Abstract
In plants as in other eukaryotes, the structural maintenance of chromosome (SMC) protein complexes cohesin, condensin and SMC5/6 are essential for sister chromatid cohesion, chromosome condensation, DNA repair and recombination. The presence of paralogous genes for various components of the different SMC complexes suggests the diversification of their biological functions during the evolution of higher plants. In Arabidopsis thaliana, we identified two candidate genes (Cap-D2 and Cap-D3) which may express conserved proteins presumably associated with condensin. In silico analyses using public databases suggest that both genes are controlled by factors acting in a cell cycle-dependent manner. Cap-D2 is essential because homozygous T-DNA insertion mutants were not viable. The heterozygous mutant showed wild-type growth habit but reduced fertility. For Cap-D3, we selected two homozygous mutants expressing truncated transcripts which are obviously not fully functional. Both mutants show reduced pollen fertility and seed set (one of them also reduced plant vigour), a lower chromatin density and frequent (peri)centromere association in interphase nuclei. Sister chromatid cohesion was impaired compared to wild-type in the cap-D3 mutants but not in the heterozygous cap-D2 mutant. At superresolution (Structured Illumination Microscopy), we found no alteration of chromatin substructure for both cap-D mutants. Chromosome-associated polypeptide (CAP)-D3 and the cohesin subunit SMC3 form similar but positionally non-overlapping reticulate structures in 2C-16C nuclei, suggesting their importance for interphase chromatin architecture in differentiated nuclei. Thus, we presume that CAP-D proteins are required for fertility, growth, chromatin organisation, sister chromatid cohesion and in a process preventing the association of centromeric repeats.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In plants as in other eukaryotes, the evolutionarily conserved structural maintenance of chromosome (SMC) protein complexes cohesin, condensin and SMC5/6 are involved in such basic biological processes as sister chromatid cohesion, chromosome condensation, DNA repair and recombination (Nasmyth and Haering 2005; Hirano and Hirano 2006; Palecek et al. 2006). In addition, SMC proteins control gene expression and development (Wood et al. 2010; Hirano 2012).
The two rod-like structured condensin complexes I and II, identified in vertebrates, consist of the subunits SMC2 and SMC4 which are linked by the kleisins chromosome-associated polypeptide (CAP)-H and CAP-H2, respectively. In addition, two HEAT ((Huntingtin, elongation factor 3 (EF3), protein phosphatase 2A (PP2A) and the yeast kinase TOR1)) repeat-containing proteins are connected to condensin I (CAP-D2 and CAP-G) or II (CAP-D3 and CAP-G2; Losada and Hirano 2005; Nasmyth and Haering 2005).
Although condensins are conserved in all investigated higher eukaryotes, there is evidence that they may fulfil diverse functions in different phylogenetic branches based on the evolution of paralogous genes (Peric-Hupkes and van Steensel 2008).
In higher plants, different paralogues for the condensin subunits may be combined to form various complexes. Arabidopsis thaliana L. has three genes for SMC4 and two genes each for SMC2, Cap-H, CAP-D and CAP-G (Fig. 1; Schubert 2009).
Subunit composition of condensin complexes in A. thaliana based on a model according to Nasmyth and Hearing (2005), Schubert (2009) and Sakamoto et al. (2011). The condensin complexes presumably have three alternative SMC4 subunits, two alternative SMC2 subunits, two different kleisins and also one of two different CAP-D and CAP-G subunits
The subunit variants SMC2A and SMC2B, SMC4A and SMC4B and also the kleisins CAP-H and CAP-H2 are required for chromosome condensation and segregation during mitosis, meiosis and for embryo development (Liu et al. 2002; Tzafrir et al. 2002; Siddiqui et al. 2003 2006; Fujimoto et al. 2005). Sakamoto et al. (2011) proved the participation of A. thaliana condensin subunits CAP-H2 and CAP-G2 in DNA damage repair and thus in protecting the genome against genotoxic stress.
The biological function of the two A. thaliana CAP-D homologues has not yet been determined. Here we show, based on in silico analyses, that both genes are controlled by factors acting in a cell cycle-dependent manner. The morphological characterization of T-DNA mutants suggests that CAP-D proteins are essential for plant growth and fertility. Based on fluorescent in situ hybridization (FISH), immunolabelling and superresolution microscopy, we conclude that the CAP-D proteins are involved in interphase chromatin organisation.
Materials and methods
Plant material, genotyping and phylogenetic analyses
The SALK and SAIL T-DNA insertion lines in Columbia (Col-0) were obtained from the Salk Institute, Genomic Analysis Laboratory (http://signal.salk.edu/cgi-bin/tdnaexpress; Alonso et al. 2003) and from the Syngenta collection of T-DNA insertion mutants (Sessions et al. 2002), respectively, and provided by the Nottingham Arabidopsis Stock Centre (http://nasc.nott.ac.uk).
Seeds were germinated on agar, followed by further cultivation in soil under short day condition (8 h light/16 h dark) at 21 °C. Genomic DNA was isolated from rosette leaves and used for PCR-based genotyping to identify heterozygous and homozygous T-DNA insertion mutants. The PCR primers used for genotyping are listed in Electronic supplementary material (ESM) Table S3 and their positions are shown together with the corresponding gene structure (http://mips.gsf.de/; MAtDB Version 10) in Fig. 2a. PCR using the gene-specific primer sets yielded DNA fragments of ∼1 kb representing the wild-type alleles. The PCR fragments specific for the disrupted allele yielded PCR products of ∼ 0.5 kb. The positions of T-DNA insertion were confirmed by sequencing the PCR-amplified T-DNA junction fragments (ESM Table S4).
A. thaliana Cap-D2 and Cap-D3 gene and protein structures (mips.helmholtz-muenchen.de, Version 10; ncbi.nlm.nih.gov; pfam.sanger.ac.uk). a Schematic view of the gene structures and expressed truncated transcripts. Exons are shown as blue boxes. UTRs are visible in grey. T-DNA insertions (SALK and SAIL lines) are indicated. The positions and directions of primers are shown as horizontal arrows. Arabic numbers indicate gene-specific primers used for genotyping; Roman numbers denote primers applied for RT and real-time PCR. The position of the peptide used to raise antibodies is localised in the first exon of Cap-D3. The CAP-D3 mutants SAIL_826_B06 and SALK_094776 express two and one truncated transcripts (green), respectively. b Schematic view of the protein structures. Cnd1 (conserved non-SMC condensin domain), COG5098 (condensin domain D2) and HEAT domains are indicated
Phylogenetic trees were generated from a search using the CAP-D2 and CAP-D3 proteins against all non-redundant GenBank CDS translations + PDB + SwissProt + PIR + PRF, excluding environmental samples, from WGS projects (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The BLAST tree was created from genetic distances calculated using Kimura’s standard method for proteins (Kimura, 1983). The trees themselves are then built from these distance matrices using the Fast Minimum Evolution method (Desper et al. 2002).
Gene expression analyses
Total RNA was isolated from pooled seedlings using the RNeasy plant mini kit (QIAGEN) according to manufacturer’s instructions. Reverse transcription was performed using a First-Strand cDNA Synthesis Kit (Fermentas) and 1 μg of total RNA as starting material. RT-PCR and real-time RT-PCR primers used to amplify transcripts are shown in Fig. 2a and ESM Table S5.
For quantitative real-time RT-PCR, an iCycler iQ (Bio-Rad) and an iQ SYBR Green Supermix (Bio-Rad) were used. Each transcript was quantified three times using three independent biological replicates. A fragment of Actin2 cDNA was amplified for data normalisation. The cDNA equivalent to 50 ng of total RNA was used in a 25 μl PCR reaction to amplify Actin2 cDNA or Cap-D2 cDNA, respectively. The following PCR program was used: initial denaturation for 5 min, then 40 cycles with 10 s denaturation at 95 °C, 20 s annealing at 60 °C and 20 s elongation at 72 °C.
For semi-quantitative RT-PCR, the following program was used: 5 min initial denaturation, and then 35 cycles with 45 s denaturation at 95 °C, 45 s annealing at 59 °C, 1 min 20 s elongation at 72 °C. Elongation factor 1α served as standard.
Antibody production and peptide competition assay
Polyclonal guinea pig anti-Cap-D3 and rabbit anti-SMC3 antibodies were produced and purified by Eurogentec (http://www.eurogentec.com/). The synthetic Cap-D3 YLKRRDDSAGQGSNS (corresponding amino acids 111–125; Fig. 2a, ESM Fig. S1) and SMC3 RIKAPRVNYPKDSDA (corresponding amino acids 583–597) peptides were conjugated with keyhole limpet haemocyanin and used to immunise a guinea pig and a rabbit, respectively. The serum was affinity-purified over a mixture of ACH and CNBr–sepharose covalently coupled with the peptide.
Three samples each with 5 μl of CAP-D3 antibodies (5.6 μg/μl) were prepared. Each sample was diluted in 100 μl of MTSB buffer (50 mM PIPES, 5 mM MgSO4, 5 mM EGTA, pH 6.9). One sample was used as a control without incubation with the antigenic peptide and to the other two samples, 188 or 376 μg peptide for blocking CAP-D3 antibodies was added. The volume of each sample was adjusted to 200 μl with MTSB. Samples were incubated at room temperature for 1 h. After incubation, the volume of each sample was adjusted to 1 ml with antibody dilution buffer. All three antibody samples (with and without peptide) were used for immunostaining according to the protocol described below. For blocking of unspecific antibodies, 8 % BSA in 0.1 % Triton X100/MTSB was used. Similarly, the specificity of the SMC3 antibodies was confirmed.
Preparation of nuclei, probe labelling, FISH and immunolocalization
Nuclei of differentiated cells which do no longer cycle were isolated and flow-sorted according to their endopolyploidy level from rosette leaves after formaldehyde fixation using a FACS Aria (BD Biosciences) as described (Pecinka et al. 2004). The 2C nuclei contain two unreplicated, 4C nuclei two replicated, 8C, 16C, 32C nuclei further endoreduplicated chromosome sets (ESM Fig. S2).
Due to the tiny Arabidopsis meristems, sorting of sufficient numbers of nuclei from cycling cells in G1, S or G2 phase is not feasible. To analyse anaphase bridge frequencies, fresh cotyledons of 3-day-old seedlings were mounted in 2 μg/ml DAPI diluted in antifade (Vectashield) and immediately evaluated under the fluorescence microscope.
The 178-bp centromeric repeat probe (pAL; Martinez-Zapater et al. 1986) was generated by PCR as previously described (Kawabe and Nasuda 2005). A. thaliana BACs were obtained from the Arabidopsis Biological Resource Center (Columbus, OH, USA).
For painting of the chromosome 1 top arm (CT1top), 17 pools of a total of 87 BACs (from T25K16 to F12K21) and for the chromosome 4 bottom arm (CT4bottom) 15 pools of a total of 60 BACs (from T25P22 to T19P19) were labelled with biotin-dUTP and digoxigenin-dUTP, respectively (Pecinka et al. 2004).
BAC DNA from single positions along chromosomes 1and 4 (Fig. 4a and 6a) was labelled by nick translation with digoxigenin-dUTP, biotin-dUTP or Cy3-dUTP according to Ward (2002). Biotin was detected by avidin conjugated with TexasRed (1:1,000; Vector Laboratories), goat anti-avidin conjugated with biotin (1:200; Vector Laboratories) and again with avidin conjugated with Texas Red; digoxigenin was detected by mouse anti-digoxigenin (1:250; Roche) and goat-anti-mouse conjugated with Alexa-488 (1:200; Molecular Probes). Cy3 was observed directly. FISH was performed according to Schubert et al. (2001). Nuclei and chromosomes were counterstained with DAPI (1 μg/ml) in Vectashield (Vector Laboratories).
For co-localisation of CAP-D3 and SMC3 immunosignals, nuclei were fixed in 4 % paraformaldehyde/3.6 % sucrose and immunostaining was performed as described (Jasencakova et al. 2000). CAP-D3 was detected with guinea pig polyclonal antibodies (1:200) and goat anti-guinea pig Alexa 488 (1:250; Invitrogen), SMC3 with rabbit polyclonal antibodies (1:250) and goat anti-rabbit rhodamine (1:200; Jackson Immuno Research Laboratories). CENH3 was detected using antibodies against A. thaliana CENH3 (1:500; Nagaki et al. 2004) and goat anti-rabbit rhodamine (1:100; Jackson Immuno Research Laboratories).
Microscopic evaluation, image processing and statistics
Analysis of FISH signals was performed with an epifluorescence microscope (Zeiss Axiophot) using a 100x/1.45 Zeiss α plan-fluar objective and a 3-chip Sony (DXC-950P) colour camera. The microscope was integrated into a Digital Optical 3D Microscope system (Confovis, Jena, Germany). To achieve an optical resolution of ∼100 nm, we applied Structured Illumination Microscopy (SIM) using a C-Apo 63x/1.2 W Korr objective of an Elyra microscope system and the software ZEN (Zeiss, Germany).
Images were captured separately for each fluorochrome using appropriate excitation and emission filters. The images were merged using Adobe Photoshop 6.0 software (Adobe Systems, San Jose, USA). Embryo development (Jenik et al. 2007) was analysed using differential interference contrast microscopy.
In 4C nuclei, positionally separated sister chromatids are indicated by three or four FISH signals (Fig. 4b). The evaluation followed the criteria described by Schubert et al. (2008). Centromeric (pAL) and pericentromeric (a ~113 kb segment cloned in BAC F28D6) repeats together covering more than 70% and CTs covering more than 50% of the nucleus area were regarded as dispersed (Figs. 4c1 , 4c3 and 6b).
The frequencies of seed abortion, euchromatin condensation, sister chromatid cohesion and of anaphase bridges in cotyledons of 3-day-old seedlings observed for mutants in comparison to wild type were compared by the two-sided Fisher’s exact test. The differences of centromeric FISH signals per nucleus were tested with the Kruskal–Wallis test (SigmaStat 3.1) and multiply compared against Columbia wild-type with Dunn’s method at P < 0.01 level.
Results
In silico studies revealed two conserved A. thaliana Cap-D genes that are regulated in a cell cycle-dependent manner
To retrieve available knowledge about structure, phylogenetic relationship and expression regulation of putative AtCap-D genes, database searches were performed. Searching A. thaliana databases (arabidopsis.org; mips.helmholtz-muenchen.de) for the homologues of Cap-D genes, two candidates were identified and designated as AtCap-D2 (AT3G57060) and AtCap-D3 (AT4G15890) because sequence comparisons annotated them as homologues of subunits of the vertebrate condensin I and II complexes, respectively (http://blast.ncbi.nlm.nih.gov/; www.uniprot.org; Fig. 1). Gene models suggest 16 exons for Cap-D2 and ten exons for Cap-D3 (http://mips.helmholtz-muenchen.de/plant/athal/; Fig. 2a).
Sequence comparison of CAP-D2 and CAP-D3 proteins revealed 13.4 % identical and 38.6 % similar amino acid residues. In silico analysis predicted a HEAT domain, the condensin subunit domain COG5098 and the conserved non-SMC condensin subunit domain Cnd1 for both proteins (ncbi.nlm.nih.gov; pfam.sanger.ac.uk; Fig. 2b, ESM Fig. S1). Similar to human CAP-D2 protein, the CAP-D2 homologue of Arabidopsis contains additionally a Cnd1-N domain at its N-terminus, while in CAP-D3 homologues this domain is absent (Fig. 2b; http://www.ncbi.nlm.nih.gov/cdd/).
Phylogenetic analysis of protein sequence suggests a relatively high conservation of both Cap-D genes (ESM Fig. S3 and ESM Fig. S4). A similar expression behaviour (with peaks at bolting, flowering and seed formation) during plant development of the Cap-D genes and other condensin subunit candidate genes supports a synchronised activity (ESM Fig. S5). However, it is not clear whether they act separately or as multi-subunit complexes in various subunit combinations.
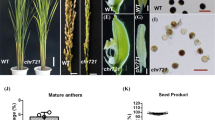
Impaired plant growth and fertility of the cap-D3 mutants. a Reduced plant size and pollen fertility of line SAIL_826_B06 compared to wild type. b Reduced seed set in siliques of both cap-D3 lines SAIL_826_B06 and SALK_094776. c Similar as in wild type, the SALK line showed normal embryo development during the first cell divisions until the heart stage. In the SAIL line in addition to ovules containing normal embryos (left), translucent ovules without (middle) or with abnormal embryos before the heart-shaped stage (right) were observed
In the genevestigator.com database, we found that the COP9 signalosome (CSN) subunits 3, 4, 5, the retinoblastoma-related protein (RBR), the transcription factor E2Fa-DPa heterodimer and the histone monoubiquitination1 (HUB1) protein influence the expression of both Cap-D genes (ESM Table S1).
A. thaliana E2Fa is a transcription activator of genes that are important for cell division (De Veylder et al. 2002; Magyar et al. 2005; Sozzani et al. 2006). E2Fa binds E2F promoter motifs as a heterodimers with DPa in a cell cycle-dependent manner. This complex also transiently includes the RBR protein, which binds in a hypophosphorylated state to E2Fs, thereby inhibiting their transcription factor activity (Luo et al. 1998; van den Heuvel and Dyson 2008; Magyar 2008; Heckmann et al. 2011; Magyar et al. 2012).
The upregulation of Cap-D2 and Cap-D3 in plants overexpressing E2Fa-DPa and RNAi-depleted for RBR suggests a regulatory capacity of E2Fa-DPa heterodimers and RBR for both genes. Computer analysis of the presumed promoter regions of Cap-D2 and Cap-D3 genes using the ATCOECIS database (http://bioinformatics.psb.ugent.be/ATCOECIS/) revealed two putative E2F binding sites for both genes: at positions -345 (ATTGGCCG) and -114 (ATTCCCGC) from the ATG start codon for the Cap-D2 and at positions -397 (CCGCCAAA) and -84 (GCGGGAAA) for the Cap-D3 gene.
The A. thaliana COP9 signalosome complex regulates protein degradation. A transcriptional repression ability of the CSN subunits of the COP9 complex is suggested by the observed upregulation of both Cap-D genes in csn3, csn4 and csn5 mutants (reviewed in Chamovitz et al. 2009).
HUB1 mediates gene activation and cell cycle regulation (Fleury et al. 2007). The downregulation of both Cap-D genes in hub1 mutants indicates HUB1 as a possible gene activator for condensin subunits.
Analysing Affymetrix array datasets (http://www.cyclebase.org/), we found that the expression of Cap-D2 and Cap-D3 increases during G2 phase, culminates in the middle of G2 and drops during mitosis (ESM Fig. S6). Thus, both genes show a cell cycle-dependent expression. Csn3 and Csn4 genes display a similar cell cycle-dependent expression pattern. Also the Hub1 gene follows a cell-cycle-dependent expression pattern with a peak shift toward the beginning of M phase.
In addition, we analyzed tissue-specific, hormone- and stress-induced coexpression pattern of CAP-D, E2Fs, RBR1, Csn3, 4, 5 and HUB1 genes of A. thaliana using the software CORNET (De Bodt et al. 2012) and a predefined set of arrays (ESM Table S2). We found a clearly correlated expression of the selected genes with expression of CAP-D2 and CAP-D3 at almost all selected stages (development, flower, seed, whole plant, but not in roots and leaves), after hormone treatment and after application of biotic and abiotic stress. All in all, we conclude that the activity of both conserved Cap-D genes is regulated by factors acting in a cell cycle-dependent manner.
CAP-D2 is essential for viability, fertility and euchromatin organisation
From the A. thaliana SALK T-DNA insertion mutant collection, a mutant of Cap-D2 was identified. By PCR using gene-specific and T-DNA specific primers (Fig. 2a; ESM Table S3) and by sequencing the PCR products, we confirmed the mutant carrying the inserted T-DNA. The insertion was found in the 15th intron (ESM Table S4). Only heterozygous mutants could be selected and the progeny segregated into heterozygous and wild-type plants. This indicates the requirement of the gene for plant viability. Compared to wild type, the transcript level of heterozygotes reached only 48.6 % as detected by real-time RT-PCR using gene-specific primers (Fig. 2a; ESM Table S5). Nevertheless, the mutants showed a wild-type growth habit. However, significantly reduced pollen quantity and low seed set (35.2 % aborted seeds) indicate the importance of Cap-D2 for fertility (Table 1). The aborted seeds might represent the segregating homozygous progeny.
Next, we studied effects of reduced Cap-D2 on interphase chromosome arrangement and found a significantly higher number of 4C, 8C and 16C (but not 2C) nuclei that show increased CT1top arm (labelled by FISH; Figs. 4a and c3) dispersion, suggesting a possible role of Cap-D2 for maintenance of normal euchromatin density. Because no aberrant mitoses were detectable, the decreased euchromatin density does either not impair nuclear division, or the euchromatin density decrease is restricted to differentiated cells starting from the 4C level on, while G2 cells are rare among cells possessing 4C nuclei (see “Material and methods” section). Positional sister chromatid cohesion at ∼100 kb euchromatic mid-arm segments (labelled by FISH) of chromosome 1 (BACs T2P11 and F11P17; Fig. 4a) did not deviate significantly from that of wild type (Table 1). Thus, Cap-D2 seems to be essential for interphase euchromatin organisation and fertility but not for sister chromatid cohesion.
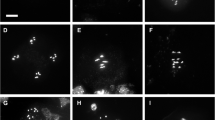
Centromere association, euchromatin condensation and positional sister chromatid cohesion/separation in differentiated 2C, 4C, 8C nuclei and meristematic interphase nuclei of cap-D T-DNA insertion mutants compared to wild type. a Location of FISH probes at chromosome 1 detecting centromeric 178-bp repeats (pAL), top arm territory (CT1top), and ∼100 kb mid-arm segments (BACs T2P11 and F11P17). pAL and CENH3 localise to all 10 centromeres. b Wild-type 4C nuclei (from left to right) with eight centromeric (pAL) signals, two compact CT1top arms, two associated homologous CT1top arms with associated ∼100 kb chromatin segments (T2P11) inside and a compact cohesive CT1top homologue with punctual sister chromatin separation (left) and two separated sister CT1top arms each of them containing a non-cohesive segment (right). c capD3 mutant 4C nuclei and capD2 mutant 8C nucleus. C 1 associated centromeric repeats (left), CT1top chromatin decondensation accompanied by punctual T2P11 segment separation at both homologues (right) in mutant SAIL_826_B06; C 2 associated centromeric repeats together with three (left) and two (right) F11P12 signals indicating punctual separation at one of the two CT1bottom homologues and cohesion at both homologues, respectively, in line SALK_094776; C 3 CT1top chromatin decondensation accompanied by associated centromeric repeats in a 8C nucleus of the heterozygous capD2 mutant. d Examples of 2C and 4C nuclei showing association of CENH3 signals. e Ten individual centromeric pAL signals in a meristematic wild-type nucleus and examples of meristematic SAIL_826_B06 mutant nuclei showing centromere clustering at the nuclear periphery (left), and pAL association to only two (middle) or one (right) centromeric cluster
CAP-D3 is required for plant vigour, fertility, chromatin condensation and sister chromatid cohesion
From A. thaliana SALK and Syngenta SAIL collections, two homozygous Cap-D3 T-DNA insertion mutants were selected. We confirmed the mutants carrying the inserted T-DNA in the first (SAIL_826_B06) and fourth (SALK_094776) exon, respectively, by PCR using gene-specific and T-DNA specific primers (Fig. 2a; ESM Table S3) and by sequencing the obtained PCR products (ESM Table S4).
Semiquantitative RT-PCR analysis was performed on RNA isolated from pools of seedlings to test for the presence of full length or truncated transcripts of the CapD3 gene in both selected mutants (ESM Fig. S7a). In both cases, no PCR products were detected with primer pairs adjacent to the T-DNA insertion borders (III + IV for SAIL_826_B06 and VII + VIII for SALK_094776) indicating that T-DNA insertions disturb synthesis of full length transcripts. However, in the SAIL_826_B06 mutant, two truncated transcripts of a higher level than in the wild-type were detected: a transcript upstream of the T-DNA insertion (V + VI) and also a transcript downstream of the T-DNA (5 + 6), the latter possibly driven by the strong 35S promoter within the T-DNA (Lermontova et al. 2008). Thus, we cannot exclude that an almost complete transcript (without first exon) in the SAIL_826_B06 mutant might be translated into an N-terminally truncated partially functional protein. In case of the SALK_094776 mutant with the T-DNA insertion within the fourth exon, a relatively long transcript upstream of the T-DNA might yield in a C-terminally truncated, partially functional protein.
Immunostaining with antibodies against an N-terminal peptide of Cap-D3 (Fig. 2a) on nuclei of squashed root tips of both mutants showed labelling within the nucleoplasm, similar to the immunolabelling pattern of the wild-type (Fig. 8). After blocking of antibodies with the peptide used for the immunisation, immunosignals disappeared from the nuclei of the wild-type and both mutants (ESM Fig. S7b), indicating that immunosignals against CAP-D3 are specific and in the cap-D3 mutants are indeed caused by the N-terminus of the C-terminally truncated proteins.
The SAIL_826_B06 mutants are smaller (Fig. 3a) whereas the SALK_094776 mutants show wild-type habit. This suggests an essential function of an uninterrupted N-terminal part of Cap-D3 for plant vigour. Significantly reduced pollen fertility and seed set of both mutants (Fig. 3a, b; Table 1) indicate the importance of intact CAP-D3 for fertility. While the SALK line embryos developed normally, translucent ovules and ovules containing abnormal embryos were observed in the SAIL line in addition to normally developed embryos (Fig. 3c).
Significantly decreased sister chromatid cohesion in 4C nuclei at chromosome 1 mid-arm positions (FISH with BACs T2P11 and F11P17) was found for both homozygous mutants (Fig. 4a, b; Table 1) and even in heterozygous plants of line SAIL_826_B06 with 36.7 % (n = 1,996, P < 0.001) and 41.9 % (n = 4,600, P < 0.001) of homologues showing sister cohesion at BAC positions T2P11 and F11P17, respectively. Chromosome 1 top arm territories were significantly more often dispersed, covering >50 % of the nuclear area, in both mutants compared to wild-type nuclei (Table 1). It is possible that more frequent chromosome territory dispersion is correlated with the frequency of separated ∼100 kb chromatin segments such as T2P11 and F11P17. However, the CAP-D2 mutant although showing increased CT1top dispersion did not show decreased positional sister chromatid cohesion.
We found previously (Schubert et al. 2009) that sister chromatid cohesion along the chromosome arm may be low, while centromeres still remain stably cohered. Also in the Cap-D3 mutants, stable centromere cohesion seems to prevent segregation problems. We conclude that CAP-D3 is needed for correct chromatin condensation and full sister chromatid cohesion which may when disturbed impinge on normal plant growth and fertility.
Both CAP-D proteins prevent centromeric and pericentromeric heterochromatin association
Loss of sister centromere cohesion on the one hand, and association of centromeres on the other, may occur in nuclei of higher endopolyploidy level of A. thaliana and A. lyrata, as shown by numbers of centromeric FISH signals deviating from the number of centromeres per nucleus (Berr et al. 2006; Schubert et al. 2006). To test the impact of CAP-D2 and CAP-D3 on interphase centromere arrangement, we studied 2-16C leaf nuclei by FISH with the 178 bp pAL (Fig. 4a). In most mutants and ploidy levels we found a highly significant (P < 0.01) increase of centromere association into one or few clusters. Only in 4C nuclei of the heterozygous cap-D3 mutant SAIL_826_B06 and in 16C nuclei of the heterozygous cap-D2 mutant SALK_077796 the deviation of centromere signal numbers between mutant and wild-type was not significant at P < 0.05 (Fig. 5). Wild-type nuclei never showed only one centromeric signal; however, mutant nuclei frequently did. In accordance with these results, also immunosignals for the centromeric histone variant CENH3 and FISH signals for (peri)centromeric sequences were more often associated in the mutant than in wild-type nuclei (Figs. 4b–d and 6). The increased frequency of centromere association was found in differentiated as well as in meristematic nuclei (Fig. 4e). We conclude that both CAP-D proteins prevent the spontaneous association of centromeric and pericentromeric repeats but do not influence the nucleolus positioning.
CENH3 (marked by arrows) and centromeric FISH (not marked) signal frequencies in 2–16C nuclei of homozygous (ho) and heterozygous (he) cap-D mutants compared to wild-type (number of investigated nuclei in parentheses; significant differences at * P<0.05, ** P<0.01). The distribution of CENH3 and centromeric FISH signal frequencies is similar in 2C and 4C nuclei of the Cap-D3 SALK mutant
(Peri)centromeric heterochromatin and euchromatin arrangement in 2C and 4C nuclei of the homozygous cap-D3 T-DNA insertion mutant SAIL_826_B06 compared to wild-type. a Location of FISH probes at chromosome 4 detecting euchromatic bottom arm territories (CT4bottom), centromeric 178-bp repeats (pAL), and a ∼113 kb segment (BAC F28D6). pAL and F28D6 label all 10 heterochromatic centromeres and pericentromeres, respectively. b 2C wild-type nucleus: nine compact centromeric signals (two of them associated) co-localise to mainly compact pericentromeric signals. Both compact CT4bottom arms associate. 4C wild-type nucleus: 10 compact centromeric signals co-localise to mainly compact pericentromeric signals. Both compact CT4bottom arms are separated. Upper 2C cap-D3 mutant nucleus: all centromeres and pericentromeres are compact, associate and co-localise. Both compact CT4bottom arms associate. Lower 2C cap-D3 mutant nucleus: compact centromeres and pericentromeres associate to three signals and co-localise. Both associated CT4bottom arms are decondensed. 4C cap-D3 mutant nuclei: the compact centromeres associate to one (top) and four (bottom) signals, while the pericentromeric heterochromatin and the CT4bottom euchromatin are decondensed
CAP-D3 supports typical euchromatin and heterochromatin density
Similar as the cap-D2 mutant, both cap-D3 mutants show a significantly increased frequency of CT1top arm euchromatin dispersion in 2-16C nuclei but no increase of anaphase bridge frequencies compared to wild type (Fig. 4a, c1 , c3 ; Table 1). To compare in parallel centromere, pericentromere and euchromatin structures in both cap-D3 mutants versus wild-type, we performed multicolor FISH on 2C and 4C nuclei with probes for pAL, pericentromeres (BAC F28D6; Tessadori et al. 2007) and for the entire chromosome 4 bottom arm (CT4bottom; Fig. 6a).
In wild-type nuclei centromeres, pericentromeres and the euchromatic part of CT4bottom arm appeared mainly compact. The SAIL_826_B06, but not the SALK_094776 mutant showed more often dispersion of all three chromatin domains, while in both mutants dispersion at CT1top was more frequent than in wild type (Fig. 6b; Tables 1 and 2). Thus, it seems that euchromatic chromosome arms, although of similar size (CT1 top = 14.2 Mb, CT4 bottom = 13.5 Mb; Kotani 2002), may display a varying sensitivity regarding CAP-D3 mutations. However, the density and distribution, at least of the truncated CAP-D3 is not correlated with the different degree of chromosome arm dispersion because the distribution of CAP-D3 immunosignals is similar in wild type and mutant nuclei (Fig. 8). In wild type as well as in the cap-D3 mutants, the degree of euchromatic CT4bottom arm dispersion was clearly higher in 2C than in 4C nuclei (Table 2). This was not the case for CT1top (Table 1).
Along interphase chromosomes, starting from the centromere via the pericentromere towards the euchromatic CT4bottom arm, the degree of dispersion increased in wild type but also in both cap-D3 mutants. In most nuclei, dispersion or compactness varied in parallel at all the three chromatin domains. In 16–32 % of wild-type and mutant nuclei the degree of compaction between these domains did not vary in parallel (Table 2). We conclude that especially the N-terminal part of CAP-D3 seems to be involved in realising chromatin density along chromosomes.
The CAP-D proteins do not influence the chromatin substructure
To test in more detail whether the increased centromere association also influences the chromatin substructure in the cap-D mutants, we applied SIM. At a resolution of ∼100 nm, chromatin networks containing centromeric repeats appear as spherical structures at the nuclear periphery in wild type and in the mutants, even in endopolyploid nuclei when several identical chromatids are involved. When centromeres associate, centromeric repeats seem to coalesce resulting in bigger spheres showing similar network-like structures. Euchromatic CT1top arm sequences painted with BAC contigs showed, despite being more dispersed in the mutants, a similar chromatin substructure as the centromeric sequences although not forming spheres (Fig. 7; Figure/Movie in ESM S8).
Centromeric and euchromatic chromatin structures in nuclei of cap-D mutants compared to wild-type analysed by SIM. Left 4C wild-type nucleus: a chromatin network forms 10 spheric structures (pAL) at ten cohesive centromeres (each containing two sister chromatids) localised close to the nuclear periphery. Right 4C wild-type nucleus: similar structures of the eight centromeric signals indicate that associated centromeres also form fused spheres. Both homologous CT1top arms are mainly compact and separated. 4C cap-D3 mutant nucleus: the centromere coalescence yields three spheres, one of them not completely fused (arrow). Both CT1top arms are associated. Left 16C cap-D2 mutant nucleus: the 10 centromeres (each containing eight chromatids) form spherical structures of different size, the big ones obviously due to association (asterisks), the smaller possibly due to sister centromere separation (arrows). The inset demonstrates at higher magnification that the spherical structures are formed by a network of chromatin. Both CT1top arms are mainly compact and associated. Right 16C cap-D2 mutant nucleus: 12 centromeric signals suggest that not all of them are cohesive. The associated CT1top arms are less condensed than in the left 16C nucleus. The inset shows a comparison between heterochromatic centromeric and euchromatic CT1top structure at higher magnification
We suggest that the CAP-D proteins do not alter the chromatin substructure but influence the higher order organisation of interphase nuclei by inducing chromatin compactness and preventing (peri)centromeric heterochromatin association.
CAP-D3 and SMC3 show separate but adjacent distribution throughout euchromatic nucleoplasm
We tried to raise antibodies against most condensin (including CAP-D2) and cohesin subunits. However, only antibodies against CAP-D3 and the cohesin subunit SMC3 were useful for localization in situ (Fig. 8). To study the presence and distribution of CAP-D3, we applied simultaneously antibodies against CAP-D3 and against SMC3 to 2-16C interphase nuclei. At all ploidy levels, both proteins are homogeneously distributed with similar density throughout the nucleoplasm except for the nucleolus and the heterochromatic chromocenters. Also Lam et al. (2005) found SMC3 to be absent from nucleoli of somatic interphase nuclei. SIM revealed looped fibre networks of both proteins within euchromatic domains which only occasionally attach each other. The CAP-D3 network is more extended to the nuclear periphery than that of SMC3 which concentrates more in the nuclear interior (Fig. 8). CAP-D3 was not present at metaphase chromosomes, while SMC3 signals were weak along chromosome arms and enriched at centromeres. These observations suggest that cohesin and condensin form distinct reticulate structures to organize interphase chromatin in differentiated nuclei of all endopolyploidy levels.
CAP-D3 and SMC3 distribution in 2–16C nuclei of Columbia wild-type and a cap-D3 mutant expressing a truncated protein. Both proteins visible as looped fibres form differential reticulate structures sometimes in contact with each other and with chromatin. The proteins are not present in the nucleolus and at heterochromatin. For comparison to the SIM images, the 2C and 16C (scaled down) nuclei are also shown in wide-field (WF) illumination
Discussion
The conserved CAP-D proteins fulfil essential functions during plant development
In addition to the well-known functions in chromosome condensation and sister chromatid cohesion, condensin and cohesin complexes and their subunits control genome organisation, gene expression, development and meiosis (reviewed in Hudson et al. 2009; Wood et al. 2010).
Here, we show that, also in higher plants, two conserved HEAT repeat containing condensin CAP-D subunits are essential for chromatin density, cohesion, normal growth and fertility. The highly degenerated repeated motifs of the HEAT domain mediate protein–protein interactions and are involved in chromosome-related functions (Neuwald and Hirano 2000).
Based on in silico analysis, we propose that both CAP-D genes are regulated, similar as other condensin subunit candidate genes, in a cell cycle-dependent manner by COP9 signalosome subunits, the RBR, the transcription factor E2Fa-DPa heterodimer and the HUB1 protein.
This assumption is supported by data available for non-plant organisms. Drosophila and human RBR family proteins interact with the condensin II CAP-D3 subunit to realise chromatin compaction and cohesion during mitosis (Longworth et al. 2008; Manning et al. 2010). Drosophila RBR and CAP-D3 co-localise at polytene chromosomes and alter the expression of cell cycle-independent developmentally regulated genes with tissue-specific functions. This suggests additional functions for both proteins in nuclei of differentiated cells (Longworth et al. 2012). In differentiated endopolyploid A. thaliana nuclei, CAP-D3 has apparently similar functions.
Manning et al. (2010) showed that the RBR homologue of Drosophila promotes cohesin complex loading onto DNA and that CAP-D3 loading on chromatin was decreased in RBR-deficient cells. Moreover, knock down of CAP-D3 caused similar mitotic defects as seen after knockdown of the RB1 gene which encodes RBR. Based on sequence homology, we assigned AtCAP-D2 and AtCAP-D3 as homologues of vertebrate CAP-D2 (condensin I) and CAP-D3 (condensin II), respectively. AtCAP-D3 is present in interphase nuclei, similar as condensin II in HeLa cells (Ono et al. 2004), supporting that AtCAP-D3 corresponds to vertebrate CAP-D3.
The reduced fertility of the A. thaliana mutants suggests a role of the Cap-D genes during meiosis. CAP-D is also necessary for correct Drosophila (Hartl et al. 2008b) and porcine (Liskova et al. 2010) meiosis.
Green et al. (2012) concluded that condensin II of vertebrates is required to establish the axis of mitotic chromosomes whereas condensin I arranges interphase chromatin loops. However, we could not observe immunolabelling of metaphase chromosomes applying Cap-D3 antibodies. Either Arabidopsis metaphase chromosomes are less accessible to the antibodies or metaphase chromosome organisation differs between animals and plants.
Condensin dysfunction induces mitotic anaphase bridges in human (Ono et al. 2004; Gerlich et al. 2006; Samoshkin et al. 2012) and chicken (Green et al. 2012) cells. We did not find an increased number of anaphase bridges as a consequence of CAP-D depletion. Possibly, the remaining CAP-D2 and truncated CAP-D3 proteins are still able to prevent such disorders, or plants are less sensitive to CAP-D attenuation than vertebrates. We conclude that, similar as in animals, higher plants contain two different CAP-D proteins to organize mitosis, meiosis and interphase chromatin architecture in a similar but not identical manner as in animals.
Suppressing repeat clustering—an essential task to maintain correct chromatin organisation during interphase and early meiosis?
The clustering of repeats localised at centromeres, heterochromatin and telomeres is a common phenomenon observed in fungi, animals and plants. Indications from the literature suggest that such associations may have no functional meaning but nevertheless seem to be regulated in a development- and tissue-specific manner.
Active non-homologous centromere associations in early meiotic prophase, possibly as a prerequisite to induce homologous pairing, has been suggested for yeast (Tsubouchi and Roeder 2005), Drosophila (Takeo et al. 2011; Tanneti et al. 2011), mouse (Brinkley et al. 1986), cereals (Bennett et al. 1979; Maestra et al. 2002) and allium (Church and Moens 1976; reviewed by Stewart and Dawson 2008). In fungi and animals, the clustering of telomeres during leptotene/zygotene into a “bouquet” promotes the alignment of homologues (reviewed by Scherthan 2007). Arabidopsis centromeres and telomeres cluster at leptotene/zygotene (Armstrong et al. 2001; Da Ines et al. 2012) and the meiotic cohesin subunit SYN1 has been shown to mediate homologous centromere pairing in plants (Cai et al. 2003).
In rice, association occurs between homologous telomeres and centromeres in somatic nuclei (Prieto et al. 2004). Preferential homologous and heterologous associations may also occur at transgenic tandem repetitive sequences, at subtelomeres and pericentromeres in differentiated Arabidopsis nuclei of different endopolyploidy level (Jovtchev et al. 2008, 2011; Pecinka et al. 2005; Watanabe et al. 2005; Schubert et al. 2012). Here, we show that A. thaliana centromeres often associate because only in ∼2–8 % of the 2–8C wild-type nuclei were all ten centromeres separated. Whether centromere association is functional is not yet clear. At least the findings of Fang and Spector (2005), showing a cell type-dependent distribution of A. thaliana centromeres in endoreduplicated nuclei with predominant clustering in root epidermal cells and dispersion in leaf epidermal cells, suggest tissue-specific centromere localisation. Also the negative correlation between heterochromatin clustering and transcriptional activity, proven for several plant species (Ceccarelli et al. 1998), indicates that such a clustering may be a dynamic process related to cell differentiation and development.
The spatial distribution of chromatin segments and its dynamics in interphase nuclei can be explained by models based on self-organisation and fractal globule formation (Hancock 2004; McNally and Mazza 2010; Misteli 2007, 2009; Rajapakse et al. 2009; Nishino et al. 2012). Specific protein–protein, protein–DNA and DNA–DNA interactions restrict a random spatial arrangement of chromatin (Lanctot et al. 2007; Misteli 2007). However, De Nooijer et al. (2009) conclude that non-specific interactions leading to entropic effects, such as, e.g. depletion attraction (Marenduzzo et al. 2006a, b), are sufficient to explain the position of heterochromatic chromocenters at the nuclear periphery and of nucleoli in the centre of Arabidosis interphase nuclei. Consistent with the Rosette model for Arabidopsis chromatin organisation (Fransz et al. 2002; Fransz and De Jong 2011), 15 chromocenter-associated loops would suppress completely the chromocenter clustering, while fewer loops (<3) would allow chromocenters to cluster. The looped arms with chromocenters (LAC) model in which loops do not attach to chromocenters requires specific interactions for a central nucleolus positioning but promotes the association of chromocenters to one or two clusters (De Nooijer et al. 2009). Here and previously (Schubert et al. 2009), we showed that A. thaliana 2–8C wild-type nuclei contain ∼4 to 10 centromere clusters. We also found that only ∼37 % of the telomeres in 2C nuclei associate to centromeres indicating that not all chromatin fibres are chromocenter-associated (Schubert et al. 2012). In the CAP-D mutants, the nucleolus stays mostly in the centre of the nucleus. Thus, a regulated dynamic switch of chromatin organisation between the rosette and LAC model-predicted structures is likely. The increased centromere association up to only one cluster in the cap-D mutants strongly supports the assumption that the tendency of repeat clustering by self-organisation via entropic forces is regulated by specific factors such as CAP-D proteins.
In plants, factors regulating centromere association are known. The wheat pairing homoeologous1 locus reduces meiotic and somatic non-homologous centromere associations (Martinez-Perez et al. 2001) by suppressing the cyclin-dependent kinase activity (Greer et al. 2012).
In addition to its well-known functions in chromatin condensation along the chromosome axis during nuclear division and in interphase, condensin complexes inhibit associations between homologues (reviewed by Wood et al. 2010). Drosophila condensin II antagonizes the pairing of homologues (Joyce et al. 2012) and reduces transvection by limiting interactions between homologues during interphase. Furthermore, the condensin II subunits SMC4, Cap-H2 and Cap-D3 are necessary to disassemble aligned polytene chromosomes (Hartl et al. 2008a). Drosophila condensin II also induces the formation of compact CTs and is required to disperse heterologous centromeres (Bauer et al. 2012). It has been speculated that condensin-mediated supercoiling (Hirano 2006) occurs in interphase nuclei and can disrupt tight polytene chromosome alignment to enable gene expression (Hartl et al. 2008a). The presence of the Arabidopsis condensin subunit CAP-D3 in highly endopolyploid cells might be necessary to maintain the previously described non-cohesive chromatid arrangement in differentiated leaf nuclei >16C (Schubert et al. 2012).
Furthermore, cohesin subunits can increase as well as decrease centromere association in 4C A. thaliana nuclei (Schubert et al. 2009). Here, we show in cap-D mutants a strong increase in centromere clustering. Thus, it becomes obvious that cohesin and condensin play a role in regulating interphase chromatin architecture by either inducing or suppressing the clustering of “sticky” repeats.
Cohesin and condensin proteins form reticulate structures to organize interphase chromatin architecture
SMC complexes interact with DNA to organize higher order chromosome topology (reviewed by Carter and Sjögren 2012). The retinoblastoma protein-mediated association of human and Drosophila CAP-D3 to chromatin is important for its condensation (Longworth et al. 2008, 2012). Condensins hold looped chromatin fibres around the length axis of human mitotic chromosomes (Nishino et al. 2012) and in yeast cohesin and condensin complexes may form chromatin fibre linkages to organize centromeres (Stephens et al. 2011; Haering and Jessberger 2012).
Here, we show that the CAP-D proteins mediate condensation along interphase chromosomes and prevent interchromosomal association, but seem not to alter the shape of chromatin substructures that possibly correspond to topologically associating domains (TADs). TADs were proposed by Gibcus and Dekker (2013) to represent the basic chromosomal structure and to mediate interactions in cis connected with long-range gene regulations. Thus, the CAP-D proteins seem to influence the higher-order chromatin organisation of euchromatin and heterochromatin domains by controlling chromatin density and sister chromatid cohesion. Both CAP-D3 and the cohesion subunit SMC3 form distinct reticulate structures potentially based on proposed di- and/or multimerisation of cohesin and condensin subunits and/or complexes (Thadani et al. 2012) in interphase nuclei of different ploidy level in wild type as well as in mutants. However, the task to prevent clustering of (peri)centromeric heterochromatin by counteracting entropic forces (see above) is no longer sufficiently performed in the mutants. Either there are not enough molecules available (hemizygous cap-D2/Cap-D2) or, in case of the truncated versions of CAP-D3, necessary protein interaction(s) cannot take place properly. Both cohesin and condensin components, although not displaying identical localisation, possibly cooperate to fulfil similar functions, e.g. sister chromatid cohesion and chromatin compaction in interphase nuclei independent of their endopolyploidy level.
References
Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, Gadrinab C, Heller C, Jeske A, Koesema E, Meyers CC, Parker H, Prednis L, Ansari Y, Choy N, Deen H, Geralt M, Hazari N, Hom E, Karnes M, Mulholland C, Ndubaku R, Schmidt I, Guzman P, Aguilar-Henonin L, Schmid M, Weigel D, Carter DE, Marchand T, Risseeuw E, Brogden D, Zeko A, Crosby WL, Berry CC, Ecker JR (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301:653–657
Armstrong SJ, Franklin FC, Jones GH (2001) Nucleolus-associated telomere clustering and pairing precede meiotic chromosome synapsis in Arabidopsis thaliana. J Cell Sci 114:4207–4217
Bauer CR, Hartl TA, Bosco G (2012) Condensin II promotes the formation of chromosome territories by inducing axial compaction of polyploid interphase chromosomes. PLoS Genet 8:e1002873
Bennett MD (1979) Centromere arrangements in Triticum aestivum and their relation to synapsis. Heredity 43:157–157
Berr A, Pecinka A, Meister A, Kreth G, Fuchs J, Blattner FR, Lysak MA, Schubert I (2006) Chromosome arrangement and nuclear architecture but not centromeric sequences are conserved between Arabidopsis thaliana and Arabidopsis lyrata. Plant J 48:771–783
Brinkley BR, Brenner SL, Hall JM, Tousson A, Balczon RD, Valdivia MM (1986) Arrangements of kinetochores in mouse cells during meiosis and spermiogenesis. Chromosoma 94:309–317
Cai X, Dong F, Edelmann RE, Makaroff CA (2003) The Arabidopsis SYN1 cohesin protein is required for sister chromatid arm cohesion and homologous chromosome pairing. J Cell Sci 116:2999–3007
Carter SD, Sjogren C (2012) The SMC complexes, DNA and chromosome topology: right or knot? Crit Rev Biochem Mol Biol 47:1–16
Ceccarelli M, Morosi L, Cionini PG (1998) Chromocenter association in plant cell nuclei: determinants, functional significance, and evolutionary implications. Genome 41:96–103
Chamovitz DA (2009) Revisiting the COP9 signalosome as a transcriptional regulator. EMBO Rep 10:352–358
Church K, Moens PB (1976) Centromere behavior during interphase and meiotic prophase in Allium fistulosum from 3-D E.M. reconstruction. Chromosoma 56:249–263
Da Ines O, Abe K, Goubely C, Gallego ME, White CI (2012) Differing requirements for RAD51 and DMC1 in meiotic pairing of centromeres and chromosome arms in Arabidopsis thaliana. PLoS Genet 8:e1002636
De Bodt S, Hollunder J, Nelissen H, Meulemeester N, Inze D (2012) CORNET 2.0: integrating plant coexpression, protein–protein interactions, regulatory interactions, gene associations and functional annotations. New Phytol 195:707–720
De Nooijer S, Wellink J, Mulder B, Bisseling T (2009) Non-specific interactions are sufficient to explain the position of heterochromatic chromocenters and nucleoli in interphase nuclei. Nucl Acids Res 37:3558–3568
De Veylder L, Beeckman T, Beemster GT, De Almeida-Engler J, Ormenes S, Maes S, Naudts M, Van Der Schueren E, Jacqmard A, Engler G, Inze D (2002) Control of proliferation, endoreduplication and differentiation by the Arabidopsis E2Fa-DPa transcription factor. EMBO J 21:1360–1368
Desper R, Gascuel O (2002) Fast and accurate phylogeny reconstruction algorithms based on the minimum-evolution principle. J Comput Biol 9:687–705
Fang Y, Spector DL (2005) Centromere positioning and dynamics in living Arabidopsis plants. Mol Biol Cell 16:5710–5718
Fleury D, Himanen K, Cnops G, Nelissen H, Boccardi TM, Maere S, Beemster GT, Neyt P, Anami S, Robles P, Micol JL, Inze D, Van Lijsebettens M (2007) The Arabidopsis thaliana homolog of yeast BRE1 has a function in cell cycle regulation during early leaf and root growth. Plant Cell 19:417–432
Fransz P, de Jong H (2011) From nucleosome to chromosome: a dynamic organization of genetic information. Plant J 66:4–17
Fransz P, De Jong JH, Lysak M, Castiglione MR, Schubert I (2002) Interphase chromosomes in Arabidopsis are organized as well defined chromocenters from which euchromatin loops emanate. Proc Nat Acad Sci USA 99:14584–14589
Fujimoto S, Yonemura M, Matsunaga S, Nakagawa T, Uchiyama S, Fukui K (2005) Characterization and dynamic analysis of Arabidopsis condensin subunits, AtCAP-H and AtCAP-H2. Planta 222:293–300
Gerlich D, Hirota T, Koch B, Peters JM, Ellenberg J (2006) Condensin I stabilizes chromosomes mechanically through a dynamic interaction in live cells. Curr Biol 16:333–344
Gibcus JH, Dekker J (2013) The hierarchy of the 3D genome. Mol Cell 49:773–782
Green LC, Kalitsis P, Chang TM, Cipetic M, Kim JH, Marshall O, Turnbull L, Whitchurch CB, Vagnarelli P, Samejima K, Earnshaw WC, Choo KH, Hudson DF (2012) Contrasting roles of condensin I and condensin II in mitotic chromosome formation. J Cell Sci 125:1591–1604
Greer E, Martin AC, Pendle A, Colas I, Jones AM, Moore G, Shaw P (2012) The Ph1 locus suppresses Cdk2-type activity during premeiosis and meiosis in wheat. Plant Cell 24:152–162
Haering CH, Jessberger R (2012) Cohesin in determining chromosome architecture. Exp Cell Res 318:1386–1393
Hancock R (2004) Internal organisation of the nucleus: assembly of compartments by macromolecular crowding and the nuclear matrix model. Biol Cell 96:595–601
Hartl TA, Smith HF, Bosco G (2008a) Chromosome alignment and transvection are antagonized by condensin II. Science 322:1384–1387
Hartl TA, Sweeney SJ, Knepler PJ, Bosco G (2008b) Condensin II resolves chromosomal associations to enable anaphase I segregation in Drosophila male meiosis. PLoS Genet 4:e1000228
Heckmann S, Lermontova I, Berckmans B, De Veylder L, Bäumlein H, Schubert I (2011) The E2F transcription factor family regulates CENH3 expression in Arabidopsis thaliana. Plant J 68:646–656
Hirano T (2006) At the heart of the chromosome: SMC proteins in action. Nat Rev Mol Cell Biol 7:311–322
Hirano T (2012) Condensins: universal organizers of chromosomes with diverse functions. Genes Dev 26:1659–1678
Hirano M, Hirano T (2006) Opening closed arms: long-distance activation of SMC ATPase by hinge–DNA interactions. Mol Cell 21:175–186
Hudson DF, Marshall KM, Earnshaw WC (2009) Condensin: architect of mitotic chromosomes. Chromosome Res 17:131–144
Jasencakova Z, Meister A, Walter J, Turner BM, Schubert I (2000) Histone H4 acetylation of euchromatin and heterochromatin is cell cycle dependent and correlated with replication rather than with transcription. Plant Cell 12:2087–2100
Jenik PD, Gillmor CS, Lukowitz W (2007) Embryonic patterning in Arabidopsis thaliana. Ann Rev Cell Dev Biol 23:207–236
Jovtchev G, Watanabe K, Pecinka A, Rosin FM, Mette MF, Lam E, Schubert I (2008) Size and number of tandem repeat arrays can determine somatic homologous pairing of transgene loci mediated by epigenetic modifications in Arabidopsis thaliana nuclei. Chromosoma 117:267–276
Jovtchev G, Borisova BE, Kuhlmann M, Fuchs J, Watanabe K, Schubert I, Mette MF (2011) Pairing of lacO tandem repeats in Arabidopsis thaliana nuclei requires the presence of hypermethylated, large arrays at two chromosomal positions, but does not depend on H3-lysine-9-dimethylation. Chromosoma 120:609–619
Joyce EF, Williams BR, Xie T, Wu CT (2012) Identification of genes that promote or antagonize somatic homolog pairing using a high-throughput FISH-based screen. PLoS Genet 8:e1002667
Kawabe A, Nasuda S (2005) Structure and genomic organization of centromeric repeats in Arabidopsis species. Mol Genet Genomics 272:593–602
Kimura M (1983) The neutral theory of molecular evolution. Cambridge Univ Press, Cambridge, p 367
Kotani H (2002) The size and genome organization of Arabidopsis thaliana. In: Murata M and Sakamoto W (eds) Structure and function of plant centromeres: a challenge to the orthodoxy pp 35–39
Lam WS, Yang X, Makaroff CA (2005) Characterization of Arabidopsis thaliana SMC1 and SMC3: evidence that AtSMC3 may function beyond chromosome cohesion. J Cell Sci 118:3037–3048
Lanctot C, Cheutin T, Cremer M, Cavalli G, Cremer T (2007) Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nature Rev 8:104–115
Lermontova I, Fuchs J, Schubert I (2008) The Arabidopsis checkpoint protein Bub3.1 is essential for gametophyte development. Front Biosci 13:5202–5211
Liskova L, Susor A, Pivonkova K, Saskova A, Karabinova P, Kubelka M (2010) Detection of condensin I and II in maturing pig oocytes. Reprod Fertil Dev 22:644–652
Liu CM, McElver J, Tzafrir I, Joosen R, Wittich P, Patton D, Van Lammeren AA, Meinke D (2002) Condensin and cohesin knockouts in Arabidopsis exhibit a titan seed phenotype. Plant J 29:405–415
Longworth MS, Herr A, Ji JY, Dyson NJ (2008) RBF1 promotes chromatin condensation through a conserved interaction with the condensin II protein dCAP-D3. Genes Dev 22:1011–1024
Longworth MS, Walker JA, Anderssen E, Moon NS, Gladden A, Heck MM, Ramaswamy S, Dyson NJ (2012) A shared role for RBF1 and dCAP-D3 in the regulation of transcription with consequences for innate immunity. PLoS Genet 8:e1002618
Losada A, Hirano T (2005) Dynamic molecular linkers of the genome: the first decade of SMC proteins. Genes Dev 19:1269–1287
Luo RX, Postigo AA, Dean DC (1998) Rb interacts with histone deacetylase to repress transcription. Cell 92:463–473
Maestra B, Hans de Jong J, Shepherd K, Naranjo T (2002) Chromosome arrangement and behaviour of two rye homologous telosomes at the onset of meiosis in disomic wheat-5RL addition lines with and without the Ph1 locus. Chromosome Res 10:655–667
Magyar Z (2008) Keeping the balance between proliferation and differentiation by the E2F transcriptional regulatory network is central to plant growth and development. In: Bogre L, Beemster GT (eds) Plant growth and signaling. Springer, Berlin
Magyar Z, De Veylder L, Atanassova A, Bako L, Inze D, Bogre L (2005) The role of the Arabidopsis E2FB transcription factor in regulating auxin-dependent cell division. Plant Cell 17:2527–2541
Magyar Z, Horvath B, Khan S, Mohammed B, Henriques R, De Veylder L, Bako L, Scheres B, Bogre L (2012) Arabidopsis E2FA stimulates proliferation and endocycle separately through RBR-bound and RBR-free complexes. EMBO J 31:1480–1493
Manning AL, Longworth MS, Dyson NJ (2010) Loss of pRB causes centromere dysfunction and chromosomal instability. Genes Dev 24:1364–1376
Marenduzzo D, Finan K, Cook PR (2006a) The depletion attraction: an underappreciated force driving cellular organization. J Cell Biol 175:681–686
Marenduzzo D, Micheletti C, Cook PR (2006b) Entropy-driven genome organization. Biophys J 90:3712–3721
Martinez-Perez E, Shaw P, Moore G (2001) The Ph1 locus is needed to ensure specific somatic and meiotic centromere association. Nature 411:204–207
Martinez-Zapater J, Estelle A, Sommerville C (1986) A highly repeated DNA sequence in Arabidopsis thaliana. Mol Gen Genet 204:4417–4423
McNally JG, Mazza D (2010) Fractal geometry in the nucleus. EMBO J 29:2–3
Misteli T (2007) Beyond the sequence: cellular organization of genome function. Cell 128:787–800
Misteli T (2009) Self-organization in the genome. Proc Nat Acad Sci USA 106:6885–6886
Nagaki K, Cheng Z, Ouyang S, Talbert PB, Kim M, Jones KM, Henikoff S, Buell CR, Jiang J (2004) Sequencing of a rice centromere uncovers active genes. Nat Genet 36:138–145
Nasmyth K, Haering CH (2005) The structure and function of SMC and kleisin complexes. Annu Rev Biochem 74:595–648
Neuwald AF, Hirano T (2000) HEAT repeats associated with condensins, cohesins, and other complexes involved in chromosome-related functions. Genome Res 10:1445–1452
Nishino Y, Eltsov M, Joti Y, Ito K, Takata H, Takahashi Y, Hihara S, Frangakis AS, Imamoto N, Ishikawa T, Maeshima K (2012) Human mitotic chromosomes consist predominantly of irregularly folded nucleosome fibres without a 30-nm chromatin structure. EMBO J 31:1644–1653
Ono T, Fan Y, Spector DL, Hirano T (2004) Spatial and temporal regulation of condensins I and II in mitotic chromosome assembly in human cells. Mol Biol Cell 15:3296–3308
Palecek J, Vidot S, Feng M, Doherty AJ, Lehmann AR (2006) The Smc5-Smc6 DNA repair complex: bridging of the Smc5-Smc6 heads by the KLEISIN, Nse4, and non-Kleisin subunits. J Biol Chem 281:36952–36959
Pecinka A, Schubert V, Meister A, Kreth G, Klatte M, Lysa MA, Fuchs J, Schubert I (2004) Chromosome territory arrangement and homologous pairing in nuclei of Arabidopsis thaliana are predominantly random except for NOR-bearing chromosomes. Chromosoma 113:258–269
Pecinka A, Kato N, Meister A, Probst AV, Schubert I, Lam E (2005) Tandem repetitive transgenes and fluorescent chromatin tags alter local interphase chromosome arrangement in Arabidopsis thaliana. J Cell Sci 118:3751–3758
Peric-Hupkes D, Van Steensel B (2008) Linking cohesin to gene regulation. Cell 132:925–928
Prieto P, Santos AP, Moore G, Shaw P (2004) Chromosomes associate premeiotically and in xylem vessel cells via their telomeres and centromeres in diploid rice (Oryza sativa). Chromosoma 112:300–307
Rajapakse I, Perlman MD, Scalzo D, Kooperberg C, Groudine M, Kosak ST (2009) The emergence of lineage-specific chromosomal topologies from coordinate gene regulation. Proc Nat Acad Sci USA 106:6679–6684
Sakamoto T, Inui YT, Uraguchi S, Yoshizumi T, Matsunaga S, Mastui M, Umeda M, Fukui K, Fujiwara T (2011) Condensin II alleviates DNA damage and is essential for tolerance of boron overload stress in Arabidopsis. Plant Cell 23:3533–3546
Samoshkin A, Dulev S, Loukinov D, Rosenfeld JA, Strunnikov AV (2012) Condensin dysfunction in human cells induces nonrandom chromosomal breaks in anaphase, with distinct patterns for both unique and repeated genomic regions. Chromosoma 121:191–199
Scherthan H (2007) Telomere attachment and clustering during meiosis. Cell Mol Life Sci 64:117–124
Schubert V (2009) SMC proteins and their multiple functions in higher plants. Cytogenet Genome Res 124:202–214
Schubert I, Fransz PF, Fuchs J, de Jong JH (2001) Chromosome painting in plants. Meth Cell Sci 23:57–69
Schubert V, Klatte M, Pecinka A, Meister A, Jasencakova Z, Schubert I (2006) Sister chromatids are often incompletely aligned in meristematic and endopolyploid interphase nuclei of Arabidopsis thaliana. Genetics 172:467–475
Schubert V, Kim YM, Schubert I (2008) Arabidopsis sister chromatids often show complete alignment or separation along a 1.2-Mb euchromatic region but no cohesion “hot spots”. Chromosoma 117:261–266
Schubert V, Weissleder A, Ali H, Fuchs J, Lermontova I, Meister A, Schubert I (2009) Cohesin gene defects may impair sister chromatid alignment and genome stability in Arabidopsis thaliana. Chromosoma 118:591–605
Schubert V, Berr A, Meister A (2012) Interphase chromatin organisation in Arabidopsis nuclei: constraints versus randomness. Chromosoma 121:369–387
Sessions A, Burke E, Presting G, Aux G, McElver J, Patton D, Dietrich B, Ho P, Bacwaden J, Ko C, Clarke JD, Cotton D, Bullis D, Snell J, Miguel T, Hutchison D, Kimmerly B, Mitzel T, Katagiri F, Glazebrook J, Law M, Goff SA (2002) A high-throughput Arabidopsis reverse genetics system. Plant Cell 14:2985–2994
Siddiqui NU, Stronghill PE, Dengler RE, Hasenkampf CA, Riggs CD (2003) Mutations in Arabidopsis condensin genes disrupt embryogenesis, meristem organization and segregation of homologous chromosomes during meiosis. Development 130:3283–3295
Siddiqui NU, Rusyniak S, Hasenkampf CA, Riggs CD (2006) Disruption of the Arabidopsis SMC4 gene, AtCAP-C, compromises gametogenesis and embryogenesis. Planta 223:990–997
Sozzani R, Maggio C, Varotto S, Canova S, Bergounioux C, Albani D, Cella R (2006) Interplay between Arabidopsis activating factors E2Fb and E2Fa in cell cycle progression and development. Plant Physiol 140:1355–1366
Stephens AD, Haase J, Vicci L, Taylor RM 2nd, Bloom K (2011) Cohesin, condensin, and the intramolecular centromere loop together generate the mitotic chromatin spring. J Cell Biol 193:1167–1180
Stewart MN, Dawson DS (2008) Changing partners: moving from non-homologous to homologous centromere pairing in meiosis. Trends Genet 24:564–573
Takeo S, Lake CM, Morais-de-Sa E, Sunkel CE, Hawley RS (2011) Synaptonemal complex-dependent centromeric clustering and the initiation of synapsis in Drosophila oocytes. Curr Biol 21:1845–1851
Tanneti NS, Landy K, Joyce EF, McKim KS (2011) A pathway for synapsis initiation during zygotene in Drosophila oocytes. Curr Biol 21:1852–1857
Tessadori F, Chupeau MC, Chupeau Y, Knip M, Germann S, van Driel R, Fransz P, Gaudin V (2007) Large-scale dissociation and sequential reassembly of pericentric heterochromatin in dedifferentiated Arabidopsis cells. J Cell Sci 120:1200–1208
Thadani R, Uhlmann F, Heeger S (2012) Condensin, chromatin crossbarring and chromosome condensation. Curr Biol 22:R1012–R1021
Tsubouchi T, Roeder GS (2005) A synaptonemal complex protein promotes homology-independent centromere coupling. Science 308:870–873
Tzafrir I, McElver JA, C-M L, Yang LJ, Wu JQ, Martinez A, Patton DA, Meinke DW (2002) Diversity of TITAN functions in Arabidopsis seed development. Plant Physiol 128:38–51
Van den Heuvel S, Dyson NJ (2008) Conserved functions of the pRB and E2F families. Nat Rev Mol Cell Biol 9:713–724
Ward P (2002) FISH probes and labelling techniques. In: FISH. Beatty B, Squire J (eds). Oxford Univ Press: Oxford. pp 5–28
Watanabe K, Pecinka A, Meister A, Schubert I, Lam E (2005) DNA hypomethylation reduces homologous pairing of inserted tandem repeat arrays in somatic nuclei of Arabidopsis thaliana. Plant J 44:531–540
Wood AJ, Severson AF, Meyer BJ (2010) Condensin and cohesin complexity: the expanding repertoire of functions. Nature Rev 11:391–404
Acknowledgments
We thank Jörg Fuchs for flow sorting of nuclei, Martina Kühne, Andrea Kunze, Andrea Weißleder, Joachim Bruder and Rita Schubert for excellent assistance, Armin Meister for help with statistics and Swetlana Friedel for help with analysis of coexpression by CORNET program.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Schubert, V., Lermontova, I. & Schubert, I. The Arabidopsis CAP-D proteins are required for correct chromatin organisation, growth and fertility. Chromosoma 122, 517–533 (2013). https://doi.org/10.1007/s00412-013-0424-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00412-013-0424-y