical maize using linked molecular markers and environmental covariables. Theor Appl Genet 99:611–625
Crossa J, Vargas M, Joshi AK (2010) Linear, bilinear, and linear-bilinear fixed and mixed models for analyzing genotype × environment interaction in plant breeding and agronomy. Can J Plant Sci 90:561–574
De Mendiburu MF (2016) Package “Agricolae”, statistical procedures for agricultural research. Version 1.3.0
De Leon N, Jannink JL, Edwards JW, Kaeppler SM (2016) Introduction to a special issue on genotype by environment interaction. Crop Sci 56:2081–2089
Elias AA, Robbins KR, Doerge RW, Tuinstra MR (2016) Half a century of studying genotype× environment interactions in plant breeding experiments. Crop Sci 56:2090–2105
Fusi N, Lippert C, Borgwardt K, Lawrence ND, Stegle O (2013) Detecting regulatory gene–environment interactions with unmeasured environmental factors. Bioinformatics 29:1382–1389
Gororo NN, Flood RG, Eastwood RF, Eagles HA (2001) Photoperiod and vernalization responses in Triticum turgidum × T. tauschii synthetic hexaploid wheats. Ann Bot 88:947–952
Herrera JM, Levy Häner L, Mascher F, Hiltbrunner J, Fossati D, Brabant C, Charles R, Pellet D (2020) Lessons from 20 years of studies of wheat genotypes in multiple environments and under contrasting production systems. Front Plant Sci 10:1745
Heslot N, Akdemir D, Sorrells ME, Jannink JL (2014) Integrating environmental covariates and crop modeling into the genomic selection framework to predict genotype by environment interactions. Theor Appl Genet 127:463–480
Iqbal M, Shahzad A, Ahmed I (2011) Allelic variation at the Vrn-A1, Vrn-B1, Vrn-D1, Vrn-B3 and Ppd-D1a loci of Pakistani spring wheat cultivars. Electron J Biotechn 14:1–2
Jambhulkar NN, Bose LK, Singh ON (2014) AMMI stability index for stability analysis. In: Mohapatra T (ed) CRRI Newsletter, Jan–Mar 2014, 15. Central Rice Research Institute, Cuttack
Kamran A, Iqbal M, Spaner D (2014) Flowering time in wheat (Triticum aestivum L.): a key factor for global adaptability. Euphytica 197:1–26
Kang MS (1988) A rank-sum method for selecting high-yielding, stable corn genotypes. Cereal Res Commun 16:113–115
Kassambara A, Mundt F (2017) Package “factoextra”. https://cran.r676project.org/web/packages/factoextra/index.html
Kondić-Špika A, Mladenov N, Grahovac N, Zorić M, Mikić S, Trkulja D, Marjanović-Jeromela A, Miladinović D, Hristov N (2019) Biometric analyses of yield, oil and protein contents of wheat (Triticum aestivum L.) genotypes in different environments. Agron J 9:270
Likhenko IE, Stasyuk AI, Zyryanova AF, Likhenko NI, Salina EA (2015) Study of allelic composition of Vrn-1 and Ppd-1 genes in early-ripening and middle-early varieties of spring soft wheat in siberia. Russ J Genet Appl Res 5:198–207
Macholdt J, Honermeier B (2019) Stability analysis for grain yield of winter wheat in a long-term field experiment. Arch Agron Soil Sci 65:686–699
Malosetti M, Ribaut JM, Van Eeuwijk FA (2013) The statistical analysis of multi-environment data: modeling genotype-by-environment interaction and its genetic basis. Front Physiol 4:44
Mansour E, Moustafa ES, El-Naggar NZ, Abdelsalam A, Igartua E (2018) Grain yield stability of high-yielding barley genotypes under Egyptian conditions for enhancing resilience to climate change. Crop Pasture Sci 69:681–690
Mohammadi R (2017) Interpretation of genotype× year interaction in rainfed durum wheat under moderate cold conditions of Iran. New Zeal J Crop Hort 45:55–74
Monteverde E, Gutierrez L, Blanco P, De Vida FP, Rosas JE, Bonnecarrère V, Quero G, Mccouch S (2019) Integrating molecular markers and environmental covariates to interpret genotype by environment interaction in Rice (Oryza sativa L) grown in subtropical areas. Genome 9:1519–1531
Olivoto T, Lúcio ADC (2020) metan: An R package for multi-environment trial analysis. Methods Ecol Evol 11:783–789
Olivoto T, Lúcio AD, da Silva JA, Marchioro VS, de Souza VQ, Jost E (2019) Mean performance and stability in multi-environment trials I: Combining features of AMMI and BLUP techniques. Agron J 111:2949–2960
Pacheco A, Vargas M, Alvarado G (2015) GEA-R (Genotype by environment analysis with R for Windows) Version 3.0—CIMMYT Research Software Dataverse—CIMMYT Dataverse Network
Pour-Aboughadareh A, Yousefian M, Moradkhani H, Poczai P, Siddique KH (2019) STABILITYSOFT: a new online program to calculate parametric and nonparametric stability statistics for crop traits. Appl Plant Sci 7:e01211
Purchase JL (1997) Parametric analysis to describe genotype × environment interaction and yield stability in winter wheat. Ph.D. thesis, Department of Agronomy, Faculty of Agriculture of the University of the Free State, Bloemfontein, South Africa
Ramburan S, Zhou M, Labuschagne M (2012) Integrating empirical and analytical approaches to investigate genotype× environment interactions in sugarcane. Crop Sci 52:2153–2165
Resende MDV (2007) Matemática e Estatística na Análise de Experimentos e No Melhoramento Genético. Embrapa Florestas, Colombo
Reynolds MP, Trethowan R, Crossa J, Vargas M, Sayre KD (2002) Physiological factors associated with genotype by environment interaction in wheat. Field Crops Res 75:139–160
Richards RA (1982) Breeding and selection for drought resistant wheat. In: Drought resistance in crops with emphasis on Rice. International Rice Research Institute, Manila
Rosado RDS, Rosado TB, Cruz CD, Ferraz AG, Laviola BG (2019) Genetic parameters and simultaneous selection for adaptability and stability of macaw palm. Sci Hortic 248:291–296
Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RWL (1984) Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proc Nati Acad Sci 81:8014–8018
Schmidt J, Tricker PJ, Eckermann P, Kalambettu P, Garcia M, Fleury DL (2019) Novel alleles for combined drought and heat stress tolerance in wheat. Front Plant Sci 10:1800
Shukla GK (1972) Some statistical aspects of partitioning genotype environmental components of variability. Heredity 29:237–245
Sneller CH, Kilgore-Norquest L, Dombek D (1997) Repeatability of yield stability statistics in soybean. Crop Sci 37:383–390
Taranto F, Nicolia A, Pavan S, De Vita P, D’agostino N, (2018) Biotechnological and digital revolution for climate-smart plant breeding. Agron J 8:277
Vaezi B, Pour-Aboughadareh A, Mohammadi R, Mehraban A, Hossein-Pour T, Koohkan E, Ghasemi S, Moradkhani H, Siddique KH (2019) Integrating different stability models to investigate genotype × environment interactions and identify stable and high-yielding barley genotypes. Euphytica 215:63
Voltas J, Lopez-Corcoles H, Borras G (2005) Use of biplot analysis and factorial regression for the investigation of superior genotypes in multi-environment trials. Eur J Agron 22:309–324
Whittal A, Kaviani M, Graf R, Humphreys G, Navabi A (2018) Allelic variation of vernalization and photoperiod response genes in a diverse set of North American high latitude winter wheat genotypes. PLoS ONE 13:e0203068–e0203068
Wricke G (1962) Uber eine Methode zur Erfassung der okologischen Streubreite in Feldverzuchen. Zeitschr F Pflanzenz 47:92–96
Xu Y (2016) Envirotyping for deciphering environmental impacts on crop plants. Theor Appl Genet 129:653–673
Zali H, Farshadfar E, Sabaghpour SH, Karimizadeh R (2012) Evaluation of genotype× environment interaction in chickpea using measures of stability from AMMI model. Ann Biol Res 3:3126–3136
Zorić M, Terzić S, Sikora V, Brdar-Jokanović M, Vassilev D (2017) Effect of environmental variables on performance of Jerusalem artichoke (Helianthus tuberosus L.) cultivars in a long term trial: a statistical approach. Euphytica 213:23
Funding
The authors received no financial support for the research, authorship and publication of this article.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by A. Mohan.
Supplementary Information
Rights and permissions
About this article
Cite this article
Alipour, H., Abdi, H., Rahimi, Y. et al. Genotype-by-year interaction for grain yield of Iranian wheat cultivars and its interpretation using Vrn and Ppd functional markers and environmental covariables. CEREAL RESEARCH COMMUNICATIONS 49, 681–690 (2021). https://doi.org/10.1007/s42976-021-00130-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42976-021-00130-8