Abstract
A fundamental requirement in nature is for a cell to correctly package and divide its replicated genome. Condensin is a mechanical multisubunit complex critical to this process. Condensin uses ATP to power conformational changes in DNA to enable to correct DNA compaction, organization, and segregation of DNA from the simplest bacteria to humans. The highly conserved nature of the condensin complex and the structural similarities it shares with the related cohesin complex have provided important clues as to how it functions in cells. The fundamental requirement for condensin in mitosis and meiosis is well established, yet the precise mechanism of action is still an open question. Mutation or removal of condensin subunits across a range of species disrupts orderly chromosome condensation leading to errors in chromosome segregation and likely death of the cell. There are divergences in function across species for condensin. Once considered to function solely in mitosis and meiosis, an accumulating body of evidence suggests that condensin has key roles in also regulating the interphase genome. This review will examine how condensin organizes our genomes, explain where and how it binds the genome at a mechanical level, and highlight controversies and future directions as the complex continues to fascinate and baffle biologists.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Faithful segregation of the genome during cell division is a prerequisite for the life of all organisms. A major class of proteins controlling this task during the cell division of all species are the structural maintenance of chromosome (SMC) proteins. SMC proteins are ABC ATPases, capable of altering DNA topology through ATP hydrolysis (Losada and Hirano 2005). Although structurally similar, different combinations of SMC subunits form different complexes. SMC1 and SMC3 are the core subunits of cohesin that coordinates sister chromatid cohesin, while SMC2 and SMC4 are the core subunits of condensin and the main focus of this review. SMC5 and SMC6 form an as yet unnamed complex important for DNA replication and chromosome segregation (Kegel et al. 2011; Bermudez-Lopez and Aragon 2016; Pryzhkova and Jordan 2016).
All organisms contain some form of condensin. The complex is both highly conserved and ancient, predating histones. The bacterial SMC complex is considered a condensin and not a cohesin, with replicated sister chromatids of Muk− cells showing increased rather than decreased cohesion (Danilova et al. 2007). Bacteria and archaea contain the condensin SMC-ScpAB, most commonly studied in Bacillus subtilis, while the condensin MukBEF is present in ϒ-proteobacteria and includes Escherichia coli. The newest addition to the condensin family is MksBEF, which is distantly related to MukBEF and often coexists in bacteria with SMC-ScpAB and or MukBEF (Petrushenko et al. 2011). Animals and plants have two condensins, condensins I and II. They differ from their prokaryotic counterparts by having an SMC heterodimer (SMC2/SMC4) instead of a homodimer, and three non-SMC subunits (CAP-D2, CAP-H, CAP-G for condensin I and CAP-D3, CAP-H2, CAP-G2 for condensin II) instead of two. Fungi and the ciliate Tetrahymena thermophila lack condensin II, which was presumably lost in evolution of these organisms, while all metazoans possess condensins I and II (Hirano 2016).
This review will focus on the many roles of the condensin complexes and present a view as to how they might act at a mechanistic level. With advances in sequencing technologies, proteomics, and super-resolution microscopy, we have learnt a great deal about condensins across a variety of species. However, there are still a number of key questions and controversies that need to be resolved.
Discovery and early history of condensin
A pioneering series of biochemical experiments led to condensin first being described around the same time as a genetic screen in the fission yeast Schizosaccharomyces pombe identified mutants in complex members. In 1994, Hirano and Mitchison isolated a heterodimer XCAP-C and E (later SMC4 and SMC2) purified from Xenopus egg extracts capable of assembling and maintaining the structure of sperm DNA (Hirano and Mitchison 1994). These genes were cloned, and antibodies raised against the proteins displayed mitotic chromosome localization. This breakthrough study was followed in 1997 by the Hirano lab that described a 13S complex containing XCAP-C, E, and further subunits (CAP-D2, H, and G), in addition to an 8S complex containing XCAP-C and E (Hirano et al. 1997). The study was the first full description of the condensin I complex and where the collective term “condensin” first appeared. Concurrently in 1994, the Earnshaw laboratory independently identified scaffold protein 2 (ScII) in chicken cells using an antibody raised to the bulk chromosome scaffold (Saitoh et al. 1994) that proved to be SMC2. The protein was cloned, and specific antibodies demonstrated a staining along the chromosome axis during mitosis. This study was one of the earliest to recognize that each SMC subunit might self-fold intra-molecularly along the coiled coil region. The first in vivo description of a condensin subunit came from S. pombe in 1993, using a genetic screen that identified a cell untimely torn (cut) phenotype, describing mutants that bisected the nucleus during cytokinesis (Samejima et al. 1993). One of these genes was cut14, later identified as SMC2, and in 1994 cut3/SMC4 and cut14/SMC2 were further characterized as genes required for chromosome condensation and segregation during mitosis (Saka et al. 1994). Almost a decade after the discovery of condensin I, condensin II was first described by the Hirano lab after identifying a set of proteins that had high homology to the non-SMC subunits of condensin I (Ono et al. 2003). These proteins were the condensin II subunits CAP-D3, H2, and G2, which were then cloned and purified.
Structure and organization of condensin, rod-like or open?
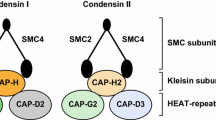
The SMC2/4 core condensin subunits are long polypeptides (1200 and 1500 amino acids) consisting of 3 distinct domains: (1) the head region where the SMC dimers come together to form an ATPase capable of binding and hydrolyzing two ATP molecules, (2) the long coiled-coil region of about 50 nm, and (3) a hinge region connected to coiled coils, which mediates SMC dimerization and binds DNA (Hirano 2012) (Fig. 1).
Schematic of vertebrate condensin I and II structure and organization. The pentameric condensin complex consists of three domains: (1) the head where the SMC subunits form an ATPase that in turn binds the auxiliary subunits (CAP-H, D2, and G for condensin I and CAP-H, D3, and G2 for condensin II), (2) the long coiled-coil region, and (3) the hinge that facilitates SMC2/4 dimerization
The non-SMC or auxiliary subunits (CAP-D2, G, H for condensin I and D3, G2, H2 for condensin II) bind the head domain of SMC2 and SMC4. CAP-D2/G and CAP-D3/G2 subunits contain HEAT (Huntingtin, elongation factor 3, the A subunit of protein phosphatase 2A, TOR lipid kinase) repeats that are thought to elicit DNA binding activity, at least for condensin I (Piazza et al. 2014). The HEAT domains are also thought to be necessary for formation of the chromosome axis, as these domains in CAP-D2 and CAP-G subunits act antagonistically in the assembly of chromosome axes (Kinoshita et al. 2015).
Cross-linking mass spec analyses and arrangement of condensin
CAP-H/H2, like its SccI counterpart in cohesin, is the third member of the pentameric condensin complex and considered the kleisin (Greek for closure) subunit of each pentameric condensin complex, with the ability to connect to both SMC2 and SMC4. Cross-linking mass spectrometry of condensin I in chicken DT40 supported this view and suggested a tight apposition along the coiled coils of SMC2 and SMC4 leading to a rod-like appearance (Barysz et al. 2015). This appearance was also evident in early electron micrographs of purified condensin (Anderson et al. 2002).
The cross-linking mass spectrometry analyses of purified condensin I in chicken DT40 (Barysz et al. 2015) and also purified non-SMC subunits of condensin in yeast (Ycg1/CAP-G, Ycs4/CAP-D2, Brn1/CAP-H) (Piazza et al. 2014) provide a key framework for condensin subunit interactions. Together with immunoprecipitation assays using pairwise expression of fragments of human condensin I subunits in baculovirus (Onn et al. 2007), a consensus for the geometry and juxtaposition of condensin emerged (reflected in Fig. 1), albeit with some differences.
Available data suggest that the kleisin CAP-H platforms the SMC subunits, with SMC2 interacting with the N-terminus of CAP-H and SMC4 with the C-terminus (Onn et al. 2007; Barysz et al. 2015). These studies found that the HEAT subunits CAP-D2 and CAP-G do not interact with either SMC2 or SMC4. Cross-linking mass spec analyses from yeast and chicken and immunoprecipitation data show that CAP-D2 and CAP-G have minimal or weak interaction with each other (Onn et al. 2007; Piazza et al. 2014; Barysz et al. 2015). Immunoprecipitation analyses in baculovirus show CAP-D2 binding the N-terminus of CAP-H and CAP-G with the C-terminus of CAP-H. Yeast cross-linking data of the non-SMC subunits of condensin also show that the majority of cross-links occur between the N-terminus of CAP-H and CAP-D2 and the C-terminus of CAP-H and CAP-G (Piazza et al. 2014), whereas chicken cross-linking data somewhat differs with cross-links found between the middle and C-terminus parts of CAP-H for CAP-G and CAP-D2 (Barysz et al. 2015).
A similar set of experiments using baculovirus expressed subunits was set up for human condensin II, and the arrangement was the same, with the position of hCAP-D3 and hCAP-G2 in condensin II equivalent to that of hCAP-D2 and hCAP-G (Onn et al. 2007). Although much has been learnt from the described experiments, the data are largely based on free/not DNA-bound condensin; so whether the same arrangement exists for DNA bound condensin is not yet clear, although this seems likely.
Crystallographic studies and high resolution imaging of condensin
Crystallographic studies on bacterial SMC complexes (Burmann et al. 2013) and eukaryotic cohesin (Gligoris et al. 2014; Huis in ‘t Veld et al. 2014) have shown that, although the CAP-H kleisin C-terminus makes contact with the SMC head, the N-terminus associates with the coiled coil to form the putative entry/exit gate. Furthermore, recent crystal structures of prokaryote condensin and also yeast SMC2/SMC4 showed a rod-like structure, but demonstrated that ATP binding of the hinge to DNA leads to a more open conformation (rod-to-ring transition) (Soh et al. 2015).
The notion of condensin appearing as predominately rod-like has again been challenged with a combination of “dry” and real-time “liquid” and high-resolution imaging of SMC2/4 conformational dynamics in the yeast S. cerevisiae (Eeftens et al. 2016). The results, based on the purified SMC2/4 dimer, showed a somewhat different picture from corresponding earlier rotary shadowing analyses (Anderson et al. 2002). In the novel study, only ~1/3 of condensin dimers had tightly apposed coiled coils in an I- or Y-like structure, with the majority displaying more open V- or O-like structures. These “dry analyses” were then followed by high-speed liquid atomic force microscopy to create real-time images of SMC2/SMC4 dimers in liquid. The real-time analyses also confirmed a majority of V and O structures and identified two additional confirmations: a P-shaped dimer where only one of the heads engages with the hinge and a B structure where both heads engage with the hinge. The accumulated data present a picture where condensin likely exists in a rod-like structure before becoming more open after DNA contact.
Species differences and localization of condensins in eukaryotes
The long evolutionary history of the condensin complex has resulted in some divergent localizations among different species, suggesting some evolution of additional functions to cope with different genome sizes and modes of division.
Perhaps the most common cytological image that springs to mind when biologists think about condensin localization is its dotted appearance along the central axes of swollen mitotic chromosomes. Condensin I is notably enriched at centromeres, which is apparent both cytologically and also through ChIP-seq analyses (Gerlich et al. 2006; Kim et al. 2013), while cytological and live cell imaging analyses suggest that condensin II is also enriched at the centromere (Ono et al. 2004; Gerlich et al. 2006). In vertebrates, fluorescence recovery after photobleaching (FRAP) analyses show that condensin II is highly stable while condensin I is more mobile during mitosis (Gerlich et al. 2006). Cross sections of mitotic chromosomes revealed condensin, as well as topoisomerase II and KIF4A, running through a central core of the chromosome, giving rise to the notion that a central “scaffold” network of proteins is key to organizing and folding chromatin (Saitoh et al. 1994; Maeshima and Laemmli 2003; Kireeva et al. 2004; Belmont 2006; Hudson et al. 2009; Samejima et al. 2012; Poonperm et al. 2015). Experiments from the Marko Lab have challenged the idea of an interconnected protein network supporting mitotic chromosomes, showing that structural links in mitotic chromosomes are maintained by chromatin fibers and not by a scaffold network (Poirier and Marko 2002). However, there is no doubt that condensins and other scaffold proteins occupy an inner central structure of the chromatid.
The cytological localization of condensin in eukaryotes has been studied extensively over many years in a variety of systems revealing important clues. In vertebrates, condensin I dramatically concentrates on chromosomes following nuclear envelope breakdown (NEBD) after prophase and begins to disappear from chromosomes during telophase (Hirota et al. 2004; Ono et al. 2004; Gerlich et al. 2006; Zhang et al. 2016). Condensin II is present on DNA throughout interphase and notably concentrates on prophase chromosomes (Hirota et al. 2004; Gerlich et al. 2006; Zhang et al. 2016), suggesting it is important in establishing the initial chromatid axis in mitosis. Condensin II interphase dynamics have recently been studied using quantitative live-cell imaging in chicken cells. This study showed that the complex is dramatically enriched in G1 DNA after cells exit mitosis (Zhang et al. 2016), and a number of studies suggest a key role for condensin II in interphase nuclear architecture (Bauer et al. 2012).
Vertebrates
In metozoans, the common view is that condensin I is cytoplasmic during interphase and is only present on chromosomes following NEBD after prophase (Hirota et al. 2004; Gerlich et al. 2006). However, recent data suggest that the situation may not be quite as clear-cut. Using a combination of fixed and live cell imaging and immunoblots to isolate chromatin in chicken DT40 cells, a small pool of condensin I was found to persist in the nucleus during G1, which was gradually lost during S and G2 phases (Zhang et al. 2016). Very early cytological studies in human cells correspondingly detected a small pool of CNAP1/CAP-D2 localizing to G1 nuclei that disappeared as cells progressed into S and G2 phases (Schmiesing et al. 2000). Further evidence for condensin I in interphase nuclei was provided at a molecular level from ChIP-seq analyses. In human MCF breast cancer cells synchronized to 80–95% G0/G1, condensin I was predominately found at intergenic and intronic regions and notably at ER-α-binding sites, occupying 2916 sites genome-wide, increasing to 7292 after estradiol treatment (Li et al. 2015). Knockdown of condensin I in G0/G1 MCF cells and asynchronous DT40 cells caused gene misregulation, and 3D-FISH in DT40 revealed a loss of compaction of ribosomal DNA (rDNA) and Z-Rep loci following condensin I removal, suggesting that the pool of condensin I in interphase might not merely be a passive carryover from mitosis (Li et al. 2015; Zhang et al. 2016). The data also provide some rationale why a mitotic complex (condensin I) predominately binds at gene regulatory regions (Kim et al. 2013). In vertebrates, while it is true that condensin I is most strongly enriched on mitotic chromosomes, a growing body of evidence questions the notion that it is completely excluded from interphase nuclei.
Fungi and algae
Unicellular organisms show quite a divergence in condensin behavior. In fungi, which only contain condensin I, the yeasts S. cerevisiae and S. pombe vary in condensin dynamics. Similar to vertebrates, condensin I in S. pombe appears cytoplasmic during interphase and then accumulates on chromosomes in mitosis after NEBD (Sutani et al. 1999). In contrast, S. cerevisiae condensin is present in the nucleus throughout the cell cycle (Freeman et al. 2000). The red alga Cyanidioschyzon merolae has not lost condensin II, unlike many unicellular organisms. Condensin II in C. merolae is present in low levels from S phase to prophase and then concentrates on centromeres during metaphase, whereas condensin I is barely detectable until metaphase where it then accumulates on chromosome arms and persists through to telophase (Fujiwara et al. 2013).
Worms
Caenorhabditis elegans has condensins I and II but additionally a third condensin referred to as the dosage compensation complex (DCC), which has a specialized role in balancing gene expression between XX hermaphrodites and XO males (Csankovszki et al. 2009; Crane et al. 2015). C. elegans chromosomes are holocentric with centromeres dotted along the arms. Condensin II localizes on the outward face of chromosomes in parallel axes, whereas condensin I coats mitotic chromatids more inwardly with little condensin II colocalization (Hagstrom et al. 2002). An interesting observation in C. elegans is that some condensin I colocalizes with Aurora B (AIR-2) at the spindle midzone during anaphase (Collette et al. 2011), which to date has not been described in other species.
Flies
Drosophila condensin II is significantly different from vertebrate condensin II. There is no known CAP-G2 gene present in the fly genome, although it is worth noting that CAP-G in worm was not initially found using homology-dependent searches and was only identified when proteomic data became available (Csankovszki et al. 2009). In fly, loss-of-function mutations for Cap-D3 and Cap-H2 genes do not affect viability, although they produce male sterility, suggesting condensin II is dispensable for mitosis (Savvidou et al. 2005; Hartl et al. 2008). This somewhat parallels the primitive red alga C. merolae where condensin I plays a key role in mitosis while condensin II is dispensable in mitosis but required for interphase nuclear structure and meiosis (Fujiwara et al. 2013). A recent study further questioned the existence of an abundant and soluble condensin II complex in flies (Herzog et al. 2013). Using GFP condensin I transgenes in flies, the study also found that Cap-G behavior resembled condensin II localization in vertebrates with strong interphase and prophase localization, while the other condensin I subunits Cap-H and Cap-D2 were predominately cytoplasmic in interphase and only associated strongly with chromatin after NEBD (Herzog et al. 2013). There was, however, no biochemical evidence that Cap-G can substitute for Cap-G2.
Condensin ratios
A further difference between species is the ratio of condensin I/II. The ratio of condensin I/II is 1:1 in human HeLa cells, but 5:1 in Xenopus egg extracts, while isolated mitotic chromosomes from chicken DT40 cells have the condensin I/II ratio of 10:1 (Ono et al. 2003; Ohta et al. 2010; Shintomi and Hirano 2011). However, it is not clear whether the differences relate to species or cell type or both. For instance, the above analyses came from oocytes, lymphoma B, and cervical cancer cells in addition to being from different species. While it is tempting to link the ratio of condensin I/II to the particular requirements and sizes of genomes, comparisons at this stage can only be tentative. A unified pan-analysis using quantitative proteomics across different cell types in diverse species assessing total, cytoplasmic, and chromosome-bound ratios of condensin I/II would be of great interest to define the balance for mitotic shaping of chromosomes.
Is condensin binding sequence dependent or epigenetic?
The initial cytological localization of condensin on metaphase chromosomes showed an axial staining pattern along the length of each chromatid. From such patterns, one could predict that condensin complexes would be evenly distributed to gather up loops of chromatin to form the condensed metaphase chromosome. Genome-wide analyses of condensin-binding sites in bacteria and yeast confirmed that the complex binds along the length of the chromosome (Wang et al. 2005; D'Ambrosio et al. 2008; Gruber and Errington 2009).
Closer inspection of the binding sites started to reveal some interesting characteristics. In the bacterium B. subtilis, condensin localized to specific sequences such as the parS centromere-like sequences, around replication origins and highly expressed genes like rDNAs and tRNAs (Gruber and Errington 2009). Similar patterns were observed in single-celled budding and fission yeasts (Wang et al. 2005; D'Ambrosio et al. 2008; Nakazawa et al. 2015; Sutani et al. 2015). A common feature of condensin binding from genome-wide distribution is the localization to transcription start and termination sites.
Strikingly, in multicellular eukaryotes like worm, chicken, and mouse, these binding patterns were conserved suggesting that condensin may have additional roles other than genome packaging for efficient chromosome segregation (Dowen et al. 2013; Kim et al. 2013; Kranz et al. 2013).
Genome-wide analyses have shown that condensin has a preference for specific DNA regions such as centromeres, telomeres, genes, and sites of initiation and termination of DNA replication (Dowen et al. 2013; Kim et al. 2013; Kranz et al. 2013). The DCC in worm that contains all condensin I subunits except for SMC4 (replaced with its variant DPY27) shows sequence specificity for the X chromosome (McDonel et al. 2006; Ercan et al. 2007; Jans et al. 2009). However, there is little evidence to show that either the canonical condensin I or II complexes show a strict DNA sequence motif dependency. One example of this feature is in budding yeast where it binds to promoter motifs of RNA Pol III transcribed genes (D'Ambrosio et al. 2008); however, other evidence is accumulating in favor of a chromatin conformation dependency together with other chromatin-binding factors (Kimura and Hirano 1997; Liu et al. 2010). Furthermore, condensin I binds directly to histones H2A and H4 during mitosis (Tada et al. 2011; Barysz et al. 2015). Recent experimental data showed that condensin bound to unwound DNA structures to keep the transcription machinery off the chromatin during mitosis (Sutani et al. 2015) (see also ATP independent DNA reannealing below). This activity implies that condensin has a clearing function before chromosome segregation and a potential bookmarking role to ensure genes are ready for transcription in the subsequent interphase.
Mechanism of action of condensin
Condensin is an enzyme with two well-described activities: (1) ATPase-dependent positive supercoiling of closed circular double-stranded DNA (dsDNA) in the presence of topoisomerase I and (2) ATP-independent DNA reannealing activity (Hirano 2005). How do these activities relate to chromosome condensation?
ATPase-dependent positive supercoiling
The ability of condensin to introduce positive supercoils into DNA has been shown in vitro (Kimura et al. 1999) and presents an attractive model for DNA loop organization (Hirano 2012), with evidence that condensin can organize multiple supercoils into a solenoidal structure (Kimura et al. 1999). Large-scale folding of chromatin by condensin-mediated positive writhing increases torsional stress in the DNA (Gilbert and Allan 2014). This torsional stress needs to be relieved to allow continuous folding (Farr et al. 2014), explaining why dsDNA passing type IV topoisomerases, like human topoisomerase IIα (Top2), are essential for hypercompaction of chromosomes (Samejima et al. 2012; Farr et al. 2014). Conversely, inactivation of condensin impedes chromosome segregation (Lukas et al. 2011; Green et al. 2012). Providing an explanation for this, condensins’ ability to positively supercoil DNA stimulates decatenation of catenated DNA by Top2 in vitro (Baxter et al. 2011) and on mini-chromosomes transfected into bacterial cells (Charbin et al. 2014). In line with these observations, ChIP-qPCR showed that Top2 was recruited to chromosome arms in anaphase of budding yeast in a condensin-dependent manner (Leonard et al. 2015). The authors suggested that the overwinding or positive writhing activity of condensin on DNA attracts Top2. Thus, mitotic DNA condensation and decatentation appear to be two sides of the same process governed by type IV topoisomerases and condensin complexes (Baxter and Aragon 2012).
ATP independent DNA reannealing
Recent data suggest that the reannealing activity of condensin is necessary to reduce unwound DNA segments generated by transcription at protein encoding genes (Sutani et al. 2015). This elimination of unwound DNA is believed to be a prerequisite for supercoiling activity (Hirano 2012; Leonard et al. 2015; Sutani et al. 2015). Presciently, Mitsuhiro Yanagida envisaged that a key function of condensin was to clear the way for mitosis (Yanagida 2009).
DNA-condensin interaction
Cohesin acts by forming a ring around DNA (Haering et al. 2002). Ideas for the mode of DNA entry into and out of cohesin have been continually evolving, but recent data provide strong evidence for cohesin ATPase hydrolysis mediating both DNA entry and exit through SMC head disengagement, with a key step being removal of the cohesin subunit WAPL from SMC3, thereby fully opening the kleisin gate (Murayama and Uhlmann 2014; Murayama and Uhlmann 2015). Does condensin also interact with DNA via a similar mechanism?
The notion of condensin encircling DNA rather than binding or sitting on top of DNA was suggested after a series of experiments in budding yeast, where tobacco etch virus (TEV) sites were incorporated into condensin at symmetrical points along the coiled coil of SMC2 and in the CAP-H/kleisin subunit. This allowed cleavage to “open” the structure (Cuylen et al. 2011). Using a mini-chromosome assay, inducible TEV cleavage in yeast released DNA and conversely linearizing DNA released condensin, providing a compelling argument that condensin also entraps DNA.
Various studies point to ATPase activity being essential for condensin binding to chromosomes. Based on highly conserved ATPase mutants first described in B. subtilis condensin (Hirano et al. 2001), two independent studies in chicken DT40 and Xenopus using analogous ATPase mutations drew the same conclusion. Introduction of ATP binding mutations in the highly conserved Walker A and Walker B motifs (K-I and D-A respectively) in DT40 SMC2 prevented condensin loading onto chromosomes (Hudson et al. 2008), while a reconstituted condensin complex containing the analogous K-I ATP binding mutation in the SMC4 complex was unable to bind chromosomes (Kinoshita et al. 2015). Furthermore, Walker B E-Q ATP hydrolysis mutations in DT40 SMC2 and Xenopus SMC4 permit condensin to load onto mitotic chromosomes but failed to support proper chromosome structure with localization appearing diffuse. Together, these data indicate that ATP binding is essential for condensin loading onto chromosomes, while ATP hydrolysis of condensin mediates correct localization of the complex and formation of a higher-order structure (for putative model, see Fig. 2).
Model for condensin binding DNA and mitotic chromosome organization. i ATPase-mediated binding of condensin to DNA. Condensin comes into contact with DNA via the hinge region following ATP binding (Hudson et al. 2008; Kinoshita et al. 2015). Based on recent cohesin studies (Murayama and Uhlmann 2014; Murayama and Uhlmann 2015), we speculate condensin then opens the head region allowing embracement of DNA. ii Possible model for condensin organization of DNA. The model assumes that condensin is able to interact with neighboring condensin to form multimers
In contrast, the analogous ATP hydrolysis mutants in SMC1 and SMC3 prevented the cohesin complex from loading onto DNA (Arumugam et al. 2003; Weitzer et al. 2003), in line with recent data showing that cohesin ATP hydrolysis is necessary for both entry and exit from DNA (Murayama and Uhlmann 2014; Murayama and Uhlmann 2015). The data to date imply some mechanistic loading differences between cohesin and condensin.
Available data suggest that the HEAT repeat subunits are exposed and could provide potential contact points with DNA. Pertinently, a study found that the purified non-SMC subunits from yeast showed a significant DNA binding activity and demonstrated mutations in yeast and human Brn1/CAP-H that disrupt interactions with Ycg1/CAP-G which inhibit condensin chromosome association (Piazza et al. 2014). The DNA binding properties of the SMC2/4 hinge (Griese et al. 2010; Akai et al. 2011) and also non-SMC subunits (Piazza et al. 2014) further underscore the highly mechanical nature of the condensin complex. It will be of great interest to definitively map all the precise points of contact between DNA and condensin using proteomic approaches while also dissecting out the various functional implications of the different protein-DNA contact points.
Condensin-condensin interaction
The idea of condensin being able to multimerize with adjacent condensins presents an attractive model for the complex being able to compact chromosomes and regulate a higher-order structure (Fig. 2 (ii)). What is the evidence for multimerization? In E. coli, single-molecule millisecond multicolor fluorescence microscopy of the SMC complex MukBEF showed that 8–10 complexes accumulate in spots and measuring intensities of mCherry and GFP fusions pairs to MukB, E, and F showed a stoichiometry periodicity of 4:4:2 (Badrinarayanan et al. 2012). Furthermore, condensin I pulldowns under non-denaturing conditions showed evidence of multimer formation, with a 650-kDa band reflecting the canonical complex but additionally a smear present from 700 kDa to above 1236 kDa (Barysz et al. 2015). Understanding the clustering nature of condensin in chromosomes and providing definitive evidence that the complex can bind adjacent condensin neighbors in vivo remain a high priority.
Model for condensin action
The precise mechanism that allows condensin to facilitate mitotic chromosome condensation is still a hotly debated area, and any discussion needs to take into account the known activities of the complex (see Fig. 3). A favored mechanism for condensin action is the loop extrusion model, which suggests that a single condensin molecule can bind at two nearby points on DNA and slide to generate a progressively larger loop (Nasmyth 2001). Using polymer simulations of chromosome dynamics and simplified synthetic models of condensin, a study showed the ability of condensin to bind to two adjacent loci and slide the two contact points in opposite directions, creating an extruded loop (Goloborodko et al. 2016). A difficulty with this model is that there is no known activity or mechanism for condensin to extrude DNA loops. An alternative model suggests that the positive supercoiling activity of condensin promotes the formation of chiral loops in chromatin (Hirano 2012). A third model is that condensin acts as a cross-linker capable of bringing distant chromatin segments together (Cuylen et al. 2011; Hirano 2016; Piskadlo and Oliveira 2016). We speculate a two-step process with localized chiral looping or loop extrusion followed by cross-linking activities that generate multimerized condensins (Fig. 2 (ii)).
Differential roles of condensin I and II in mitotic chromosome formation
During mitotic chromosome condensation, chromosomes transform from an amorphous mass of DNA in interphase into the cytologically individualized X-shaped structures known as mitotic chromosomes. The process involves a thickening of the 10-nm fiber of interphase chromosomes to approximately 700 nm by metaphase in higher eukaryotes (Nishino et al. 2012). Somewhat surprisingly, the actual volume reduction as measured using GFP-H2B is only 2–3-fold (Vagnarelli 2012). Rather than being a smooth linear progression, 4D live cell microscopy and snapshot deconvolution fluorescence imaging suggest that progression to the metaphase chromosome state is a discontinuous process involving expansion and contraction (Liang et al. 2015).
There are two main theories describing mitotic chromosome condensation: (1) the hierarchical model where DNA is folded into increasingly higher-order structures and (2) the radial loop model where chromosome loops are attached and regulated by a central network of proteins referred to as the mitotic scaffold. Chromatin capture (Hi-C) data favors formation of consecutive chromatin loops (Naumova et al. 2013). Experiments using small-angle X-ray scattering (SAXS) further support the scaffold model and also imply a dynamic and flexible mitotic chromosome structure rather than a regular static one (Nishino et al. 2012). It is important to note two overlapping and contributing factors to mitotic chromosome condensation: (1) histone modifications (for further details see, Wilkins et al. 2014; Antonin and Neumann 2016) and (2) non-histone proteins discussed in here and reviewed in detail in Belmont (2006). The interplay between histones and condensin is not yet fully understood; however, a recent study has shed new light showing that nucleosome eviction in mitosis is necessary to start condensin binding and looping (Toselli-Mollereau et al. 2016).
Early cell-free experiments using Xenopus egg extract showed that individualized chromosomes were no longer seen when condensin was immunodepleted (Hirano and Mitchison 1994). This notion has since been challenged with removal of both condensin via SMC2 or SMC4 causing structurally aberrant but cytologically visible “fuzzy” mitotic chromosomes in a number of animals and also cell culture models (Hudson et al. 2003; Ono et al. 2003; Samejima et al. 2012).
Condensin-null phenotypes have been analyzed extensively for somatic cell mitosis. Various vertebrate tissue culture systems have found specific differences for condensins I and II in mitotic chromosome formation. When condensin II is removed, a loss of axial condensation is evident with mitotic chromosomes longer and less rigid and frequently appearing “curly” along the chromatid axis (Ono et al. 2003; Abe et al. 2011; Green et al. 2012) (Fig. 4). 3D-SIM of CAP-D3/condensin II-depleted mitotic chromosomes showed that chromatids often lost their parallel register and appeared to cross over or entangle (Green et al. 2012) and cells in anaphase displayed a dramatic “bulky” chromatin bridging phenotype, which may be the result of whole chromosome entanglements. A further common defect in metazoans without condensin II is a dramatically shortened prophase, with classical prophase configurations nearly absent (Hirota et al. 2004; Gerlich et al. 2006; Csankovszki et al. 2009; Abe et al. 2011).
Contrasting effects of condensins I and II in mitotic chromosome formation using tet/OFF conditional DT40 condensin knockouts (KO). i Examples of metaphase chromosome spreads (hypotonic swelling, methanol/acetic acid fixed) from chicken DT40 wild type (WT), SMC2 KO (removes both condensins I and II), CAP-H KO (removes condensin I), CAP-D3 KO (removes condensin II), and CAP-D3T1403A (CAP-D3/condensin II Cdk1 site mutant causing chromosome hypercondensation). ii Quantification of length and width of chromosome 1 from each of DT40 WT, SMC2 KO, CAP-H KO, and CAP-D3T1403A. A minimum of 30 chromosomes for each line were quantitated
In contrast, lateral condensation is altered and mitotic chromosomes are significantly shorter and wider when condensin I is removed (Green et al. 2012; Ono et al. 2013) (Fig. 4). Although cytokinesis fails and cells become polyploid after condensin I depletion, defects in the preceding anaphases appear less severe. Instead of the bulky DNA bridges evident in condensin II knockouts, finer more fiber-like DNA bridges are seen (Green et al. 2012).
Lateral chromosome compaction—is there an upper limit?
If the prevailing model for metaphase chromosomes is built upon a central axis with spiraling radial loops, then is there an upper limit on the lateral width of a compacted chromosome ready for segregation? In order to investigate this limit, the chromosome widths of two fish species, Tetraodon nigroviridis (pufferfish) and Protopterus aethiopicus (marbled lung fish), with estimated genome sizes of 340 and 130,000 Mb, respectively, were examined (Locket 1970; Grutzner et al. 1999). These species have a 380-fold genome size difference, and each fish needs to package its DNA into metaphase chromosomes for efficient segregation to daughter cells. Surprisingly, there is only a modest difference in average chromosome widths of the pufferfish and lung fish, 1.6 and 2.3 μm, respectively. Similar small chromosome width differences were observed in two plant species of the Melanthiaceae family, Chionographis japonica and Paris japonica, with genome sizes of 1500 and 150,000 Mb; however, their average chromosome widths were 0.9 and 2.5 μm, respectively. These observations suggested that there is an upper chromosome width, perhaps to prevent the lateral radial loops from becoming too long and less rigid.
A possible clue to the control of metaphase chromosome width may lie in how much condensins I and II are loaded along the chromosomal axis. We can artificially change the ratio of condensins I to II by introducing a CDK1 phospho-mutant site in the condensin II subunit, CAP-D3 (Bakhrebah et al. 2015). This mutation decreases the ratio of condensin I/II that then increases the lateral width of metaphase chromosomes (see Fig. 4). Likewise, the importance of a set balance between condensins I and II has elegantly been shown in vitro by altering the ratio of condensins I and II using a quantitative immune-depletion approach in Xenopus egg cell-free extracts (Shintomi and Hirano 2011). This approach again clearly demonstrated a predominant role of condensin I in lateral compaction compared to condensin II in axial compaction.
Condensin in meiosis
Meiosis is a specialized cell division where germ cells undergo two rounds of chromosome segregation without DNA replication. After meiosis I, there is a short interphase, also known as interkinesis. Some germ cells show very little decompaction between these two phases (Yun et al. 2014). In mouse, condensin I and II-specific subunits, Ncaph and Ncaph2, have been conditionally deleted in oocytes before the onset of the meiotic cell divisions (Houlard et al. 2015). Surprisingly, only condensin II (Ncaph2) is needed for functional compaction and segregation of chromosomes. Ncaph-depleted oocytes show shorter and rounder chromosomes similar to mitotic knockouts but can segregate without any arrest or errors. In contrast, when Ncaph2 is conditionally knocked out, chromosomes do not fully condense and form multiple chromatin bridges during anaphase I. Strikingly, chromosomes resembled the more amorphous structures originally described using immunodepletion when both condensins were depleted (Hirano and Mitchison 1994), compared to more “mild” fuzzy chromosomes seen in SMC2/4 KO tissue culture systems (Hudson et al. 2003; Hirota et al. 2004; Samejima et al. 2012). This raises the important query whether the somatic RNAi or transcriptional repression systems described above are limited as cells enter mitosis with residual amounts of condensin (Houlard et al. 2015; Hirano 2016). While it is true that the mouse oocyte system represents a powerful and definitive means of depleting all proteins before chromosome compaction, difficulties arise when trying to compare somatic cell mitosis versus mouse meiosis, due to inherent differences in mechanics and proteins involved in the two processes.
Little functional information is known about condensins’ role during male meiosis. Ncaph has been shown to localize to the chromosome axis with enrichment at the centromere and telomeric ends (Viera et al. 2007). A brief decompaction period occurs between anaphase I and metaphase II. In this period, Ncaph is found as punctuate signals within the nucleus and the nucleoli (Viera et al. 2007).
In Arabidopsis, several mutants of condensin subunit genes have been identified. Some mutations affect the viability of germ cells, and these fail to form compacted chromosomes, but mutations of CAP-D2 and CAP-D3 have little effect on chromosome compaction (Siddiqui et al. 2003; Schubert et al. 2013). In the worm C. elegans, condensin controls the distribution of double-stranded DNA breaks and meiotic crossover recombination. The condensin complex that controls meiotic crossovers contains a subunit of the SMC dosage compensation complex plus condensin I (Mets and Meyer 2009).
Future questions and concluding remarks
The field of cohesinopathy has gained considerable pace over the last 10 years, with a number of cohesin mutations now directly linked to developmental disorders, most notably Roberts syndrome/SC-phocomelia (RBS) and Cornelia de Lange Syndrome (CdLS) and also acute myeloid leukemia (AML) (Tonkin et al. 2004; Vega et al. 2005; Kon et al. 2013). Are we likely to see analogous “condensinopathies” as sequencing technology increasingly reveals new disease-causing mutations?
After many years of circumstantial evidence suggesting that condensin mutations are linked to cancer, the first direct evidence that condensin mutations cause cancer was recently provided (Woodward et al. 2016), with mice bearing missense mutations in the condensin II subunit CAP-H2 developing T cell lymphomas. This follows a pan-bioinformatics analysis showing that mutations in condensin subunits are directly associated with multiple cancers (Leiserson et al. 2015).
Condensinopathies were also recently identified in a groundbreaking study showing that biallelic mutations of the condensin subunits CAP-H, D2, and D3 cause microcephaly (Martin et al. 2016). The condensin mutations caused chromosome entanglements during mitosis resulting in aneuploidy and some cell death, suggesting that the smaller brain size was the result of reduced cell proliferation.
Another emerging field is the role of condensin in interphase structure and gene regulation. A number of studies aided by genomics technology are finding key roles for condensin outside mitosis, and it will not be surprising if some of these functions are linked to diseases. In addition, it will be of great interest to see if condensin has specialized roles and binding sites in different cell types and imparts epigenetic information for gene regulation. Posttranslational modifications of condensin also appear essential for regulating function of the complex and will no doubt continue to provide vital functional clues (Bazile et al. 2010; Abe et al. 2011).
The mechanism of action of condensin remains an area of debate and in particular how its in vitro activities relate to chromosome condensation. A key challenge is to relate ATPase-mediated structural changes of condensin to binding and shaping of DNA and in turn what role the head, hinge, and arm domains play in the process. Furthermore, there is little data on the structural and mechanistic differences between condensins I and II and how they relate to chromosome condensation and whether the complexes share the same enzymatic properties.
Although condensin still remains somewhat enigmatic, a clearer picture is beginning to unfold. Though the various SMC complexes have fundamental differences in function, underpinned by their structural and mechanistic variability, it would not be surprising if additional commonalities are found given their common ancestral history. Eventually, a unified picture of SMC modes of action is bound to emerge.
Abbreviations
- DCC:
-
Dosage compensation complex
- dsDNA:
-
Double-stranded DNA
- HEAT:
-
Huntington elongation factor 3, the A subunit of protein phosphatase 2A TOR lipid kinase
- KO:
-
Knockout
- NEBD:
-
Nuclear envelope breakdown
- ScII:
-
Scaffold protein 2
- SMC:
-
Structural maintenance of chromosomes
- TEV:
-
Tobacco etch virus
- WT:
-
Wild type
References
Abe S, Nagasaka K, Hirayama Y et al (2011) The initial phase of chromosome condensation requires Cdk1-mediated phosphorylation of the CAP-D3 subunit of condensin II. Genes Dev 25:863–874. doi:10.1101/gad.2016411
Akai Y, Kurokawa Y, Nakazawa N et al (2011) Opposing role of condensin hinge against replication protein A in mitosis and interphase through promoting DNA annealing. Open biology 1:110023. doi:10.1098/rsob.110023
Anderson DE, Losada A, Erickson HP, Hirano T (2002) Condensin and cohesin display different arm conformations with characteristic hinge angles. J Cell Biol 156:419–424. doi:10.1083/jcb.200111002
Antonin W, Neumann H (2016) Chromosome condensation and decondensation during mitosis. Curr Opin Cell Biol 40:15–22. doi:10.1016/j.ceb.2016.01.013
Arumugam P, Gruber S, Tanaka K, Haering CH, Mechtler K, Nasmyth K (2003) ATP hydrolysis is required for cohesin’s association with chromosomes. Curr Biol 13:1941–1953
Badrinarayanan A, Reyes-Lamothe R, Uphoff S, Leake MC, Sherratt DJ (2012) In vivo architecture and action of bacterial structural maintenance of chromosome proteins. Science 338:528–531. doi:10.1126/science.1227126
Bakhrebah M, Zhang T, Mann JR, Kalitsis P, Hudson DF (2015) Disruption of a conserved CAP-D3 threonine alters condensin loading on mitotic chromosomes leading to chromosome hypercondensation. J Biol Chem 290:6156–6167. doi:10.1074/jbc.M114.627109
Barysz H, Kim JH, Chen ZA, Hudson DF, Rappsilber J, Gerloff DL, Earnshaw WC (2015) Three-dimensional topology of the SMC2/SMC4 subcomplex from chicken condensin I revealed by cross-linking and molecular modelling. Open biology 5:150005. doi:10.1098/rsob.150005
Bauer CR, Hartl TA, Bosco G (2012) Condensin II promotes the formation of chromosome territories by inducing axial compaction of polyploid interphase chromosomes. PLoS Genet 8:e1002873. doi:10.1371/journal.pgen.1002873
Baxter J, Aragon L (2012) A model for chromosome condensation based on the interplay between condensin and topoisomerase II. Trends in genetics : TIG 28:110–117. doi:10.1016/j.tig.2011.11.004
Baxter J, Sen N, Martinez VL, De Carandini ME, Schvartzman JB, Diffley JF, Aragon L (2011) Positive supercoiling of mitotic DNA drives decatenation by topoisomerase II in eukaryotes. Science 331:1328–1332. doi:10.1126/science.1201538
Bazile F, St-Pierre J, D’Amours D (2010) Three-step model for condensin activation during mitotic chromosome condensation. Cell Cycle 9:3243–3255. doi:10.4161/cc.9.16.12620
Belmont AS (2006) Mitotic chromosome structure and condensation. Curr Opin Cell Biol 18:632–638. doi:10.1016/j.ceb.2006.09.007
Bermudez-Lopez M, Aragon L (2016) Smc5/6 complex regulates Sgs1 recombination functions. Curr Genet. doi:10.1007/s00294-016-0648-5
Burmann F, Shin HC, Basquin J et al (2013) An asymmetric SMC-kleisin bridge in prokaryotic condensin. Nat Struct Mol Biol 20:371–379. doi:10.1038/nsmb.2488
Charbin A, Bouchoux C, Uhlmann F (2014) Condensin aids sister chromatid decatenation by topoisomerase II. Nucleic Acids Res 42:340–348. doi:10.1093/nar/gkt882
Collette KS, Petty EL, Golenberg N, Bembenek JN, Csankovszki G (2011) Different roles for Aurora B in condensin targeting during mitosis and meiosis. J Cell Sci 124:3684–3694. doi:10.1242/jcs.088336
Crane E, Bian Q, McCord RP et al (2015) Condensin-driven remodelling of X chromosome topology during dosage compensation. Nature 523:240–244. doi:10.1038/nature14450
Csankovszki G, Collette K, Spahl K et al (2009) Three distinct condensin complexes control C. elegans chromosome dynamics. Curr Biol 19:9–19. doi:10.1016/j.cub.2008.12.006
Cuylen S, Metz J, Haering CH (2011) Condensin structures chromosomal DNA through topological links. Nat Struct Mol Biol 18:894–901. doi:10.1038/nsmb.2087
D'Ambrosio C, Schmidt CK, Katou Y, Kelly G, Itoh T, Shirahige K, Uhlmann F (2008) Identification of cis-acting sites for condensin loading onto budding yeast chromosomes. Genes Dev 22:2215–2227. doi:10.1101/gad.1675708
Danilova O, Reyes-Lamothe R, Pinskaya M, Sherratt D, Possoz C (2007) MukB colocalizes with the oriC region and is required for organization of the two Escherichia coli chromosome arms into separate cell halves. Mol Microbiol 65:1485–1492. doi:10.1111/j.1365-2958.2007.05881.x
Dowen JM, Bilodeau S, Orlando DA, Hubner MR, Abraham BJ, Spector DL, Young RA (2013) Multiple structural maintenance of chromosome complexes at transcriptional regulatory elements. Stem cell reports 1:371–378. doi:10.1016/j.stemcr.2013.09.002
Eeftens JM, Katan AJ, Kschonsak M et al (2016) Condensin Smc2-Smc4 dimers are flexible and dynamic. Cell Rep 14:1813–1818. doi:10.1016/j.celrep.2016.01.063
Ercan S, Giresi PG, Whittle CM, Zhang X, Green RD, Lieb JD (2007) X chromosome repression by localization of the C. elegans dosage compensation machinery to sites of transcription initiation. Nat Genet 39:403–408. doi:10.1038/ng1983
Farr CJ, Antoniou-Kourounioti M, Mimmack ML, Volkov A, Porter AC (2014) The alpha isoform of topoisomerase II is required for hypercompaction of mitotic chromosomes in human cells. Nucleic Acids Res 42:4414–4426. doi:10.1093/nar/gku076
Freeman L, Aragon-Alcaide L, Strunnikov A (2000) The condensin complex governs chromosome condensation and mitotic transmission of rDNA. J Cell Biol 149:811–824
Fujiwara T, Tanaka K, Kuroiwa T, Hirano T (2013) Spatiotemporal dynamics of condensins I and II: evolutionary insights from the primitive red alga Cyanidioschyzon merolae. Mol Biol Cell 24:2515–2527. doi:10.1091/mbc.E13-04-0208
Gerlich D, Hirota T, Koch B, Peters JM, Ellenberg J (2006) Condensin I stabilizes chromosomes mechanically through a dynamic interaction in live cells. Curr Biol 16:333–344. doi:10.1016/j.cub.2005.12.040
Gilbert N, Allan J (2014) Supercoiling in DNA and chromatin. Current opinion in genetics & development 25:15–21. doi:10.1016/j.gde.2013.10.013
Gligoris TG, Scheinost JC, Burmann F et al (2014) Closing the cohesin ring: structure and function of its Smc3-kleisin interface. Science 346:963–967. doi:10.1126/science.1256917
Goloborodko A, Imakaev MV, Marko JF, Mirny L (2016) Compaction and segregation of sister chromatids via active loop extrusion. eLife 5:e14864. doi:10.7554/eLife.14864
Green LC, Kalitsis P, Chang TM et al (2012) Contrasting roles of condensin I and condensin II in mitotic chromosome formation. J Cell Sci 125:1591–1604. doi:10.1242/jcs.097790
Griese JJ, Witte G, Hopfner KP (2010) Structure and DNA binding activity of the mouse condensin hinge domain highlight common and diverse features of SMC proteins. Nucleic Acids Res 38:3454–3465. doi:10.1093/nar/gkq038
Gruber S, Errington J (2009) Recruitment of condensin to replication origin regions by ParB/SpoOJ promotes chromosome segregation in B. subtilis. Cell 137:685–696. doi:10.1016/j.cell.2009.02.035
Grutzner F, Lutjens G, Rovira C, Barnes DW, Ropers HH, Haaf T (1999) Classical and molecular cytogenetics of the pufferfish Tetraodon nigroviridis. Chromosom Res 7:655–662
Haering CH, Lowe J, Hochwagen A, Nasmyth K (2002) Molecular architecture of SMC proteins and the yeast cohesin complex. Mol Cell 9:773–788
Hagstrom KA, Holmes VF, Cozzarelli NR, Meyer BJ (2002) C. elegans condensin promotes mitotic chromosome architecture, centromere organization, and sister chromatid segregation during mitosis and meiosis. Genes Dev 16:729–742. doi:10.1101/gad.968302
Hartl TA, Sweeney SJ, Knepler PJ, Bosco G (2008) Condensin II resolves chromosomal associations to enable anaphase I segregation in Drosophila male meiosis. PLoS Genet 4:e1000228. doi:10.1371/journal.pgen.1000228
Herzog S, Nagarkar Jaiswal S, Urban E, Riemer A, Fischer S, Heidmann SK (2013) Functional dissection of the Drosophila melanogaster condensin subunit Cap-G reveals its exclusive association with condensin I. PLoS Genet 9:e1003463. doi:10.1371/journal.pgen.1003463
Hirano T (2005) Condensins: organizing and segregating the genome. Curr Biol 15:R265–R275
Hirano T (2012) Condensins: universal organizers of chromosomes with diverse functions. Genes Dev 26:1659–1678. doi:10.1101/gad.194746.112
Hirano T (2016) Condensin-based chromosome organization from bacteria to vertebrates. Cell 164:847–857. doi:10.1016/j.cell.2016.01.033
Hirano T, Mitchison TJ (1994) A heterodimeric coiled-coil protein required for mitotic chromosome condensation in vitro. Cell 79:449–458
Hirano T, Kobayashi R, Hirano M (1997) Condensins, chromosome condensation protein complexes containing XCAP-C, XCAP-E and a Xenopus homolog of the Drosophila barren protein. Cell 89:511–521
Hirano M, Anderson DE, Erickson HP, Hirano T (2001) Bimodal activation of SMC ATPase by intra- and inter-molecular interactions. EMBO J 20:3238–3250. doi:10.1093/emboj/20.12.3238
Hirota T, Gerlich D, Koch B, Ellenberg J, Peters JM (2004) Distinct functions of condensin I and II in mitotic chromosome assembly. J Cell Sci 117:6435–6445. doi:10.1242/jcs.01604
Houlard M, Godwin J, Metson J, Lee J, Hirano T, Nasmyth K (2015) Condensin confers the longitudinal rigidity of chromosomes. Nat Cell Biol 17:771–781. doi:10.1038/ncb3167
Hudson DF, Vagnarelli P, Gassmann R, Earnshaw WC (2003) Condensin is required for nonhistone protein assembly and structural integrity of vertebrate mitotic chromosomes. Dev Cell 5:323–336
Hudson DF, Ohta S, Freisinger T et al (2008) Molecular and genetic analysis of condensin function in vertebrate cells. Mol Biol Cell 19:3070–3079. doi:10.1091/mbc.E08-01-0057
Hudson DF, Marshall KM, Earnshaw WC (2009) Condensin: architect of mitotic chromosomes. Chromosom Res 17:131–144
Huis in ‘t Veld PJ, Herzog F, Ladurner R et al (2014) Characterization of a DNA exit gate in the human cohesin ring. Science 346:968–972. doi:10.1126/science.1256904
Jans J, Gladden JM, Ralston EJ et al (2009) A condensin-like dosage compensation complex acts at a distance to control expression throughout the genome. Genes Dev 23:602–618. doi:10.1101/gad.1751109
Kegel A, Betts-Lindroos H, Kanno T et al (2011) Chromosome length influences replication-induced topological stress. Nature 471:392–396. doi:10.1038/nature09791
Kim JH, Zhang T, Wong NC et al (2013) Condensin I associates with structural and gene regulatory regions in vertebrate chromosomes. Nat Commun 4:2537. doi:10.1038/ncomms3537
Kimura K, Hirano T (1997) ATP-dependent positive supercoiling of DNA by 13S condensin: a biochemical implication for chromosome condensation. Cell 90:625–634
Kimura K, Rybenkov VV, Crisona NJ, Hirano T, Cozzarelli NR (1999) 13S condensin actively reconfigures DNA by introducing global positive writhe: implications for chromosome condensation. Cell 98:239–248
Kinoshita K, Kobayashi TJ, Hirano T (2015) Balancing acts of two HEAT subunits of condensin I support dynamic assembly of chromosome axes. Dev Cell 33:94–106. doi:10.1016/j.devcel.2015.01.034
Kireeva N, Lakonishok M, Kireev I, Hirano T, Belmont AS (2004) Visualization of early chromosome condensation: a hierarchical folding, axial glue model of chromosome structure. J Cell Biol 166:775–785. doi:10.1083/jcb.200406049
Kon A, Shih LY, Minamino M et al (2013) Recurrent mutations in multiple components of the cohesin complex in myeloid neoplasms. Nat Genet 45:1232–1237. doi:10.1038/ng.2731
Kranz AL, Jiao CY, Winterkorn LH, Albritton SE, Kramer M, Ercan S (2013) Genome-wide analysis of condensin binding in Caenorhabditis elegans. Genome Biol 14:R112. doi:10.1186/gb-2013-14-10-r112
Leiserson MD, Vandin F, Wu HT et al (2015) Pan-cancer network analysis identifies combinations of rare somatic mutations across pathways and protein complexes. Nat Genet 47:106–114. doi:10.1038/ng.3168
Leonard J, Sen N, Torres R, Sutani T, Jarmuz A, Shirahige K, Aragon L (2015) Condensin relocalization from centromeres to chromosome arms promotes Top2 recruitment during anaphase. Cell Rep 13:2336–2344. doi:10.1016/j.celrep.2015.11.041
Li W, Hu Y, Oh S et al (2015) Condensin I and II complexes license full estrogen receptor alpha-dependent enhancer activation. Mol Cell 59:188–202. doi:10.1016/j.molcel.2015.06.002
Liang Z, Zickler D, Prentiss M, Chang FS, Witz G, Maeshima K, Kleckner N (2015) Chromosomes progress to metaphase in multiple discrete steps via global compaction/expansion cycles. Cell 161:1124–1137. doi:10.1016/j.cell.2015.04.030
Liu W, Tanasa B, Tyurina OV et al (2010) PHF8 mediates histone H4 lysine 20 demethylation events involved in cell cycle progression. Nature 466:508–512. doi:10.1038/nature09272
Locket NA (1970) Mitosis in mature lungfish retina. Zool J Linnean Soc 49:4. doi:10.1111/j.1096-3642.1970.tb00726.x
Losada A, Hirano T (2005) Dynamic molecular linkers of the genome: the first decade of SMC proteins. Genes Dev 19:1269–1287. doi:10.1101/gad.1320505
Lukas C, Savic V, Bekker-Jensen S et al (2011) 53BP1 nuclear bodies form around DNA lesions generated by mitotic transmission of chromosomes under replication stress. Nat Cell Biol 13:243–253. doi:10.1038/ncb2201
Maeshima K, Laemmli UK (2003) A two-step scaffolding model for mitotic chromosome assembly. Dev Cell 4:467–480
Martin CA, Murray JE, Carroll P et al (2016) Mutations in genes encoding condensin complex proteins cause microcephaly through decatenation failure at mitosis. Genes Dev 30:2158–2172. doi:10.1101/gad.286351.116
McDonel P, Jans J, Peterson BK, Meyer BJ (2006) Clustered DNA motifs mark X chromosomes for repression by a dosage compensation complex. Nature 444:614–618. doi:10.1038/nature05338
Mets DG, Meyer BJ (2009) Condensins regulate meiotic DNA break distribution, thus crossover frequency, by controlling chromosome structure. Cell 139:73–86. doi:10.1016/j.cell.2009.07.035
Murayama Y, Uhlmann F (2014) Biochemical reconstitution of topological DNA binding by the cohesin ring. Nature 505:367–371. doi:10.1038/nature12867
Murayama Y, Uhlmann F (2015) DNA entry into and exit out of the cohesin ring by an interlocking gate mechanism. Cell 163:1628–1640. doi:10.1016/j.cell.2015.11.030
Nakazawa N, Sajiki K, Xu X, Villar-Briones A, Arakawa O, Yanagida M (2015) RNA pol II transcript abundance controls condensin accumulation at mitotically up-regulated and heat-shock-inducible genes in fission yeast. Genes Cells 20:481–499. doi:10.1111/gtc.12239
Nasmyth K (2001) Disseminating the genome: joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu Rev Genet 35:673–745. doi:10.1146/annurev.genet.35.102401.091334
Naumova N, Imakaev M, Fudenberg G, Zhan Y, Lajoie BR, Mirny LA, Dekker J (2013) Organization of the mitotic chromosome. Science 342:948–953. doi:10.1126/science.1236083
Nishino Y, Eltsov M, Joti Y et al (2012) Human mitotic chromosomes consist predominantly of irregularly folded nucleosome fibres without a 30-nm chromatin structure. EMBO J 31:1644–1653. doi:10.1038/emboj.2012.35
Ohta S, Bukowski-Wills JC, Sanchez-Pulido L et al (2010) The protein composition of mitotic chromosomes determined using multiclassifier combinatorial proteomics. Cell 142:810–821. doi:10.1016/j.cell.2010.07.047
Onn I, Aono N, Hirano M, Hirano T (2007) Reconstitution and subunit geometry of human condensin complexes. EMBO J 26:1024–1034. doi:10.1038/sj.emboj.7601562
Ono T, Losada A, Hirano M, Myers MP, Neuwald AF, Hirano T (2003) Differential contributions of condensin I and condensin II to mitotic chromosome architecture in vertebrate cells. Cell 115:109–121
Ono T, Fang Y, Spector DL, Hirano T (2004) Spatial and temporal regulation of condensins I and II in mitotic chromosome assembly in human cells. Mol Biol Cell 15:3296–3308. doi:10.1091/mbc.E04-03-0242
Ono T, Yamashita D, Hirano T (2013) Condensin II initiates sister chromatid resolution during S phase. J Cell Biol 200:429–441. doi:10.1083/jcb.201208008
Petrushenko ZM, She W, Rybenkov VV (2011) A new family of bacterial condensins. Mol Microbiol 81:881–896. doi:10.1111/j.1365-2958.2011.07763.x
Piazza I, Rutkowska A, Ori A et al (2014) Association of condensin with chromosomes depends on DNA binding by its HEAT-repeat subunits. Nat Struct Mol Biol 21:560–568. doi:10.1038/nsmb.2831
Piskadlo E, Oliveira RA (2016) Novel insights into mitotic chromosome condensation. F1000Res 5 doi: 10.12688/f1000research.8727.1
Poirier MG, Marko JF (2002) Mitotic chromosomes are chromatin networks without a mechanically contiguous protein scaffold. Proc Natl Acad Sci U S A 99:15393–15397. doi:10.1073/pnas.232442599
Poonperm R, Takata H, Hamano T, Matsuda A, Uchiyama S, Hiraoka Y, Fukui K (2015) Chromosome scaffold is a double-stranded assembly of scaffold proteins. Scientific reports 5:11916. doi:10.1038/srep11916
Pryzhkova MV, Jordan PW (2016) Conditional mutation of Smc5 in mouse embryonic stem cells perturbs condensin localization and mitotic progression. J Cell Sci 129:1619–1634. doi:10.1242/jcs.179036
Saitoh N, Goldberg IG, Wood ER, Earnshaw WC (1994) ScII: an abundant chromosome scaffold protein is a member of a family of putative ATPases with an unusual predicted tertiary structure. J Cell Biol 127:303–318
Saka Y, Sutani T, Yamashita Y, Saitoh S, Takeuchi M, Nakaseko Y, Yanagida M (1994) Fission yeast cut3 and cut14, members of a ubiquitous protein family, are required for chromosome condensation and segregation in mitosis. EMBO J 13:4938–4952
Samejima I, Matsumoto T, Nakaseko Y, Beach D, Yanagida M (1993) Identification of seven new cut genes involved in Schizosaccharomyces pombe mitosis. J Cell Sci 105(Pt 1):135–143
Samejima K, Samejima I, Vagnarelli P et al (2012) Mitotic chromosomes are compacted laterally by KIF4 and condensin and axially by topoisomerase IIalpha. J Cell Biol 199:755–770. doi:10.1083/jcb.201202155
Savvidou E, Cobbe N, Steffensen S, Cotterill S, Heck MM (2005) Drosophila CAP-D2 is required for condensin complex stability and resolution of sister chromatids. J Cell Sci 118:2529–2543. doi:10.1242/jcs.02392
Schmiesing JA, Gregson HC, Zhou S, Yokomori K (2000) A human condensin complex containing hCAP-C-hCAP-E and CNAP1, a homolog of Xenopus XCAP-D2, colocalizes with phosphorylated histone H3 during the early stage of mitotic chromosome condensation. Mol Cell Biol 20:6996–7006
Schubert V, Lermontova I, Schubert I (2013) The Arabidopsis CAP-D proteins are required for correct chromatin organisation, growth and fertility. Chromosoma 122:517–533. doi:10.1007/s00412-013-0424-y
Shintomi K, Hirano T (2011) The relative ratio of condensin I to II determines chromosome shapes. Genes Dev 25:1464–1469. doi:10.1101/gad.2060311
Siddiqui NU, Stronghill PE, Dengler RE, Hasenkampf CA, Riggs CD (2003) Mutations in Arabidopsis condensin genes disrupt embryogenesis, meristem organization and segregation of homologous chromosomes during meiosis. Development (Cambridge, England) 130:3283–3295
Soh YM, Burmann F, Shin HC et al (2015) Molecular basis for SMC rod formation and its dissolution upon DNA binding. Mol Cell 57:290–303. doi:10.1016/j.molcel.2014.11.023
Sutani T, Yuasa T, Tomonaga T, Dohmae N, Takio K, Yanagida M (1999) Fission yeast condensin complex: essential roles of non-SMC subunits for condensation and Cdc2 phosphorylation of Cut3/SMC4. Genes Dev 13:2271–2283
Sutani T, Sakata T, Nakato R et al (2015) Condensin targets and reduces unwound DNA structures associated with transcription in mitotic chromosome condensation. Nat Commun 6:7815. doi:10.1038/ncomms8815
Tada K, Susumu H, Sakuno T, Watanabe Y (2011) Condensin association with histone H2A shapes mitotic chromosomes. Nature 474:477–483. doi:10.1038/nature10179
Tonkin ET, Wang TJ, Lisgo S, Bamshad MJ, Strachan T (2004) NIPBL, encoding a homolog of fungal Scc2-type sister chromatid cohesion proteins and fly Nipped-B, is mutated in Cornelia de Lange syndrome. Nat Genet 36:636–641. doi:10.1038/ng1363
Toselli-Mollereau E, Robellet X, Fauque L et al (2016) Nucleosome eviction in mitosis assists condensin loading and chromosome condensation. EMBO J 35:1565–1581. doi:10.15252/embj.201592849
Vagnarelli P (2012) Mitotic chromosome condensation in vertebrates. Exp Cell Res 318:1435–1441. doi:10.1016/j.yexcr.2012.03.017
Vega H, Waisfisz Q, Gordillo M et al (2005) Roberts syndrome is caused by mutations in ESCO2, a human homolog of yeast ECO1 that is essential for the establishment of sister chromatid cohesion. Nat Genet 37:468–470. doi:10.1038/ng1548
Viera A, Gomez R, Parra MT, Schmiesing JA, Yokomori K, Rufas JS, Suja JA (2007) Condensin I reveals new insights on mouse meiotic chromosome structure and dynamics. PLoS One 2:e783. doi:10.1371/journal.pone.0000783
Wang BD, Eyre D, Basrai M, Lichten M, Strunnikov A (2005) Condensin binding at distinct and specific chromosomal sites in the Saccharomyces cerevisiae genome. Mol Cell Biol 25:7216–7225. doi:10.1128/mcb.25.16.7216-7225.2005
Weitzer S, Lehane C, Uhlmann F (2003) A model for ATP hydrolysis-dependent binding of cohesin to DNA. Curr Biol 13:1930–1940
Wilkins BJ, Rall NA, Ostwal Y et al (2014) A cascade of histone modifications induces chromatin condensation in mitosis. Science 343:77–80. doi:10.1126/science.1244508
Woodward J, Taylor GC, Soares DC et al (2016) Condensin II mutation causes T-cell lymphoma through tissue-specific genome instability. Genes Dev 30:2173–2186. doi:10.1101/gad.284562.116
Yanagida M (2009) Clearing the way for mitosis: is cohesin a target? Nat Rev Mol Cell Biol 10:489–496. doi:10.1038/nrm2712
Yun Y, Lane SI, Jones KT (2014) Premature dyad separation in meiosis II is the major segregation error with maternal age in mouse oocytes. Development (Cambridge, England) 141:199–208. doi:10.1242/dev.100206
Zhang T, Paulson JR, Bakhrebah M, Kim JH, Nowell C, Kalitsis P, Hudson DF (2016) Condensin I and II behaviour in interphase nuclei and cells undergoing premature chromosome condensation. Chromosom Res 24:243–269. doi:10.1007/s10577-016-9519-7
Acknowledgements
This work was supported by the National Health and Medical Research Council (nhmrc.gov.au) Australia Project Grants GNT1069223, GNT1047009, a Carlsberg Foundation Fellowship CF15-0905, and the Victorian Government’s Operational Infrastructure Support Program (vic.gov.au).
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editors: Nick Gilbert and Davide Marenduzzo
Rights and permissions
About this article
Cite this article
Kalitsis, P., Zhang, T., Marshall, K.M. et al. Condensin, master organizer of the genome. Chromosome Res 25, 61–76 (2017). https://doi.org/10.1007/s10577-017-9553-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10577-017-9553-0