Abstract
Condensin is a multi-subunit protein complex that belongs to the family of structural maintenance of chromosomes (SMC) complexes. Condensins regulate chromosome structure in a wide range of processes including chromosome segregation, gene regulation, DNA repair and recombination. Recent research defined the structural features and molecular activities of condensins, but it is unclear how these activities are connected to the multitude of phenotypes and functions attributed to condensins. In this review, we briefly discuss the different molecular mechanisms by which condensins may regulate global chromosome compaction, organization of topologically associated domains, clustering of specific loci such as tRNA genes, rDNA segregation, and gene regulation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Chromosome remodeling by condensin
The major molecular activity of condensins is forming chromosomal loops through their conserved SMC ring structure (Cuylen et al. 2011; van Ruiten and Rowland 2018). SMC complexes are thought to form such loops by loop extrusion, a process in which DNA is pushed through the SMC ring in an ATPase dependent manner (Nasmyth 2001; Goloborodko et al. 2016a). Loop extrusion by condensin is well supported by in vitro experiments using DNA curtains (Terakawa et al. 2017) and by single-molecule imaging (Ganji et al. 2018). In vivo evidence of loop extrusion stems mostly from the Bacillus subtilis SMC complex, which functions to juxtapose the two arms of the bacterial chromosome. By measuring the speed of this juxtaposition, the speed of B. subtilis SMC complex movement along the chromosome was estimated to be ~ 0.8 kb/s (Wang et al. 2017b). Yeast condensin extruded DNA at a similar rate in vitro, with speeds up to 1.5 kb/s depending on the concentration of ATP (Terakawa et al. 2017; Ganji et al. 2018). Accordingly, mutations that affect the ATPase activity changed the speed of the B. subtilis SMC complex (Wang et al. 2018b). In addition, the speed of extrusion was affected by transcription in B. subtilis (Wang et al. 2017b), suggesting that obstacles on the DNA, including chromatin in eukaryotes, may regulate condensin processivity.
Condensins regulate chromosome compaction for cell division
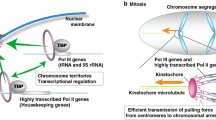
The major function of condensins in all eukaryotes is their essential role in chromosome assembly and segregation during mitosis and meiosis (Chan et al. 2004; Cuylen and Haering 2011; Houlard et al. 2015; Kinoshita and Hirano 2017; Kakui and Uhlmann 2018). In preparation for cell division, chromosomes assume a highly compacted structure, characterized by extensive looping of the chromatin fiber. Following condensin inactivation, loop formation and compaction are severely disrupted as indicated by global mapping of chromosomal interactions within mitotic chromosomes (Kakui et al. 2017; Schalbetter et al. 2017; Gibcus et al. 2018). Loop extrusion is likely one of the central drivers of chromosome compaction, and models based on loop extrusion can explain the formation of chromosomal loops in mitotic chromosomes (Fig. 1ai, ii) (Goloborodko et al. 2016a, b; Gibcus et al. 2018; Kakui and Uhlmann 2018). Interestingly, the range of loops mediated by condensin differs among organisms (Kakui and Uhlmann 2018). Unlike yeast, where there is a single condensin, metazoans contain two condensin types, named condensin I and II (Hirano 2016). Specific knockout experiments and imaging of the two condensin complexes suggest that condensin II mediates longer loops and sets up the axis of mitotic chromosomes, while condensin I mediates shorter loops thickening the mitotic chromosomes (Fig. 1aii) (Hirota et al. 2004; Green et al. 2012; Kinoshita and Hirano 2017; Walther et al. 2018).
Condensin-mediated genome organization: a Mitotic structure of chromosomes in yeast (i) and metazoans (ii). (i) In yeast, condensin compacts the rDNA and tethers the rDNA to the centromere. The centromere-proximal side of the rDNA is enriched for condensin, which compacts chromosomes through loop extrusion. (ii) In metazoans, there are two condensin complexes, which function on different scales (condensin I: shorter loops, condensin II: longer loops). Together, condensin I and II compact chromosomes through loop extrusion, forming nested loop structures. b Interphase structure of chromosomes in yeast (i) and metazoans (ii). (i) In yeast, condensin compacts chromosomes through loop extrusion and aids clustering of tRNAs and transposons by promoting long-range interactions. (ii) In metazoans, condensin compacts chromosomes through loop extrusion, clusters transposons and enhancer–promoter pairs by promoting long-range interactions, and aids the formation of TADs
The role of condensins in interphase genome organization
In addition to compacting the chromosomes in preparation for segregation, condensin function is required for interphase structures (Albritton and Ercan 2018; Yuen and Gerton 2018). In Drosophila melanogaster, condensin II promotes axial compaction of chromosomes to form chromosomal territories (Bauer et al. 2012; Smith et al. 2013; George et al. 2014; Rosin et al. 2018). In Caenorhabditis elegans, an X-chromosome specific condensin (condensin DC) is responsible for a ~ 40% reduction in the volume of the X chromosomes compared to the autosomes (Lau et al. 2014). In this context, condensin DC mediates shorter scale chromosomal interactions as well as specific long-range interactions on the X (Crane et al. 2015). Condensin depletion also caused decompaction of interphase nuclei in mouse embryonic stem cells (Fig. 1bi, ii) (Fazzio and Panning 2010).
Interphase organization of the eukaryotic nucleus involves chromatin fibers that fold into nested interaction domains termed topologically associated domains (TADs) (Sexton et al. 2012; Rao et al. 2014). It is hypothesized that TADs function to restrict the search space for enhancer–promoter interactions, allowing coherent and correct expression of genes (Lupianez et al. 2015). Establishment of TADs has primarily been linked to the function of cohesin, another SMC complex, but condensin has recently emerged as additional TAD regulator (Yuen and Gerton 2018). Insulator proteins, such as CTCF, block SMC complex movement to delimit TAD boundaries (Fudenberg et al. 2016). Condensins colocalize with cohesin at the CTCF sites found at the strongest TAD boundaries in mouse (Van Bortle et al. 2014), and were shown to regulate TAD organization in D. melanogaster and C. elegans (Fig. 1bii) (Van Bortle et al. 2014; Crane et al. 2015; Li et al. 2015a). However, in differentiated mouse hepatocytes condensin II depletion did not affect TADs, suggesting that condensins may be involved in establishment rather than maintenance of genome organization during interphase (Abdennur et al. 2018). Developing an understanding of TAD organization and the role of condensin in this process is an important next step in the field.
The role of condensins in regulating rDNA structure and organization
In addition to general roles in chromosome organization, condensins also play important roles in the organization of specific genomic elements. One of the best-understood examples in this context is the function of condensin in regulating compaction and segregation of the ribosomal DNA (rDNA). The rDNA is one of the most difficult regions of the genome to segregate during cell division (Freeman et al. 2000; D’Ambrosio et al. 2008a). The segregation problems are likely due to the high level of repetition, active transcription, and late replication (Bhalla et al. 2002; D’Ambrosio et al. 2008a). In S. cerevisiae, where most of these analyses were conducted, the rDNA is segregated by gradual extension and unzipping of the rDNA repeats followed by axial compaction (Machín et al. 2005; D’Ambrosio et al. 2008a). This multi-step process is essential, as triggering premature rDNA compaction delays segregation (de los Santos-Velázquez et al. 2017).
In addition to helping compact the rDNA array, condensin also mediates a specific long-range interaction between the rDNA and the centromere of the rDNA-carrying chromosome (Paul et al. 2018). The centromere-rDNA interaction peaks during anaphase, matching the timing of segregation (Lazar-Stefanita et al. 2017). rDNA unzipping during segregation initiates on the centromere-proximal side (Machín et al. 2005), where condensin binding is enriched (Paul et al. 2018), raising the possibility that condensin-mediated proximity of the rDNA to the centromere may play a role in rDNA segregation (Fig. 1ai). This mechanism may involve a direct centromere tether to the rDNA that helps facilitate segregation. Alternatively, the enrichment of condensin between the centromere and the rDNA may either allow more efficient decoupling of sister rDNA arrays or may help protect the DNA between the rDNA and centromere from the high tension generated by the pulling of the spindle against the entangled rDNA.
In S. cerevisiae, condensin is also essential for regulating the rDNA upon nutrient starvation (Tsang et al. 2007a; Xue and Acar 2018). rDNA compaction in response to both starvation and segregation, is accompanied by the exodus of RNA polymerase I from the nucleolus (Tsang et al. 2003; Machín et al. 2006). Thus, active transcription at the rDNA appears to have an antagonistic relationship with condensin, which is opposite to what is found in the rest of the genome (Robellet et al. 2017). There is a modest increase in interaction between the rDNA and the centromere proximal side of the rDNA during starvation, indicating that chromosome segregation and quiescence trigger a similar pattern of condensin-dependent compaction (Swygert et al. 2018). However, the mechanism of condensin regulation likely differs between the two processes. Condensin recruitment during segregation requires the replication fork barrier protein Fob1 (Johzuka et al. 2006) and inactivation of rDNA transcription by the cell-cycle phosphatase Cdc14 (D’Amours et al. 2004; Wang et al. 2006; Machín et al. 2006; Dulev et al. 2008; Clemente-Blanco et al. 2009; Matos-Perdomo and Machín 2018), whereas recruitment during starvation is initiated by Rpd3-dependent histone deacetylation (Tsang et al. 2007a). Condensin binding to the rDNA helps maintain rDNA stability during starvation stress (Tsang et al. 2007b; Tsang and Zheng 2009), possibly in part by helping establish a position-dependent structure within the rDNA, that limits activity of RNA polymerase II to a few border repeats and allows protection of the silent majority of the rDNA (Wang et al. 2016).
rDNA is fundamental to cell physiology, and the few available studies in other organisms suggest a conservation for condensin function in rDNA regulation. In human cells, condensin knockdown resulted in increased rDNA expression, suggesting the antagonistic relationship between rDNA transcription and condensin may be conserved (Huang et al. 2013). In chicken cells, condensin I is enriched at the rDNA, and depletion of condensin I results in rDNA decompaction (Zhang et al. 2016). During meiosis, condensin contributes to rDNA compaction in A. thaliana and prevents crossovers at the rDNA by suppressing programmed double strand breaks in S. cerevisiae (Li et al. 2014; Smith et al. 2014). Similarly, the human rDNA is unstable in the absence of condensin (Samoshkin et al. 2012).
Condensin-mediated clustering of specific genomic loci including tRNA genes
Condensin also drives specific interactions between distant loci. Condensin is enriched at the tRNA genes in many organisms, including S. cerevisiae, S. pombe, D. melanogaster, C. elegans, mouse and human (D’Ambrosio et al. 2008b; Kranz et al. 2013; Van Bortle et al. 2014; Iwasaki et al. 2015; Yuen et al. 2017). In S. pombe, the TATA-binding protein TBP recruits condensin to tRNA genes, while in S. cerevisiae, the TFIIIC complex was shown to be capable of recruiting condensin (D’Ambrosio et al. 2008b; Iwasaki et al. 2015). Furthermore, condensin binds to RNA polymerase III specific subunits including TFIIIB, TFIIIC, RPC82, RPC25 (Haeusler et al. 2008; Iwasaki et al. 2010). In yeasts, condensin is required to cluster tRNA genes (Haeusler et al. 2008; Iwasaki et al. 2010; Paul et al. 2018). Interestingly, in S. cerevisiae this clustering is specific for a family of tRNA genes, suggesting a possible regulatory function (Paul et al. 2018). It is important to note that the tRNA genes clustered by condensins are located on different chromosomes. Thus, a model of loop extrusion is insufficient to explain these trans interactions. Supporting the ability of condensins to make trans connections (Fig. 1Bi), S. cerevisiae condensin was shown to be able to hold two separate DNA pieces together in vitro (Terakawa et al. 2017), and interactions between different condensin molecules were needed in a computational model for the formation of the mitotic chromosome (Sakai et al. 2018).
It is possible that condensin-mediated clustering establishes structures much like ‘transcription factories’, in which co-regulated genes are clustered together to allow enrichment of regulators in one place (Branco and Pombo 2006; Du and Bai 2017). In S. pombe, reduction in tRNA gene clustering resulted in increased transcription (Iwasaki et al. 2010). Given the fact that RNA polymerase III recruits condensin, this would suggest that there is some form of negative feedback in the system. In S. cerevisiae, tRNA genes that clustered in a condensin dependent manner were bound by RNA polymerase III at a slightly higher level, suggesting a positive effect for condensin in their transcription (Paul et al. 2018). Condensin effects on clustering are not limited to interactions among tRNA genes. tRNA genes also cluster with other genomic features, including the rDNA in S. cerevisiae, and centromeres and cell-cycle regulated genes in S. pombe (Haeusler et al. 2008; Kim et al. 2016; Paul et al. 2018). Moreover, condensins also drive the association of other dispersed genomic loci. One example is centromere clustering, which is required for the repression of retrotransposons in S. pombe (Fig. 1bi) (Tanaka et al. 2012). Indeed, this function may be conserved in humans, Arabidopsis thaliana and D. melanogaster, where condensin was required to repress transposon expression (Fig. 1bii) (Wang et al. 2011, 2017a; Schuster et al. 2013). Together, these observations imply that condensin not only acts by loop-extrusion but also provides an important mechanism to selectively regulate chromatin interactions in trans.
Condensin function in gene regulation
A clear paradigm of condensin-mediated gene regulation is the C. elegans condensin DC complex, which represses both X chromosomes by a factor of two in XX hermaphrodites to equalize X chromosomal transcript levels to that of XO males. The mechanisms by which condensin DC regulates transcription has been reviewed recently (Albritton and Ercan 2018). Briefly, condensin DC specifically binds to promoters in a gene-activity dependent manner and, in turn, reduces RNA polymerase II binding to promoters, resulting in transcriptional repression. Condensin DC shares four subunits with canonical condensin I and differs from it by a single SMC4 subunit that duplicated and diverged in Caenorhabditis (Csankovszki et al. 2009). Evolution of SMC subunits for new functions is also evident in mammals, where an SMC variant named SMCHD1, which makes homodimers like prokaryotic condensins (Brideau et al. 2015), is involved in X inactivation by regulating chromosomal interactions on the inactivated X (Nozawa et al. 2013; Chen et al. 2015; Wang et al. 2018a; Jansz et al. 2018) and in Hox clusters (Jansz et al. 2018). A recent study identified an SMC like protein, SMCL1 that regulates condensin binding in C. elegans (Chao et al. 2017). Therefore, future work should focus on the evolution of SMC variants in regulating canonical SMC complex functions.
Whether and how canonical condensins directly or indirectly regulate transcription remains unclear. Two recent analyses in S. cerevisiae showed that yeast condensin does not directly regulate transcription (Paul et al. 2018; Hocquet et al. 2018). Similarly, condensin II depletion in post-mitotic mouse hepatocyte cells and condensin II mutations in T cell leukemia did not affect mRNA expression in mouse (Woodward et al. 2016; Abdennur et al. 2018). In yeast, chronic perturbation of condensin resulted in increased mRNA levels (Swygert et al. 2018; Paul et al. 2018). Some of these effects may be indirect, because in S. pombe and S. cerevisiae, increased mRNA levels upon condensin depletion were linked to chromosome segregation defects of the rDNA. These defects were linked to missegregation of the RNA exosome, leading to mRNA increase in the daughter cells (Hocquet et al. 2018). Therefore, canonical condensin function in gene regulation may be coupled to its role in cell division, such that condensin-mediated assembly of mitotic chromosome structure may regulate establishment of genome organization and gene expression patterns during interphase.
Condensin action on chromosomes and transcription
Condensin binding is enriched at the promoters and gene regulatory regions in C. elegans (Kranz et al. 2013), D. melanogaster (Wallace et al. 2015), chicken (Kim et al. 2013), and mouse (Yuen et al. 2017), and condensin knockdown and mutations have been shown to affect gene expression in several of these organisms (Longworth et al. 2012; Kranz et al. 2013; Lau and Csankovszki 2015; Schuster et al. 2015; Li et al. 2015b; Swygert et al. 2018). However, the link between widespread enrichment of canonical condensins at gene regulatory sites and transcription remains unclear. Similar to cohesin, condensin was implicated in specific long-range interaction between several enhancers and promoters in human cell lines to promote gene expression (Fig. 1bii) (Dowen et al. 2013; Li et al. 2015b). Interestingly, some of these enhancers use phase separation to mediate gene regulation (Sabari et al. 2018). One of the first nuclear bodies that was shown to undergo phase-separation was the nucleolus, a focus of condensin enrichment (Feric et al. 2016; Sawyer et al. 2018). It is conceivable that condensin binding affects establishment of liquid phase dynamics of the nucleolus and enhancer–promoter interactions.
A highly specific function of condensins in regulating chromosomal interactions for gene regulation occurs during transvection in D. melanogaster. Here, sister chromatids remain paired during interphase, allowing enhancers to act in trans to regulate promoters (Mellert and Truman 2012). Condensin antagonizes transvection, presumably by creating discrete chromosomes through loop extrusion as in mitosis (Hartl et al. 2008; Smith et al. 2013). A similar observation was made in mouse neural stem cells, where condensin II antagonizes pericentric heterochromatin clustering at chromocenters (Nishide and Hirano 2014). The involvement of condensin in both transcription activation through promoter-enhancer interactions and repression through antagonizing transvection suggests that the activity of condensins may result in different outcomes depending on the mechanism of gene regulation.
In addition to regulating chromosomal interactions, the action of condensin motors, with a ~ 50 nm ring sliding along the chromatin fiber, may create conflict with large transcription complexes (Terakawa et al. 2017). During mitosis, when condensin activity is at its highest level, transcribing RNA polymerase II complexes are removed from chromatin (Martínez-Balbás et al. 1995; Segil et al. 1996; Gottesfeld and Forbes 1997; Ginno et al. 2018), and mitotic bookmarking requires dissociation of condensin from specific loci (Xing et al. 2005, 2008). In addition to condensin ring sliding, introduction of positive supercoils to mitotic DNA in the presence of topoisomerase II (Hirano et al. 1997; Hagstrom et al. 2002; Stray and Lindsley 2003) or reannealing of single stranded DNA may help condensins repress transcription of mitotic chromosomes (Sutani et al. 2015). In S. pombe, inhibition of transcription partly suppressed condensin mutant phenotypes in cell division (Sutani et al. 2015), suggesting that condensin-mediated transcriptional silencing of mitotic chromosomes may be important for chromosome segregation.
Indirect effects of condensins on gene regulation
Beyond condensin’s direct action on DNA, condensin may regulate transcription through histone modifications. In D. melanogaster, the condensin II subunit Barren interacts with polycomb proteins to repress homeotic gene expression (Lupo et al. 2001). Likewise in C. elegans, condensin DC interacts with a H4K20me2 demethylase, DPY-21 (Brejc et al. 2017). The demethylation product is H4K20me1, which is enriched on the X chromosome and contributes to dosage compensation (Wells et al. 2012; Vielle et al. 2012; Kramer et al. 2015; Bian et al. 2017). In A. thaliana condensin meditates silencing of specific transposon families in conjunction with CG methylation by MET1, CHG methylation by CMT3, the chromatin remodeler DDM1 and H3K27 monomethylation (Wang et al. 2017a). In D. melanogaster, retrotransposon expression is blocked by condensin II mediated regulation of H3K9me3 localization (Schuster et al. 2013). Thus, it is possible that condensin also serves as a binding platform to recruit additional regulatory activities for controlling gene expression.
Finally, condensin II was reported to bind LINE-1 RNAs with Gamma-Interferon Activated Inhibitor of Translation (GAIT) in human cells (Ward et al. 2017). Together these protein complexes repressed translation of LINE-1 RNA by stopping the formation of the translation initiation complex. Additional examples of condensin interaction with RNA have yet to be found, but condensin has high affinity for RNAs in vitro (Akai et al. 2011).
Summary: condensins are integrated into fundamental chromosomal functions
Condensins were identified, because they drive the compaction of chromosome into discrete bodies that are easy to segregate (Hirano et al. 1986; Saka et al. 1994). In the absence of condensin complexes, cells fail to segregate chromosomes faithfully and daughter cells die. As a result, condensins are essential for proper growth and division in all domains of life (Hirano 2016). How then do hypomorphic mutations in condensins trigger tissue-specific problems in cell divisions leading to T-cell lymphoma or microcephaly (Martin et al. 2016; Woodward et al. 2016)? It is possible that the myriad of additional condensin functions that link cell division to cell physiology (e.g., rDNA, RNA exosome segregation, regulation of TADs or tRNA clustering) are in play. Therefore, future insights into interaction of condensins with basic nuclear processes will be important to understand condensin function and phenotypes in disease.
References
Abdennur N, Schwarzer W, Pekowska A et al (2018) Condensin II inactivation in interphase does not affect chromatin folding or gene expression. bioRxiv. https://doi.org/10.1101/437459
Akai Y, Kurokawa Y, Nakazawa N et al (2011) Opposing role of condensin hinge against replication protein A in mitosis and interphase through promoting DNA annealing. Open Biol 1:110023. https://doi.org/10.1098/rsob.110023
Albritton SE, Ercan S (2018) Caenorhabditis elegans dosage compensation: insights into condensin-mediated gene regulation. Trends Genet 34:41–53. https://doi.org/10.1016/j.tig.2017.09.010
Bauer CR, Hartl TA, Bosco G (2012) Condensin II promotes the formation of chromosome territories by inducing axial compaction of polyploid interphase chromosomes. PLoS Genet 8:e1002873. https://doi.org/10.1371/journal.pgen.1002873
Bhalla N, Biggins S, Murray A (2002) Mutation of YCS4, a budding yeast condensin subunit, affects mitotic and nonmitotic chromosome behavior. Mol Biol Cell 13:632–645. https://doi.org/10.1091/MBC.01-05-0264
Bian Q, Anderson EC, Brejc K, Meyer BJ (2017) Dynamic control of chromosome topology and gene expression by a chromatin modification. Cold Spring Harb Symp Quant Biol 82:279–291. https://doi.org/10.1101/sqb.2017.82.034439
Branco MR, Pombo A (2006) Intermingling of chromosome territories in interphase suggests role in translocations and transcription-dependent associations. PLoS Biol 4:e138. https://doi.org/10.1371/journal.pbio.0040138
Brejc K, Bian Q, Uzawa S et al (2017) Dynamic control of X chromosome conformation and repression by a histone H4K20 demethylase. Cell 171:85–102.e23. https://doi.org/10.1016/j.cell.2017.07.041
Brideau NJ, Coker H, Gendrel A-V et al (2015) Independent mechanisms target SMCHD1 to trimethylated histone H3 lysine 9-modified chromatin and the inactive X chromosome. Mol Cell Biol 35:4053. https://doi.org/10.1128/MCB.00432-15
Chan RC, Severson AF, Meyer BJ (2004) Condensin restructures chromosomes in preparation for meiotic divisions. J Cell Biol 167:613–625. https://doi.org/10.1083/jcb.200408061
Chao LF-I, Singh M, Thompson J et al (2017) An SMC-like protein binds and regulates Caenorhabditis elegans condensins. PLoS Genet 13:e1006614. https://doi.org/10.1371/journal.pgen.1006614
Chen K, Hu J, Moore DL et al (2015) Genome-wide binding and mechanistic analyses of Smchd1-mediated epigenetic regulation. Proc Natl Acad Sci USA 112:E3535–E3544. https://doi.org/10.1073/pnas.1504232112
Clemente-Blanco A, Mayán-Santos M, Schneider DA et al (2009) Cdc14 inhibits transcription by RNA polymerase I during anaphase. Nature 458:219–222. https://doi.org/10.1038/nature07652
Crane E, Bian Q, McCord RP et al (2015) Condensin-driven remodelling of X chromosome topology during dosage compensation. Nature 523:240–244. https://doi.org/10.1038/nature14450
Csankovszki G, Collette K, Spahl K et al (2009) Three distinct condensin complexes control C. elegans chromosome dynamics. Curr Biol 19:9–19. https://doi.org/10.1016/j.cub.2008.12.006
Cuylen S, Haering CH (2011) Deciphering condensin action during chromosome segregation. Trends Cell Biol 21:552–559. https://doi.org/10.1016/j.tcb.2011.06.003
Cuylen S, Metz J, Haering CH (2011) Condensin structures chromosomal DNA through topological links. Nat Struct Mol Biol 18:894–901. https://doi.org/10.1038/nsmb.2087
D’Ambrosio C, Kelly G, Shirahige K, Uhlmann F (2008a) Condensin-dependent rDNA decatenation introduces a temporal pattern to chromosome segregation. Curr Biol 18:1084–1089. https://doi.org/10.1016/j.cub.2008.06.058
D’Ambrosio C, Schmidt CK, Katou Y et al (2008b) Identification of cis-acting sites for condensin loading onto budding yeast chromosomes. Genes Dev 22:2215–2227. https://doi.org/10.1101/gad.1675708
D’Amours D, Stegmeier F, Amon A (2004) Cdc14 and condensin control the dissolution of cohesin-independent chromosome linkages at repeated DNA. Cell 117:455–469. https://doi.org/10.1016/S0092-8674(04)00413-1
de los Santos-Velázquez AI, de Oya IG, Manzano-López J, Monje-Casas F (2017) Late rDNA condensation ensures timely Cdc14 release and coordination of mitotic exit signaling with nucleolar segregation. Curr Biol 27:3248–3263.e5. https://doi.org/10.1016/J.CUB.2017.09.028
Dowen JM, Bilodeau S, Orlando DA et al (2013) Multiple structural maintenance of chromosome complexes at transcriptional regulatory elements. Stem Cell Reports 1:371–378. https://doi.org/10.1016/j.stemcr.2013.09.002
Du M, Bai L (2017) 3D clustering of co-regulated genes and its effect on gene expression. Curr Genet 63:1017–1021. https://doi.org/10.1007/s00294-017-0712-9
Dulev S, Aragon L, Strunnikov A (2008) Unreplicated DNA in mitosis precludes condensin binding and chromosome condensation in S. cerevisiae. Front Biosci 13:5838–5846
Fazzio TG, Panning B (2010) Condensin complexes regulate mitotic progression and interphase chromatin structure in embryonic stem cells. J Cell Biol 188:491–503. https://doi.org/10.1083/jcb.200908026
Feric M, Vaidya N, Harmon TS et al (2016) Coexisting liquid phases underlie nucleolar subcompartments. Cell 165:1686–1697. https://doi.org/10.1016/J.CELL.2016.04.047
Freeman L, Aragon-Alcaide L, Strunnikov A (2000) The condensin complex governs chromosome condensation and mitotic transmission of rDNA. J Cell Biol 149:811–824
Fudenberg G, Imakaev M, Lu C et al (2016) Formation of chromosomal domains by loop extrusion. Cell Rep 15:2038–2049. https://doi.org/10.1016/j.celrep.2016.04.085
Ganji M, Shaltiel IA, Bisht S et al (2018) Real-time imaging of DNA loop extrusion by condensin. Science. https://doi.org/10.1126/science.aar7831
George CM, Bozler J, Nguyen HQ, Bosco G (2014) Condensins are required for maintenance of nuclear architecture. Cells 3:865–882. https://doi.org/10.3390/cells3030865
Gibcus JH, Samejima K, Goloborodko A et al (2018) A pathway for mitotic chromosome formation. Science. https://doi.org/10.1126/science.aao6135
Ginno PA, Burger L, Seebacher J et al (2018) Cell cycle-resolved chromatin proteomics reveals the extent of mitotic preservation of the genomic regulatory landscape. Nat Commun 9:4048. https://doi.org/10.1038/s41467-018-06007-5
Goloborodko A, Imakaev MV, Marko JF, Mirny L (2016a) Compaction and segregation of sister chromatids via active loop extrusion. Elife. https://doi.org/10.7554/eLife.14864
Goloborodko A, Marko JF, Mirny LA (2016b) Chromosome compaction by active loop extrusion. Biophys J 110:2162–2168. https://doi.org/10.1016/j.bpj.2016.02.041
Gottesfeld JM, Forbes DJ (1997) Mitotic repression of the transcriptional machinery. Trends Biochem Sci 22:197–202. https://doi.org/10.1016/S0968-0004(97)01045-1
Green LC, Kalitsis P, Chang TM et al (2012) Contrasting roles of condensin I and condensin II in mitotic chromosome formation. J Cell Sci 125:1591–1604. https://doi.org/10.1242/jcs.097790
Haeusler RA, Pratt-Hyatt M, Good PD et al (2008) Clustering of yeast tRNA genes is mediated by specific association of condensin with tRNA gene transcription complexes. Genes Dev 22:2204–2214. https://doi.org/10.1101/gad.1675908
Hagstrom KA, Holmes VF, Cozzarelli NR, Meyer BJ (2002) C. elegans condensin promotes mitotic chromosome architecture, centromere organization, and sister chromatid segregation during mitosis and meiosis. Genes Dev 16:729–742. https://doi.org/10.1101/gad.968302
Hartl TA, Smith HF, Bosco G (2008) Chromosome alignment and transvection are antagonized by condensin II. Science 322:1384–1387. https://doi.org/10.1126/science.1164216
Hirano T (2016) Condensin-based chromosome organization from bacteria to vertebrates. Cell 164:847–857. https://doi.org/10.1016/j.cell.2016.01.033
Hirano T, Funahashi S, Uemura T, Yanagida M (1986) Isolation and characterization of Schizosaccharomyces pombe cutmutants that block nuclear division but not cytokinesis. EMBO J 5:2973–2979
Hirano T, Kobayashi R, Hirano M (1997) Condensins, chromosome condensation protein complexes containing XCAP-C, XCAP-E and a Xenopus homolog of the Drosophila Barren protein. Cell 89:511–521. https://doi.org/10.1016/S0092-8674(00)80233-0
Hirota T, Gerlich D, Koch B et al (2004) Distinct functions of condensin I and II in mitotic chromosome assembly. J Cell Sci 117:6435–6445. https://doi.org/10.1242/jcs.01604
Hocquet C, Robellet X, Modolo L et al (2018) Condensin controls cellular RNA levels through the accurate segregation of chromosomes instead of directly regulating transcription. Elife. https://doi.org/10.7554/eLife.38517
Houlard M, Godwin J, Metson J et al (2015) Condensin confers the longitudinal rigidity of chromosomes. Nat Cell Biol 17:771–781. https://doi.org/10.1038/ncb3167
Huang K, Jia J, Wu C et al (2013) Ribosomal RNA Gene transcription mediated by the master genome regulator protein CCCTC-binding factor (CTCF) is negatively regulated by the condensin complex. J Biol Chem 288:26067–26077. https://doi.org/10.1074/jbc.M113.486175
Iwasaki O, Tanaka A, Tanizawa H et al (2010) Centromeric localization of dispersed Pol III genes in fission yeast. Mol Biol Cell 21:254–265. https://doi.org/10.1091/mbc.E09-09-0790
Iwasaki O, Tanizawa H, Kim K-D et al (2015) Interaction between TBP and condensin drives the organization and faithful segregation of mitotic chromosomes. Mol Cell 59:755–767. https://doi.org/10.1016/j.molcel.2015.07.007
Jansz N, Keniry A, Trussart M et al (2018) Smchd1 regulates long-range chromatin interactions on the inactive X chromosome and at Hox clusters. Nat Struct Mol Biol 25:766–777. https://doi.org/10.1038/s41594-018-0111-z
Johzuka K, Terasawa M, Ogawa H et al (2006) Condensin loaded onto the replication fork barrier site in the rRNA gene repeats during S phase in a FOB1-dependent fashion to prevent contraction of a long repetitive array in Saccharomyces cerevisiae. Mol Cell Biol 26:2226–2236. https://doi.org/10.1128/MCB.26.6.2226-2236.2006
Kakui Y, Uhlmann F (2018) SMC complexes orchestrate the mitotic chromatin interaction landscape. Curr Genet 64:335–339. https://doi.org/10.1007/s00294-017-0755-y
Kakui Y, Rabinowitz A, Barry DJ, Uhlmann F (2017) Condensin-mediated remodeling of the mitotic chromatin landscape in fission yeast. Nat Genet 49:1553–1557. https://doi.org/10.1038/ng.3938
Kim JH, Zhang T, Wong NC et al (2013) Condensin I associates with structural and gene regulatory regions in vertebrate chromosomes. Nat Commun 4:2537. https://doi.org/10.1038/ncomms3537
Kim KD, Tanizawa H, Iwasaki O, Noma K (2016) Transcription factors mediate condensin recruitment and global chromosomal organization in fission yeast. Nat Genet 48:1242–1252. https://doi.org/10.1038/ng.3647
Kinoshita K, Hirano T (2017) Dynamic organization of mitotic chromosomes. Curr Opin Cell Biol 46:46–53. https://doi.org/10.1016/j.ceb.2017.01.006
Kramer M, Kranz AL, Su A et al (2015) Developmental dynamics of X-Chromosome dosage compensation by the DCC and H4K20me1 in C. elegans. PLoS Genet 11:e1005698. https://doi.org/10.1371/journal.pgen.1005698
Kranz AL, Jiao CY, Winterkorn LH et al (2013) Genome-wide analysis of condensin binding in Caenorhabditis elegans. Genome Biol 14:R112. https://doi.org/10.1186/gb-2013-14-10-r112
Lau AC, Csankovszki G (2015) Balancing up and downregulation of the C. elegans X chromosomes. Curr Opin Genet Dev 31:50–56. https://doi.org/10.1016/j.gde.2015.04.001
Lau AC, Nabeshima K, Csankovszki G (2014) The C. elegans dosage compensation complex mediates interphase X chromosome compaction. Epigenetics Chromatin 7:31. https://doi.org/10.1186/1756-8935-7-31
Lazar-Stefanita L, Scolari VF, Mercy G et al (2017) Cohesins and condensins orchestrate the 4D dynamics of yeast chromosomes during the cell cycle. EMBO J. https://doi.org/10.15252/embj.201797342
Li P, Jin H, Yu H-GG (2014) Condensin suppresses recombination and regulates double-strand break processing at the repetitive ribosomal DNA array to ensure proper chromosome segregation during meiosis in budding yeast. Mol Biol Cell 25:2934–2947. https://doi.org/10.1091/mbc.E14-05-0957
Li L, Lyu X, Hou C et al (2015a) Widespread rearrangement of 3D chromatin organization underlies polycomb-mediated stress-induced silencing. Mol Cell 58:216–231. https://doi.org/10.1016/j.molcel.2015.02.023
Li W, Hu Y, Oh S et al (2015b) Condensin I and II complexes license full estrogen receptor α-dependent enhancer activation. Mol Cell 59:188–202. https://doi.org/10.1016/j.molcel.2015.06.002
Longworth MS, Walker JA, Anderssen E et al (2012) A Shared Role for RBF1 and dCAP-D3 in the regulation of transcription with consequences for innate immunity. PLoS Genet 8:e1002618. https://doi.org/10.1371/journal.pgen.1002618
Lupianez DG, Kraft K, Heinrich V et al (2015) Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell 161:1012–1025. https://doi.org/10.1016/j.cell.2015.04.004
Lupo R, Breiling A, Bianchi ME, Orlando V (2001) Drosophila chromosome condensation proteins topoisomerase II and Barren colocalize with Polycomb and maintain Fab-7 PRE silencing. Mol Cell 7:127–136. https://doi.org/10.1016/S1097-2765(01)00161-7
Machín F, Torres-Rosell J, Jarmuz A, Aragón L (2005) Spindle-independent condensation-mediated segregation of yeast ribosomal DNA in late anaphase. J Cell Biol 168:209. https://doi.org/10.1083/JCB.200408087
Machín F, Torres-Rosell J, Piccoli G, De et al (2006) Transcription of ribosomal genes can cause nondisjunction. J Cell Biol 173:893–903. https://doi.org/10.1083/JCB.200511129
Martin C-A, Murray JE, Carroll P et al (2016) Mutations in genes encoding condensin complex proteins cause microcephaly through decatenation failure at mitosis. Genes Dev 30:2158–2172. https://doi.org/10.1101/gad.286351.116
Martínez-Balbás MA, Dey A, Rabindran SK et al (1995) Displacement of sequence-specific transcription factors from mitotic chromatin. Cell 83:29–38. https://doi.org/10.1016/0092-8674(95)90231-7
Matos-Perdomo E, Machín F (2018) The ribosomal DNA metaphase loop of Saccharomyces cerevisiae gets condensed upon heat stress in a Cdc14-independent TORC1-dependent manner. Cell Cycle 17:200. https://doi.org/10.1080/15384101.2017.1407890
Mellert DJ, Truman JW (2012) Transvection is common throughout the drosophila genome. Genetics 191:1129–1141. https://doi.org/10.1534/genetics.112.140475
Nasmyth K (2001) Disseminating the genome: joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu Rev Genet 35:673–745. https://doi.org/10.1146/annurev.genet.35.102401.091334
Nishide K, Hirano T (2014) Overlapping and non-overlapping functions of condensins I and II in neural stem cell divisions. PLoS Genet 10:e1004847. https://doi.org/10.1371/journal.pgen.1004847
Nozawa R-S, Nagao K, Igami K-T et al (2013) Human inactive X chromosome is compacted through a PRC2-independent SMCHD1-HBiX1 pathway. Nat Struct Mol Biol 20:566–573. https://doi.org/10.1038/nsmb.2532
Paul MR, Markowitz TE, Hochwagen A, Ercan S (2018) Condensin depletion causes genome decompaction without altering the level of global gene expression in saccharomyces cerevisiae. Genetics 210(1):331–344. https://doi.org/10.1534/genetics.118.301217
Rao SS, Huntley MH, Durand NC et al (2014) A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159:1665–1680. https://doi.org/10.1016/j.cell.2014.11.021
Robellet X, Vanoosthuyse V, Bernard P (2017) The loading of condensin in the context of chromatin. Curr Genet 63:577–589. https://doi.org/10.1007/s00294-016-0669-0
Rosin LF, Nguyen SC, Joyce EF (2018) Condensin II drives large-scale folding and spatial partitioning of interphase chromosomes in Drosophila nuclei. PLoS Genet 14:e1007393. https://doi.org/10.1371/journal.pgen.1007393
Sabari BR, Dall’Agnese A, Boija A et al (2018) Coactivator condensation at super-enhancers links phase separation and gene control. Science 361:eaar3958. https://doi.org/10.1126/science.aar3958
Saka Y, Sutani T, Yamashita Y et al (1994) Fission yeast cut3 and cut14, members of a ubiquitous protein family, are required for chromosome condensation and segregation in mitosis. EMBO J 13:4938–4952. https://doi.org/10.1002/j.1460-2075.1994.tb06821.x
Sakai Y, Mochizuki A, Kinoshita K et al (2018) Modeling the functions of condensin in chromosome shaping and segregation. PLOS Comput Biol 14:e1006152. https://doi.org/10.1371/journal.pcbi.1006152
Samoshkin A, Dulev S, Loukinov D et al (2012) Condensin dysfunction in human cells induces nonrandom chromosomal breaks in anaphase, with distinct patterns for both unique and repeated genomic regions. Chromosoma 121:191–199. https://doi.org/10.1007/s00412-011-0353-6
Sawyer IA, Bartek J, Dundr M (2018) Phase separated microenvironments inside the cell nucleus are linked to disease and regulate epigenetic state, transcription and RNA processing. Semin Cell Dev Biol. https://doi.org/10.1016/j.semcdb.2018.07.001
Schalbetter SA, Goloborodko A, Fudenberg G et al (2017) SMC complexes differentially compact mitotic chromosomes according to genomic context. Nat Cell Biol. https://doi.org/10.1038/ncb3594
Schuster AT, Sarvepalli K, Murphy EA, Longworth MS (2013) Condensin II subunit dCAP-D3 restricts retrotransposon mobilization in Drosophila somatic cells. PLoS Genet 9:e1003879. https://doi.org/10.1371/journal.pgen.1003879
Schuster AT, Homer CR, Kemp JR et al (2015) Chromosome-associated protein D3 promotes bacterial clearance in human intestinal epithelial cells by repressing expression of amino acid transporters. Gastroenterology 148:1405–1416.e3. https://doi.org/10.1053/j.gastro.2015.02.013
Segil N, Guermah M, Hoffmann A et al (1996) Mitotic regulation of TFIID: inhibition of activator-dependent transcription and changes in subcellular localization. Genes Dev 10:2389–2400. https://doi.org/10.1101/GAD.10.19.2389
Sexton T, Yaffe E, Kenigsberg E et al (2012) Three-dimensional folding and functional organization principles of the Drosophila genome. Cell 148:458–472. https://doi.org/10.1016/j.cell.2012.01.010
Smith HF, Roberts MA, Nguyen HQ et al (2013) Maintenance of interphase chromosome compaction and homolog pairing in drosophila is regulated by the condensin Cap-H2 and its partner Mrg15. Genetics 195(1):127–146. https://doi.org/10.1534/genetics.113.153544
Smith SJ, Osman K, Franklin FCH (2014) The condensin complexes play distinct roles to ensure normal chromosome morphogenesis during meiotic division in Arabidopsis. Plant J 80:255–268. https://doi.org/10.1111/tpj.12628
Stray JE, Lindsley JE (2003) Biochemical analysis of the yeast condensin Smc2/4 complex: an ATPase that promotes knotting of circular DNA. J Biol Chem 278:26238–26248. https://doi.org/10.1074/jbc.M302699200
Sutani T, Sakata T, Nakato R et al (2015) Condensin targets and reduces unwound DNA structures associated with transcription in mitotic chromosome condensation. Nat Commun 6:7815. https://doi.org/10.1038/ncomms8815
Swygert SG, Kim S, Wu X et al (2018) Condensin-dependent chromatin condensation represses transcription globally during quiescence. bioRxiv. https://doi.org/10.1101/320895
Tanaka A, Tanizawa H, Sriswasdi S et al (2012) Epigenetic regulation of condensin-mediated genome organization during the cell cycle and upon DNA damage through histone H3 lysine 56 acetylation. Mol Cell 48:532–546. https://doi.org/10.1016/j.molcel.2012.09.011
Terakawa T, Bisht S, Eeftens JM et al (2017) The condensin complex is a mechanochemical motor that translocates along DNA. Science 358:672–676. https://doi.org/10.1126/science.aan6516
Tsang CK, Zheng XFS (2009) Opposing role of condensin and radiation-sensitive gene RAD52 in ribosomal DNA stability regulation. J Biol Chem 284:21908–21919. https://doi.org/10.1074/jbc.M109.031302
Tsang C, Bertram P, Ai W et al (2003) Chromatin-mediated regulation of nucleolar structure and RNA Pol I localization by TOR. EMBO J 22:6045–6056. https://doi.org/10.1093/EMBOJ/CDG578
Tsang C, Li H, Zheng X (2007a) Nutrient starvation promotes condensin loading to maintain rDNA stability. EMBO J 26:448–458. https://doi.org/10.1038/SJ.EMBOJ.7601488
Tsang CK, Wei Y, Zheng XF (2007b) Compacting DNA during the interphase: condensin maintains rDNA integrity. Cell Cycle 6:2213–2218. https://doi.org/10.4161/cc.6.18.4733
van Ruiten MS, Rowland BD (2018) SMC complexes: universal DNA looping machines with distinct regulators. Trends Genet 34:477–487. https://doi.org/10.1016/j.tig.2018.03.003
Van Bortle K, Nichols MH, Li L et al (2014) Insulator function and topological domain border strength scale with architectural protein occupancy. Genome Biol 15:R82. https://doi.org/10.1186/gb-2014-15-5-r82
Vielle A, Lang J, Dong Y et al (2012) H4K20me1 contributes to downregulation of X-linked genes for C. elegans dosage compensation. PLoS Genet 8:e1002933. https://doi.org/10.1371/journal.pgen.1002933
Wallace HA, Klebba JE, Kusch T, et al (2015) Condensin II regulates interphase chromatin organization through the Mrg-binding motif of cap-H2. G3: Genes Genom Genet 5:803–817. https://doi.org/10.1534/g3.115.016634
Walther N, Hossain MJ, Politi AZ et al (2018) A quantitative map of human condensins provides new insights into mitotic chromosome architecture. J Cell Biol. https://doi.org/10.1083/jcb.201801048
Wang B-D, Butylin P, Strunnikov A (2006) Condensin function in mitotic nucleolar segregation is regulated by rDNA transcription. Cell Cycle 5:2260. https://doi.org/10.4161/CC.5.19.3292
Wang J, Geesman GJ, Hostikka SL et al (2011) Inhibition of activated pericentromeric SINE/Alu repeat transcription in senescent human adult stem cells reinstates self-renewal. Cell Cycle 10:3016–3030. https://doi.org/10.4161/cc.10.17.17543
Wang DN, Mansisidor A, Prabhakar G, Hochwagen A (2016) Condensin and Hmo1 mediate a starvation-induced transcriptional position effect within the ribosomal DNA array. Cell Rep 17:624. https://doi.org/10.1016/j.celrep.2016.09.057
Wang J, Blevins T, Podicheti R et al (2017a) Mutation of Arabidopsis SMC4 identifies condensin as a corepressor of pericentromeric transposons and conditionally expressed genes. Genes Dev 31:1601–1614. https://doi.org/10.1101/gad.301499.117
Wang X, Brandão HB, Le TBK et al (2017b) Bacillus subtilis SMC complexes juxtapose chromosome arms as they travel from origin to terminus. Science 355:524–527. https://doi.org/10.1126/science.aai8982
Wang C-Y, Jégu T, Chu H-P et al (2018a) SMCHD1 Merges chromosome compartments and assists formation of super-structures on the inactive X. Cell 174:406–421.e25. https://doi.org/10.1016/j.cell.2018.05.007
Wang X, Hughes AC, Brandão HB et al (2018b) In Vivo evidence for ATPase-dependent DNA translocation by the Bacillus subtilis SMC condensin complex. Mol Cell 71:841–847.e5. https://doi.org/10.1016/j.molcel.2018.07.006
Ward JR, Vasu K, Deutschman E et al (2017) Condensin II and GAIT complexes cooperate to restrict LINE-1 retrotransposition in epithelial cells. PLoS Genet 13:e1007051. https://doi.org/10.1371/journal.pgen.1007051
Wells MB, Snyder MJ, Custer LM, Csankovszki G (2012) Caenorhabditis elegans dosage compensation regulates histone H4 chromatin state on X chromosomes. Mol Cell Biol 32:1710–1719. https://doi.org/10.1128/MCB.06546-11
Woodward J, Taylor GC, Soares DC et al (2016) Condensin II mutation causes T-cell lymphoma through tissue-specific genome instability. Genes Dev 30:2173–2186. https://doi.org/10.1101/gad.284562.116
Xing H, Wilkerson DC, Mayhew CN et al (2005) Mechanism of hsp70i gene bookmarking. Science 307:421–423. https://doi.org/10.1126/science.1106478
Xing H, Vanderford NL, Sarge KD (2008) The TBP-PP2A mitotic complex bookmarks genes by preventing condensin action. Nat Cell Biol 10:1318–1323. https://doi.org/10.1038/ncb1790
Xue Y, Acar M (2018) Live-cell imaging of chromatin condensation dynamics by CRISPR. iScience 4:216–235. https://doi.org/10.1016/j.isci.2018.06.001
Yuen KC, Gerton JL (2018) Taking cohesin and condensin in context. PLoS Genet 14:e1007118. https://doi.org/10.1371/journal.pgen.1007118
Yuen KC, Slaughter BD, Gerton JL (2017) Condensin II is anchored by TFIIIC and H3K4me3 in the mammalian genome and supports the expression of active dense gene clusters. Sci Adv 3:e1700191. https://doi.org/10.1126/sciadv.1700191
Zhang T, Paulson JR, Bakhrebah M et al (2016) Condensin I and II behaviour in interphase nuclei and cells undergoing premature chromosome condensation. Chromosom Res 24:243–269. https://doi.org/10.1007/s10577-016-9519-7
Author information
Authors and Affiliations
Corresponding authors
Additional information
Communicated by M. Kupiec.
Rights and permissions
About this article
Cite this article
Paul, M.R., Hochwagen, A. & Ercan, S. Condensin action and compaction. Curr Genet 65, 407–415 (2019). https://doi.org/10.1007/s00294-018-0899-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00294-018-0899-4