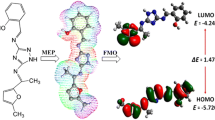
Abstract
The scientific interest in developing new complexes as inhibitors of bacterial biofilm related infections is constantly rising. The present work describes the chemical synthesis, structural and biological scrutiny of a triazole Schiff base ligand and its corresponding complexes. Triazole Schiff base, (2-methoxy-4-[(1H-1,2,4-triazol-3-ylimino)methyl]phenol) was synthesized from the condensation reaction of 3-amino-1,2,4-triazole and 4-hydroxy-3-methoxybenzaldehyde in an equimolar ratio. The triazole ligand (H2L) was characterized by physical (solubility, color, melting point), spectroscopic [UV–visible (UV–Vis), Fourier transform infrared spectroscopy (FT-IR), proton nuclear magnetic resonance (1H-NMR) and mass spectra (MS)] and micro analysis to evaluate their elemental composition. The bidentate ligand was complexed with transition metal [VO(IV), Fe(II), Co(II), Ni(II), Cu(II) and Zn(II)] in 1:2 molar ratio. The complexes were characterized by physical (color, solubility, decomposition temperature, conductance and magnetic moment), FT-IR, UV–Vis and elemental analysis. Thermal stability and fluorescence properties of the compounds were also determined. Density functional theory based theoretical calculations were accomplished to gain more insight into spectroscopic properties. The frontier molecular orbital analysis revealed that the ligand was less reactive with reduced electron donating capability and more kinetic stability than complexes. The as-synthesized compounds were scrutinized for anti-bacterial and anti-fungal activity against selected strains. Cobalt complex exhibited highest antibacterial activity against Escherichia coli and nickel complex has shown highest antifungal activity against Aspergillus niger. All the compounds also showed good antioxidant activity. The theoretical results reflect consistency with the experimental findings signifying that such compounds could be the promising chemical scaffolds in the near future against microbial infectious.
Graphic abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Chemistry and biology are now a part of our everyday environment and these two disciplines are at the crossroads of many fields and industries. Previous organic chemistry now extends to inorganic molecules, biological molecules and macromolecules (Yousef et al. 2019). Schiff bases are important organic scaffolds exhibiting various applications in the area of medicinal inorganic chemistry. Schiff bases have been playing leading role in the development of coordination chemistry (Kaushal and Thakur 2013). They have also been employed as widespread ligands owing to their structural similarity with natural biomolecule, chelated structure, synthetic flexibility and excellent electron-donor ability. The strong affinity for the chelation of the Schiff bases towards the transition metal ions is used to develop their complexes (Rambabu et al. 2020). Complexes containing triazole ligands have been widely studied for their biological action with the aim to design and discover effective and safe therapeutic agents to treat various microbial diseases (Naik et al. 2020). Pathogenic resistance coupled with various drug resistance diseases has posed researchers a severe challenge to design and develop new metal based bioactive compounds (Matshwele et al. 2020; Yasmeen et al. 2017). The essential feature of metal based drugs relies on their capability to penetrate through the microbial membrane along with their affinities towards genetic materials [ribonucleic acid (RNA) or deoxyribonucleic acid (DNA)] of these pathogenic microbes (Abu‐Dief et al. 2020).
Nowadays, extensive research work has been done on the complexes of 1,2,4-triazole Schiff bases due to their remarkable significance in medicinal chemistry (Emam et al. 2020). The literature study has revealed that triazole derived ligands and their resulting complexes have shown wide range of pharmacological and biological activities and served as antimicrobial (Utthra and Raman 2018; Bouhidel et al. 2018), anticonvulsant (Kaproń et al. 2019), anti-proliferative (Ali et al. 2020), anti-oxidant (Ommenya et al. 2020), anesthetic (Shafi et al. 2012), anticancer (Gaber et al. 2018), antidiabetic (Munawar et al. 2015) and DNA cleaving agents (Bheemarasetti et al. 2018). Complexes of 1,2,4-triazole derived Schiff bases have also been evaluated as anthelmintic and cytotoxic agents (Zafar et al. 2021). There are several triazole based antifungal drugs that are commercially available such as ravuconazole, fluconazole, terconazole and voriconazole (Nayak and Poojary 2020; Ghanaat et al. 2020). A variety of triazole-based derivatives have been synthesized and diagnosed for their anti-infective (Tarawneh et al. 2018) and anti-inflammatory actions (Shen et al. 2020; Khan et al. 2021). Triazole derived compounds are capable of inhibiting certain enzymes including tyrosinase (Akin et al. 2019), urease (Qin et al. 2014), αlpha‐amylase (Wang et al. 2019), αlpha‐glucosidase (Nasli‐Esfahani et al. 2019) and DNA-gyrase (Nastasa et al. 2018).
Latest research developments, primarily in the synthetic chemistry have coincided with computer assisted calculations to investigate and describe numerous structural and biological properties of newly synthesized chemical systems. density functional theory (DFT) has progressively emerged as a foremost methodology for the simulation and modelling of chemical compounds (Maharramov et al. 2016). These DFT based computer assisted calculations are primarily used to investigate and describe the significant structural information and biological properties by finding the insight of the electronic properties, reacting sites, chemical reaction mechanisms and stability. These computed calculations are useful in recognizing the promising noncovalent interactions within the newly synthesized chemical systems owing to the reason that these interactions improve the biological profile of bioactive compounds (Xavier and Gobinath 2012). Thus, based on the above facts and in the extension of our previous work on the synthesis, physicochemical and biological properties of triazole Schiff base complexes (Sumrra and Chohan 2012), we have merged the chemistry of 1,2,4-triazole Schiff base with transition metals to counter drug resistance. The ultimate objective of this study includes the detailed and precise account of experimental and computed data to evaluate the spectroscopic aspects of synthesized compounds. In the present work, triazole Schiff base ligand, (2-methoxy-4-[(1H-1,2,4-triazol-3-ylimino)methyl]phenol) (H2L) was synthesized from 1H-1,2,4-triazol-3-amine and 4-hydroxy-3-methoxybenzaldehyde. The triazole ligand and its corresponding transition metal chelates have been characterized using spectroscopic techniques including FT‐IR, UV–Vis, 1H-NMR and MS along with elemental, fluorescence study and thermal analysis. The experimentally obtained spectroscopic results (FR-IR and UV–Vis) were further counter-checked by DFT based theoretical calculations employing Becke, 3-parameter, Lee–Yang–Parr (B3LYP). Further, the antioxidant as well as in vitro antibacterial and antifungal screenings were also performed for the synthesized compounds. The obtained results exhibited promising results which could be further developed into efficient antimicrobial and antioxidant agents.
Experimental
All the chemicals and reagents used in laboratory during the research work were of analytical grade. The synthesis of triazole ligand and its transition metal chelates was carried out in distilled ethanol and methanol, respectively. Stuart apparatus was utilized to determine the melting point of triazole Schiff base and decomposition points of complexes by using an open capillary technique. In order to determine the characteristic absorbance bands of solid samples, infrared spectroscopic technique was used having the range of 3700–370 cm−1 by using Nicolet FT-IR Impact 400D infrared spectrometer. 1HNMR spectra of synthesized ligand and its Zn(II) complex were obtained using dimethyl sulfoxide (DMSO-d6) solvent on Bruker Advance 300 MHz instrument at room temperature with an internal standard of tetramethyl silane (TMS). For the mass spectrometric analysis, the synthesized compounds (ligand and its derived complexes) were dissolved in methanol. Mass fragmentation of synthesized compounds was recorded with LTQ XL Linear Ion Trap Mass Spectrometer (Thermo Scientific, USA) instrument equipped with an Electro Spray Ionization (ESI) probe. A high tech, Perkin Elmer 2400 series-II (CHN) analyzer based on classical organic analysis Pregl-Dumas technique was utilized to obtain the elemental (C, H, N) content of all synthesized compounds. However, metals content (Fe, Co, Ni, Cu and Zn) of complexes were determined by ethylenediaminetetraacetic acid (EDTA) complexometric titrations followed by the decomposition of complexes by fuming nitric acid. While the estimation of vanadium was also achieved volumetrically utilizing 0.1 N potassium permanganate (KMnO4) solution (oxidizing agent) in the presence of dilute sulfuric acid. UV–Vis spectra were documented on Hitachi UV-3200 spectrophotometer using DMF (dimethyl formamide) as solvent in the wavelength range of 200–1100 nm. Molar conductivity measurements of complexes were executed on an Inolab Cond 720 Conductivity Bridge using 1 mM solutions in DMF at room temperature. The magnetic moment measurements of the complexes have been executed with the Magnetic Susceptibility Balance MSB Mk1 (Sherwood, UK) by utilizing manganese(II) chloride as calibrant. The heating curves of thermogravimetric analysis (TGA) and differential thermogravimetry (DTG) were obtained from Perkin-Elmer diamond instrument under N2 atmosphere with a sample mass of 4–5 mg using a heating rate of 10 °C/min over the temperature range of 40–600 °C. The fluorescence emission spectra of triazole ligand and its resulting complexes were recorded on Shimadzu 8101AFT-IR spectrophotometer at room temperature. For fluorescent study, the solutions (10−3 M concentration) of all the compounds were prepared in DMF solvent. The antibacterial and antifungal activities were performed in the Department of Biochemistry and Biotechnology, University of Gujrat, Gujrat and antioxidant activity was done in the Department of Chemistry, University of Gujrat, Gujrat, Pakistan.
Synthesis of ligand (H 2 L)
The triazole based ligand, (2-methoxy-4-[(1H-1,2,4-triazol-3-ylimino)methyl]phenol) (H2L) was prepared as described in Scheme 1 (Jamil et al. 2018). For the synthesis of Schiff base ligand (H2L), 10 mL ethanolic solution of 3-amino-1,2,4-triazole (0.08 g, 1 mmol) was refluxed in two neck round bottom flask for 30 min. Then 10 mL ethanolic solution of 4-hydroxy-3-methoxybenzaldehyde (0.15 g, 1 mmol) was added dropwise and the resulting reaction mixture was further refluxed for 8 h. The progress of reaction was continually observed by thin layer chromatography (TLC). Light yellow precipitates of the product were formed in the flask which indicted the formation of ligand. The precipitated product was cooled, filtered, then thoroughly washed with hot ethanol and dried. After drying the precipitate, the product was weighed as well as yield was calculated.
2-Methoxy-4-[(1H-1,2,4-triazol-3-ylimino)methyl]phenol (H 2 L)
Light yellow solid; yield 92%; m.p: 135 °C; IR (KBr) νmax = 3110 m (OH), 3065 w (N–H), 2916m (OCH3), 1630 s (HC=N), 1575 m (C=N), 1372m (C–O), 1032 w (N–N) cm−1; 1H-NMR (DMSO-d6, 300 MHz): δ = 3.86 (s, OCH3), 6.91 (1H, d, J = 8.1), 7.43 (1H, dd, J = 8.1, 1.8), 7.58 (1H, d, J = 1.8), 7.91 (s, OH), 8.50 (s, HC=N), 9.07 (s, C–H), 13.95 (s, NH) ppm; LCMS m/z 219.17 ([M + 1] (42), 204.08 (17), 202.08 (77), 163.08 (7), 125.08 (98), 113.08 (100), 111.08 (9), 95.08 (76), 85.08 (4) %; Anal. Calcd. for C10H10N4O2 (218.21): C, 55.04; H, 4.62; N, 25.68. Found: C, 54.98; H, 4.58; N, 25.61%.
Synthesis of triazole based complexes (1)–(6)
Complexes were synthesized by refluxing 10 mL of methanolic solution of triazole ligand (2 mmol) for 30 min. Then added 5 mL methanolic solution of respective metallic salt (1 mmol) dropwise in 100 mL two neck round bottom flask containing the ligand solution. The resultant reaction mixture was refluxed for 4–6 h and solid precipitates were produced in the flask which indicated the formation of complex (Scheme 1). The obtained precipitates of the complexes were recrystallized using a 1:2 mixture of methanol:dioxane by evaporation at room temperature. The complexes were weighed to calculate the yields for this second synthetic step. Their solubilities and decomposition points were also determined.
Determination of stoichiometry of the complexes
Job’s method of continuous variation was followed to determine the stoichiometry of synthesized complexes (Eze et al. 2014). A mixture of ligand and each metal salt [vanadyl sulphate, hydrated iron(II) sulfate, cobalt(II) chloride, nickel(II) chloride, copper(II) chloride and zinc(II) chloride] solution was magnetically stirred for 2 h and then it was allowed to equilibrate. Then, the absorbance was recorded at the λmax of each complex using UV–Vis spectrophotometer.
Bioactivity assay
The synthesized compounds were subjected to screen for anti-bacterial, anti-fungal and anti-oxidant activities as follows:
Antibacterial activity
1H-1,2,4-Triazol-3-amine Schiff base ligand (H2L) and its corresponding metal chelates (1)–(6) were scrutinized for in-vitro antibacterial activity against two Gram +ve bacterial species i.e., Bacillus subtilis (ATCC 6051) and Staphylococcus aureus (ATCC 25923) together with two Gram −ve bacterial species i.e., Escherichia coli (ATCC 25922), Pseudomonas aeruginosa (ATCC 27853), by means of disc diffusion method (Sumrra et al. 2016). The sterilized petri dishes were filled with agar medium. After that, 100 µL of bacterial inoculums were dispersed on solidified medium surface using sterilized glass loop spreader. Discs of filter paper impregnated with 10 µL of sample solutions at 25 mg/mL concentration in DMSO solvent and one disc with only DMSO (used as negative control) were positioned at different points on the medium surface. Augmentin syrup (SD1) was used as standard clinical drug for this study owing to its wide-ranging activity. Then, these petri dishes were kept for incubation at 37 °C temperature for 1 day, later on the diameters of inhibition zone areas were noted in millimeters (mm) for samples and reference drug against each bacterial strain.
Antifungal activity
Anti-fungal action of triazole Schiff base (H2L) and its corresponding metal chelates (1)–(6) was studied against two fungal species, Aspergillus niger (ATCC 1015) and Candida albicans (ATCC 10231) by means of disc diffusion method (Sumrra et al. 2016). Sabouraud dextrose agar was seeded with 105 (CFU)/mL fungal spore suspensions and then decant to petri dishes. Strips of filter paper were saturated in 20 mL (dissolved in DMSO) of the sample solutions, then positioned at different points on the surface of semi-solidified agar. The petri dishes were kept for incubation at 37 °C for 2 days. The antifungal activity was noted as diameters of inhibition zones (mm) and then it was compared with the reference drug, fluconazole (SD2).
Antioxidant activity
Anti-oxidant profile of synthesized compounds (triazole ligand and its complexes) was determined by means of DPPH method, reducing power method and concentration of total phenolic content.
DPPH method
All the synthesized compounds were analyzed for 1,1-diphenyl-2-picrylhydrazyl (DPPH) free radical scavenging (%) action (Yakan et al. 2020). Concisely, this method included the preparation of stock solutions (1 mg/mL) of samples, which were poured into seven test tubes (for each compound) and mixed with 2.5 mL DPPH (2 mM) solution. The stock solutions of samples and DPPH were prepared in methanol. Then, these test tubes were kept in dark for half an hour. By using UV–Vis spectrophotometer, the absorbance reading of all the test samples was determined at 517 nm wavelength. And percentage inhibition of samples was obtained using following formula:
Reducing power method
Assessment of reducing power ability of all synthesized compounds was carried out in accordance with standard method (Bulut et al. 2018) by mixing 1 mL sample solution (of 1 mg/mL concentration) with 2.5 mL of both 1% potassium ferricyanide and sodium phosphate buffer (200 mM, pH 6.6). The resultant mixture was incubated at 50 °C temperature for 20 min. Afterward, 2.5 mL of 10% trichloroacetic acid was mixed in each test tube, then the resultant mix was centrifuged for 8 min at 3000 RPM. Following the centrifugation, 5 mL sample from upper layer was collected and 5 mL of deionized water was added in it, then mixed well with 1 mL of ferric chloride solution (0.1%). The reducing power ability was assessed by taking the absorbance reading of resultant mixture at 700 nm.
Concentration of total phenolic content
Total phenolic content of compounds was checked by means of standard procedure (Tadavi et al. 2017). This method included the mixing of 1 mL of sample solutions (2 mg/mL) with 1 mL of 10% Folin–Ciocalteu reagent, 2 mL of 7.5% solution of sodium carbonate along with 4 mL of methanol in different test tubes. Then, these test tubes were kept at 37 °C for half an hour after vortexing for 30 s. After that, with the help of UV–Vis spectrophotometer, the absorbance was checked at 760 nm wavelength. In this procedure, gallic acid was utilized as a reference agent. Measurements were done in triplicates for each sample and their quantifications were achieved by using gallic acid calibration curve.
Density functional theory (DFT) computations
The computational study of synthesized ligand (H2L) and its derived complexes was carried out using Gaussian program package. For ground state configuration of compounds, the optimized geometries were obtained in gas phase, neutral state, and in the absence of symmetry constraints at B3LYP level of theory in combination with 6-31G(d,p) basis set. Gauss View 5.0 has been used to prepare all the input files for DFT study by Gaussian (Frisch et al. 2009). Whereas, the Chemcraft, Gauss View 5.0, GaussSum and Avogadro programs were utilized to study the obtained output files.
Results and discussion
The preparation of 3-amino-1,2,4-triazole Schiff base endowed complexes involved two steps as presented in Scheme 1. Initially, the condensation reaction of 1H-1,2,4-triazole-3-amine with 4-hydroxy-3-methoxy benzaldehyde in ethanol gave Schiff base ligand named (2-methoxy-4-[(1H-1,2,4-triazol-3-ylimino)methyl]phenol). The synthesized ligand was yellow colored solid compound, which was stable in moisture and air as well as soluble in DMSO and DMF without heating. The ligand was characterized by means of spectroscopic methods, elemental analyses and DFT study. Moreover, the synthesized ligand was used for chelation with 3d-transition metals [VO(IV), Fe(II), Co(II), Ni(II), Cu(II) and Zn(II)] salts in 2:1 molar ratio to produce complexes having different structural geometries. The triazole Schiff base acted as bidentate ligand and form chelates by making 5 membered ring system through the coordination of deprotonated oxygen of hydroxyl group and oxygen of methoxy group with the transition metal centers. All the complexes were obtained as non-electrolytes with octahedral geometry with the exception of vanadyl complex which has a square pyramidal geometry. All the solid complexes were stable at room temperature, decomposed without melting, intensely colored except zinc complex which was of off white color and partially soluble in most organic solvents but completely soluble in DMSO and DMF (shown in Table 1). The spectroscopic data of the synthesized compounds agreed well with their proposed formula signifying the high purity of all the synthesized compounds.
Stoichiometry of complexes by Job’s method of continuous variation
Job’s plot for divalent and tetravalent complexes of triazole Schiff base is given in Fig. S1, while the data including mole fraction of complexes and their resulting absorbance values are given in Table S1. The curve showed a highest absorbance at a mole fraction Xmetal = 0.32–0.35, which suggested the synthesis of complexes having a 1:2 metal to ligand stoichiometry ratio.
FT-IR spectra
Infrared study of triazole ligand (H2L) and its derived complexes (1)–(6) has shown various peaks of different functional groups (Table S2). The disappearance of triazole NH2 and C=O band of aldehyde group at 3330 and 1720 cm−1, respectively and the occurrence of a new band at 1630 cm−1 validated the formation of azomethine linkage (HC=N) (Vinusha et al. 2019) by condensation reaction between 3-amino-1,2,4-triazole and 4-hydroxy-3-methoxybenzaldehyde. The bands of O–H, N–H and O-CH3 groups of triazole ligand appeared at 3110, 3065 and 2916 cm−1, respectively. Similarly, the C=N of triazole moiety, C-O and N–N peaks were observed at 1575, 1372 and 1032 cm−1, individually (Sumrra et al. 2015). The IR data confirmed the coordination of the divalent and tetravalent transition metal cations with ligand through coordinated sites, oxygen of OCH3 and OH groups. The band of methoxy (O-CH3) group was shifted towards lower frequencies at 2902–2906 cm−1 by 10–14. The OH band at 3110 cm−1 was totally disappeared and C-O band was shifted to 1355–1360 cm−1. Occurrence of weak band at 533–539 cm−1 was assigned to ν(M–O) also supported the coordination. A typical band at 969 cm−1 was allocated to the coordination of ligand towards the vanadium in complex (1) (Hanif and Chohan 2013a). Broader peak of H2O molecules appeared at 3470–3475 cm−1 in the complexes (2)–(6). The C=N, N–H and N–N vibrations of triazole ring and HC=N band of azomethine linkage at 3065, 1032, 1575 and 1630 cm−1, respectively remained unaffected (Hanif and Chohan 2013b) showing their non-involvement in the coordination phenomenon (Fig. S2-S4). The computational and experimental data of the ligand along with IR stretching and vibrations were preceded through DFT/B3LYP with 6-31G(d,p) basis set (Table S3). The results in Fig. S5 have shown a close agreement between experimental and computed IR spectra.
Electronic spectra
UV–visible spectra of the triazole ligand (H2L) and its derived complexes (1)–(6) were recorded using 1 mM solutions in DMF at room temperature. The ligand showed two absorption bands. First band appeared at 247 nm due to n–π* transition and gave a clue about the existence of aromatic benzene ring. Whereas, the second band appeared at 363 nm was because of π–π* transition, which indicated the occurrence of azomethine (HC=N) linkage within the synthesized triazole ligand (Joshi et al. 2019). The computational data obtained by gas phase time-dependent density-functional theory (TD-DFT) study of ligand was used for comparison between experimental work and theoretical data (Table S4). The TD-DFT data had shown the transition of ligand which occurred at 331.79 and 246.63 nm with oscillator strength of 0.2757 and 0.0271, respectively. These two excitations were also in close resemblance with experimental values that were 363 and 247 nm individually as presented in Figs. S6 and S7.
Oxovanadium(IV) complex (1) displayed three distinctive bands at 744, 550 and 378 nm due to B2 → Eπ, B2 → B1 and B2 → A1 transitions. Band at 331 nm declared the charge transfer phenomenon from metal to ligand (MLCT). All these bands indicated square pyramidal geometry of vanadium complex (Hassan et al. 2021). Octahedral geometry of Fe(II) complex (2) was confirmed by 6A1g → 4T2g(G) and 6A1g → 4Eg(G) electronic transitions at 784 and 756 nm respectively. Co(II) complex (3) presented two low energy bands at 1087 and 568 nm giving 4T1g(F) → 4T2g(F) and 4T1g(F) → 4T1g(P) electronic transitions and a absorption band of high energy at 337 nm proved their octahedral geometry with high spin. Three characteristic absorptions bands at 972, 617 and 409 nm for Ni(II) complex (4) were allocated to 3A2g(F) → 3T2g(F), 3A2g(F) → 3T1g(F) and 3T2g(F) → 3T1g(P) electronic transitions, correspondingly. Cu(II) complex (5) demonstrated octahedral geometry due to two typical bands at 666 and 514 nm as a result of 2B1g → 2A1g and 2B1g → 2Eg transitions and a highly intense band at 389 nm because of MLCT. Zn(II) complex (6) exhibited no d-d transition characteristic band due to its diamagnetic nature. Only metal to ligand charge transfer band of high intensity was recorded at 340 nm (Hanif and Chohan 2013a,b).
1H-NMR spectra
The 1H-NMR spectrum of triazole ligand (H2L) revealed characteristics peaks of various protons present in the triazole ligand. The presence of singlet peak at δ 8.50 ppm due to presence of (HC=N) azomethine proton confirmed the formation of ligand. Other singlet peaks at δ 13.95, 9.07, 7.91 and 3.86 ppm confirmed the N–H, C-H, OH and OCH3 protons, separately (Fig. S8). The aromatic protons of phenyl ring exhibiting ABX pattern were appeared at 6.91 (1H, d, J = 8.1), 7.43 (1H, dd, J = 8.1, 1.8), 7.58 (1H, d, J = 1.8) (Chaurasia et al. 2019).
The formation of Zn(II) complex was further supported by 1HNMR in which the major difference was the disappearance of OH proton from 7.91 ppm and shifting of OCH3 peak to upfield from 3.86 to 3.02 ppm (Fig. S9). The singlet peak of HC=N experienced a very minor shifting from 8.50 to 8.49 ppm signifying the non-involvement of azomethine linkage in the complexation. Similarly, all other remaining protons also underwent slight upfield shift because of increased conjugation as well as coordination. All the protons of Zn(II) complex were in close agreement with the elemental analysis and confirmed the synthesis of the Zn(II) complex.
Mass spectra
Mass spectrum of triazole ligand was obtained by liquid Chromatography with tandem mass spectrometry (LCMS)-electron ionization method. The ionization of ligand followed by cleavage of C=N, C–C and C=C in exocyclic and endocyclic way. The spectrum revealed the fragmentation pattern of ligand at various points along with specific intensity and mass to charge (m/z) ratio. The fragmentation pattern of recognized peaks was used to produce the daughter ions that confirmed the molecular identity and the anticipated formula of triazole ligand (Fig. S10). In the mass spectrum of ligand (H2L), the molecular ionic peak with molecular formula [C10H10N4O2] was appeared at m/z = 219.17 (40%) and base peak of ligand (H2L) was obtained at m/z = 113.08 agreeing well with the fragment [C6H10NO]·. The other significant fragmentation peaks along with their relative abundance were observed at 204.08 (17) due to fragment [C10H10N3O2·], 202.08 (77) because of [C9H7N4O2·] fragment, 125.08 (98) owing to the second stable fragment of [C6H7NO24·], 95.08 (76) with fragment [C3H3N4·]. While the other fragmentation peaks were observed at 111.08 (9), 163.08 (7), and 85.08 (4%).
The mass spectra of the complexes were also concurrent with the calculated masses from their intended formulae. The molecular ion peak of the vanadyl complex (C20H18N8O5V) was found to be at m/z = 502.58 (14%) and the most stable fragment [C15H14N4O5V2·] of maximum intensity at m/z = 381.71. Beside these, the most prominent fragmentation peaks were observed at 486.08 (37), 463.08 (22), 339.92 (45), 284.01 (67), 254.00 (18), 217.08 (39), 187.08 (30), 172.07 (12) and 108.04 (14%) due to fragments of [C19H15N8O5V·], [C19H20N6O5V], [C15H14NO5V2·], [C10H9N4O3V2·], [C9H8N4O2V3·], [C10H9N4O22·], [C9H7N4O·], [C9H8N4], [C4H4N42·], respectively (Fig. S11). The molecular ion peak of the zinc complex (C20H22N8O6Zn) appeared at m/z = 536.75 (22%), while the most stable (base peak) with the [C16H18N2O6Zn2·] fragment was observed at m/z = 398.08. Some major peaks having evident percentages were found at 507.33 (12), 454.08 (57), 364.92 (36), 360.08 (22), 310.05 (60), 217.08 (44), 179.17 (32) and 157.04 (10%) owing to fragments [C19H21N7O6Zn·], [C18H22N4O6Zn], [C11H16N4O6Zn2·], [C15H20O6Zn·], [C11H18O6Zn4·], [C10H9N4O22·], [C9H11N2O22·] and [C9H8N2O2·], correspondingly along with some other fragment peaks with their relative abundances at 297.00 (4), 131.08 (5%) (Fig. S12). The aspects of the triazole Schiff base ligand (H2L) and its respective complexes obtained by the mass spectra strongly validated their synthesis and agreed well with their suggested bonding patterns.
Thermal analysis
To study the thermal stability, decomposition behaviour and presence of water molecules outside or inside complexes, thermo-gravimetric analysis (TGA) together with differential thermogravimetric (DTG) analysis were performed. In this study, the thermal behaviour of the synthesized complexes was explored with the rise in temperature ranging from 40 to 600 °C. Figure 1 illustrates the TGA/DTG diagrams of the complexes with the experimental mass loss. For vanadyl complex (1), no weight loss was observed under 200 °C signifying the absence of water molecules in the complex. The decomposition take place in three steps within the temperature ranging from 210–290, 290–460 and 460–560 °C corresponding to the mass loss of 22.70, 19.30 and 4.50%, respectively. While the other two complexes have shown quite different thermal behavior. On heating the nickel and zinc complexes (4) and (6), the initial step of decomposition corresponds to the removal of two coordinated water molecules within the temperature range of 70–150 and 80–170 °C, respectively (Badea et al. 2014). The second steps refer to the decomposition of anhydrous complexes in the temperature range 150–340 and 170–430 °C with a mass loss of 16.21 and 39.91%, correspondingly. Whereas in the case of nickel complex (4), the third and last step of decomposition was observed within the temperature ranging from 340 to 510 °C with a mass loss of 27.05%.
Having in view all the above discussion, thermal curve of vanadyl complex validated the absence of coordinated water molecules in their proposed structure while the thermal data for all the other complexes correlated with the IR data and validated the presence of two water molecules in their proposed structure. And the analysis of the TG/DTG curves course of nickel and zinc complexes (4) and (6) signified that on heating both the complexes decomposed in a similar manner in two ground stages; in first stage, dehydration of the complexes was observed with formation of less stable anhydrous compounds. While in the second stage, the TG/DTG curves showed a slight but systematic mass loss as a result of the decomposition of anhydrous compounds owing to the oxidation of organic ligand.
Fluorescence study
The fluorescent action of the triazole ligand (H2L) and its derived divalent and tetravalent complexes was explored using sample solutions in DMF solvent at room temperature. To record and study the photo-luminescent properties of the investigated compounds, the wavelengths of maximum absorption have been selected as the excitation wavelengths (Fig. 2). The spectra have displayed the fluorescence within the wavelength ranging from 300 to 550 nm. From the figure, it was observed that after photoexcitation at 350 nm, the triazole ligand (H2L) have shown a strong peak at 390 nm in the emission spectra predominantly because of π–π* electronic transition, that was altered after bonding with transition metal cations. In contrast to the ligand spectra, complexes experienced slight fluorescence quenching with slightly reduced fluorescence emission. It was evident from the emission spectra that the intensity of emission for triazole ligand was much decreased on the complexation with metal(II/IV) cations. This considerable decline in the emission intensity upon coordination with metal ions was because of the decrease in the distribution of electronic charge density on the triazole ligand (H2L). The complexes presented a minor red shift in the emission wavelength subsequently with a reduction in the intensity of emission, which may be attributable to transfer of electronic charge density from ligand to metal (Patil et al. 2011). The zinc(II) complex (6) demonstrated a wide peak at 400 nm, which was to some extent lesser in comparison with triazole ligand (H2L). The overall increasing order of fluorescence quenching effect in the complexes was found as; Zn(II) < Cu(II) < VO(IV) < Fe(II) < Co(II) < Ni(II).
Molar conductance and magnetic moment
The as-synthesized complexes were dissolved in DMF solvent and their molar conductance was measured. The values of molar conductivity were observed ranging from 11.3 to 18.4 Ω−1cm2mol−1 confirming (Table S2) their non-conductor nature (Aly et al. 2015). The vanadium (1) and copper complexes (5) exhibited magnetic moments in the range of 1.79–1.81 BM showing the existence of one unpaired electron (Hassan et al. 2021). The magnetic moment of cobalt complex (3) was 4.29 BM which specified three unpaired electrons. The nickel complex (4) displayed 2.97 BM magnetic moment confirming two unpaired electrons. The magnetic moment of iron complex (2) was 5.06 BM, displaying the existence of four unpaired electrons in the d-orbitals. The zero magnetic moment of zinc complex (6) confirmed its diamagnetic nature due to fully filled d10 system (Chohan and Hanif 2013). Molar conductance along with magnetic moments reinforced the synthesis of complexes.
Optimized molecular geometries
In this study, initially the geometries of triazole ligand (H2L) and its complexes (1), (3) and (5) were completely optimized by means of B3LYP functional with 6-31G(d,p) as the basis set. Figure 3 illustrates the optimized geometries of studied compounds. The computed optimized geometrical parameters were found to be good approximation and they were further employed as a base to calculate and analyze other characteristic features in the present study. No negative frequency was obtained after calculating the frequency for all the investigated compounds. This showed that the optimized molecular geometries are true minima on the potential energy surfaces. After that, the frontier molecular orbital (FMO) energies, global reactivity descriptors (GRD), Mulliken atomic charges (MAC) as well as molecular electrostatic potentials (MEP) of investigated compounds were computed by employing the same basis set.
Molecular electrostatic potential (MEP) analysis
Three dimensional shapes of molecular electrostatic potential surface are also called MEP maps. MEP mapped surface displays molecular size, shape, charge distribution and electrostatic potential. It is reliable method to study chemical reactivity along with nucleophilic and electrophilic regions of molecular systems. In these surfaces, there are different colors which represent the electrostatic potential. The magnitude of electrostatic potential in terms of descending order was obtained as; Blue < green < yellow < orange < red. Among these colors, blue denotes the positive potential surface and corresponds to electron deficient zone, red color indicates the negative potential surface and represents the electron rich zone area, whereas the green color signifies to zone of the low positive or nearly neutral electrostatic potential. These negative and positive regions presented the attractions for nucleophilic and electrophilic attacks (Sert et al. 2020). The chemical reactivity of ligand (H2L) and its corresponding complexes (1), (3) and (5), was explored by molecular electrostatic potential (MEP) plotted on the surface of optimized geometries (Fig. 4). As shown in the figure, all the hydrogen atoms of all the compounds having blue color, signify the relative nonparticipation of the electrons around the hydrogen atoms. Whereas, the atoms of high electronegativity such as nitrogen and oxygen in red color show comparatively more magnitude of electronic charge density. Consequently, they can be demonstrated as the preferred positions, where the electrophilic attacks are most likely to occur.
Overall, the MEP analysis proposed that all the studied complexes have a significant positive electrostatic potential dispersed on their skeleton, which suggests that these complexes have the potential to strongly bind with the microorganism having the negative molecular electrostatic potential. This interpretation will provide assistance to develop a stabilized complex for a perfect drug docking within the ligand binding of the investigated microorganism. The quantum chemical specifications evidently described the biological behavior of studied compounds with their MEP surfaces.
Mulliken atomic charge (MAC) analysis
Mulliken charge analysis is an important component for molecular framework in quantum chemical operation. The Mulliken population analysis has significant contribution in the application of quantum chemical operation to molecular framework. Mulliken atomic charges analysis predicts the features of molecules which are based on atomic charges that can affect their molecular properties such as molecular polarizability and dipole moment (Machado et al. 2019). Mulliken atomic charges of ligand (H2L) were calculated (Table S5) and the histogram of Mulliken atomic charges is shown in Fig. 5. The Mulliken atomic charges demonstrated the existence of high electronegative atoms within the triazole ligand such as (20O = − 0.28596), (22O = − 0.16673) (12 N = − 0.14860), (13 N = − 0.17498) and (14 N = − 0.10747), and unequal redistribution of electronic charge density in the title compound. The more negative values were obtained for the both oxygen atoms indicating these as most electronegative atoms within the triazole ligand. These charges also validated the coordinating action of triazole ligand with metal centers via both oxygen atoms (Ohydroxy and Omethoxy). In addition, Mulliken population analysis revealed that there has been observed no discrepancy in electronic charge distribution over all the hydrogen atoms, hence confirming hydrogen charges as positive.
The MAC data of complexes revealed that the existence of more electronegative atoms in the complexes such as oxygen and nitrogen created unequal redistribution of the electronic charge density over the benzene rings. The highly electronegative atoms within the complexes were (O34 = − 1.346081), (O15 = − 0.997597) (O31 = − 0.997443) and (N5 = − 0.923134) in vanadyl complex (1), while (O2 = − 1.210704), (O1 = − 1.168419), (N7 = − 0.891375) and (N22 = − 0.881898) in cobalt complex (3) and (20O = − 0.28596), (13 N = − 0.17498), (22O = − 0.16673), (12 N = − 0.14860) and (14 N = − 0.10747) in copper complex (5). On the other hand, metals of the studied complexes (1), (3) and (5), were marked as highly electropositive atoms such as V32, Co34 and Cu34 with 3.285460, 5.983480 and 9.145800, respectively.
Frontier molecular orbital (FMO) analysis
The stability and reactivity of FMOs are based upon the energy difference. The electron donor orbitals are named as HOMO (highest occupied molecular orbital) while electron acceptor orbitals are titled as LUMO (lowest unoccupied molecular orbital). The EHOMO-LUMO gap is an important tool to obtain information on the dynamic stability and chemical reactivity of molecules (Jin et al. 2020). DFT based computed calculations have been accomplished to understand the EHOMO, ELUMO and the difference in their energies ELUMO–EHOMO of investigated triazole ligand and its resultant complexes (Table 2) and the HOMO and LUMO orbitals of ligand and complexes are demonstrated in Fig. 6. According to the DFT study, the total number of orbitals for triazole ligand was 412. Out of these orbitals, 57 and 58 were HOMO and LUMO orbitals, correspondingly. While for complexes, there were total 326, 335 and 335 orbitals. And out of these orbitals, 120, 126 and 127 were marked as HOMO whereas 121, 127 and 128 were marked as LUMO for complexes (1), (3) and (5), respectively. The positive part was shown by red while the negative part was shown by green color. In HOMO, HOMO-1 of the ligand (H2L), the charge density is distributed on the entire ligand (2-methoxy-4-[(1H-1,2,4-triazol-3-ylimino)methyl]phenol), but in LUMO and LUMO + 1, the charge density is scattered on the complete ligand excluding the methoxy (OCH3) group. In all FMO’s of complex (1), the charge density is partially dispersed on the whole complex excluding the triazole moieties of the ligand. Similarly in all the FMO’s of complexes (3) and (5), the electronic charge density is only focused on one ligand of the complex.
The energy of HOMO and LUMO orbitals was − 6.373 eV and − 2.231 eV, respectively. The energy gap between these two orbitals was 4.141 eV. The energy for HOMO-1, and LUMO+1 was found to be − 7.085 and − 0.944, respectively. The energy gaps of these two orbitals was 6.141 eV. The energy gap for HOMO–LUMO was observed as 0.706, 0.010 and 0.017 eV for complexes (1), (3) and (5), respectively. Whereas the energy difference for HOMO-1 to LUMO + 1 was found to be 0.712, 0.846 and 0.507 eV for complexes (1), (3) and (5), correspondingly. For complexes, the decreasing order of energy difference in the case of ELUMO → EHOMO was (1) > (5) > (3). A chemical system can be considered as less reactive and more stable if its ELUMO → EHOMO gap value was found to be greater. In contrast, a chemical system can be considered as a highly reactive, soft and least stable if its ELUMO → EHOMO energy difference value was observed as low. The Egap values indicated that the ligand was less reactive with less electron donating capability and more kinetic stability than the corresponding complexes. The obtained FMO results revealed that complex (1) has greater ELUMO → EHOMO energy gap than other studied complexes (3) and (5). As a result, the least energy gap interprets that the complex (3) has superior capability of electron transfer. In addition, on the basis of above discussed parameters in our investigation, complex (1) is more stable and least reactive than other studied complexes. Moreover, the FMO analysis also provided the theoretical description required to consider the investigated compounds to be non-toxic and biologically active.
Frontier molecular orbitals are significantly involved for calculating the global chemical reactivity parameters (GCRP) to understand and explain the molecular stability of studied systems. Thus, chemical reactivity softness and hardness of triazole ligand and its derived complexes have been computed using the values of EHOMO, ELUMO and ∆EHOMO–LUMO. The global hardness (η) is described as the resistance to transfer of charge. Equations S1–S5 explained in the supplementary information file were used to calculate the global chemical reactivity parameters (GCRP). The ionization potential (IP) values were found to be 6.373, 7.344, 6.515 and 6.305 eV for ligand (H2L) and complexes (1), (3), (5), respectively. The electron affinity (EA) values of ligand (H2L) and complexes (1), (3), (5) were found to be 2.231, 6.638, 6.505 and 6.288 eV, respectively. As the electron accepting and donating character of above-mentioned compounds can be examined by values of electron affinity and ionization potential. In our studied compounds, values of ionization potential (IP) were found to be greater than that of electron affinity (EA) values. These results revealed that all the compounds were declared as less reactive with decreased electron donating capabilities and more kinetic stability. Likewise, the chemical hardness (η) values of ligand (H2L) and complexes (1), (3), (5) were found in the following decreasing order; [H2L (η = 2.071 eV)] > [1 (η = 0.353 eV)] > [5 (η = 0.008 eV)] > [3 (η = 0.005 eV)] as shown in Table 2. The trend of electronegativity (χ) was found as [1 (χ = 6.991 eV)] > [3 (χ = 6.510 eV)] > [5 (χ = 6.297 eV)] > [H2L (χ = 4.302 eV)]. The reactivity as well as stability of synthesized compounds can also be calculated by values of chemical potential (μ). The decreasing order of chemical potential was [1 (μ = − 6.991 eV)] > [3 (μ = − 6.510 eV)] > [5 (μ = − 6.297 eV)] > [H2L (μ = − 4.302 eV)].
Natural bond orbital (NBO) analysis
The natural bond orbital (NBO) analysis of synthesized triazole ligand (H2L) was accomplished by DFT/B3LYP/6-31G(d,p) method to obtain the perturbation theory (Alphonsa et al. 2016). It is useful tool to analyze the interactions within the molecule. NBO analysis was achieved by optimized geometry of ligand (illustrated in Fig. 3) and the results displayed the acceptor as well as donor orbitals with all types of values (Table S6). NBO analysis provides the stabilization energy which is related to the hyper conjugative association within the molecule from donor atoms to acceptor atoms (Subashchandrabose et al. 2010). A large amount of data was obtained but here only the possible transitions are mentioned having higher value of stabilization energy. The NBO data demonstrated the charge transfer from π(C1–C6) → π*(C2–C3), π(C4–C5) → π* (C2–C3) and π(C8–N9) → π*(C2–C3) with greater stabilization energy values 210.66, 204.24 and 76.83 kcal/mol, respectively showing the delocalization within the molecular framework. Similarly, charge transfer from π(N13–C15) → π*(C11–N12), π(C8–N9) → π*(C11–N12), σ(C3–C4) → σ*(C3–C8) and σ(C1–C2) → σ*(C1–O20), gave stabilization energies 16.99, 14.96, 10.7 and 4.10 kcal/mol, correspondingly indicated the conjugative interaction of C–C and C–N. The transfer of lone pair charge also took place from lone pair O22 → π*(C4–C5), O20 → π*(C1–C6), N14 → π*(N13–C15), N13 → π*(N14–C15), N12 → π*(C11–N13) and N9 → π*(C3–C8) with the stabilization energies 17.56, 16.74, 16.76, 9.17, 7.83 and 2.79, and kcal/mol, individually indicated the strong conjugative interaction of O to C–C, N to C–N and C–C.
For complexes, the most prominent charge transfer owing to π → π* interactions were identified as π(N2–N3) → π*(C4–N5), π(N17–N18) → π*(C19–N20) with 14.40 kcal/mol stabilization energies for complex (1), π(N7–C8) → π*(N3–C6), π(N2–N5) → π*(N3–C6), π(N19–N20) → π*(C21–N22) with 19.06, 10.95 and 10.67 kcal/mol stabilization energies for complex (3), π(C12–C13) → π*(C10–C11), π(C25–C26) → π*(N18–C24), π(C27–C28) → π*(C25–C32) with 11.69, 11.44 and 11.11 kcal/mol stabilization energies for complex (5). In addition, some other transitions have been observed as π(C23–C24) → π*(N16–C22), π(C8–C9) → π*(N1–C7) with 8.17 and 7.32 kcal/mol stabilization energies, respectively for complex (1), π(C29–C30) → π*(C25–C26), π(C14–C15) → π*(C10–C11) and π(N18–C24) → π*(C21–N22) with 9.76, 8.10 and 6.79 kcal/mol stabilization energies, respectively for complex (3), π(N19–N20) → π*(C21–N22), π(O31–Cu34) → π*(O31–C32), π(C29–C30) → π*(C25–C26), and π(C14–C15) → π*(C10–C11) with 10.87, 10.45, 9.47 and 8.09 kcal/mol stabilization energies, respectively for complex (5) (as depicted in Tables S7–S9).
The transfer of charge on the basis of LP → π* interactions were obtained as C6 → π*(N2–N3) and C21 → π*(N17–N18) with greater stabilization energies i.e., 73.56 and 74.01 kcal/mol, respectively for complex (1), C23 → π*(N19–N20) and O33 → π*(O1–Co34) with greater stabilization energies of 55.84 and 8.88 kcal/mol, correspondingly for complex (3), C23 → π*(N19–N20), O31 → π*(O31–O32) and O1 → π*(O2–Cu34) with greater stabilization energies of 59.56, 21.22 and 17.46 kcal/mol, respectively for complex (5). The NBO findings have revealed that intra-molecular charge transfer (ICT), hyper-conjugative interactions in addition to conjugation were found in studied compounds. The values of stabilization energy are greater for ligand than that of complexes, signifying that the intramolecular electrostatic interactions are stronger for ligand in comparison with complexes. The NBO results also supported the greater stability and less reactivity of ligand in contrast to complexes. The charge transfer mechanism arising from bonding to anti-bonding orbitals involves the donation of electron from the good antioxidant agents to the free radical, causing the radical anion to be persistent enough for the enhanced radical scavenging activity. In this way, the antioxidant ability of any chemical system can be evaluated by employing second order petulance, which is a physical measurement that provides the range for electron transfer, calculated by using NBO analysis (Ngo et al. 2017). The radical scavenging activity will be greater for the compounds having smaller E(2) values. Therefore in the present investigation, NBO analysis revealed that the studied compounds have good antioxidant properties, which is promising in terms of evaluating the radical scavenging capacity. All the studied complexes have enhanced anti-oxidant activity as compared to triazole ligand which can easily be visualized by smaller values of E(2) for complexes as compared to the free ligand.
Antibacterial activity
The studied compounds have shown variable antibacterial activities as illustrated in Fig. 7. The results of antibacterial activity were compared with standard drug, augmentin (SD1). All the compounds exhibited weak to significant antibacterial potential but were found to be less active than standard drug. But the standard has exhibited the maximum inhibitory activity of 21, 20, 19 and 18 mm against Pseudomonas aeruginosa, Staphylococcus aureus, E. coli and Bacillus subtilis, respectively. The ligand (H2L) has shown least antibacterial activity with 10, 09, 08 and 07 mm inhibition zone against Bacillus subtilis, Pseudomonas aeruginosa, E. coli and Staphylococcus aureus, respectively. Among all the synthesized compounds, the Co(II) complex (3) exhibited maximum antibacterial activity with 16, 15 and 14 mm zone of inhibtion against E. coli, Staphylococcus aureus and Bacillus subtilis, correspondingly. While remaining complexes possessed 11–14 mm zone of inhibition (Table S10). All the complexes revealed enhanced antibacterial activities in contrast to free ligand due to chelation.
Antifungal activity
The results of antifungal activity have shown that the compounds displayed moderate to significant activity against selected fungal species (Fig. 7). The results of antifungal activity were recorded and compared with standard drug, fluconazole (SD2). And this comparison revealed that all the synthesized compounds were less active than that of standard drug. The standard has exhibited the maximum inhibitory activity of 20 and 18 mm against C. albicans and A. niger, respectively. The ligand (H2L) has shown the minimum antifungal activity with 9 and 11 mm inhibition zone against A. niger and C. albicans, respectively. While the complexes demonstrated 11–16 mm inhibition zone against both fungal strains (Table S10). The Ni(II) complex (4) showed the greatest fungal inhibitory activity with 16 and 15 mm inhibition zones against the A. niger and C. albicans, respectively. Among the complexes, Fe(II) complex has shown least inhibitory activity with 11 mm zone against C. albicans.
Antioxidant activity
The anti-oxidant activity of ligand (H2L) and its transition complexes (1)–(6) was calculated by DPPH radical scavenging assay, reducing power assay and total phenolic contents.
DPPH radical scavenging assay
The antiradical scavenging action of all the synthesized compounds was determined by spectrophotometry. For this assay, DPPH was used as the free radical while the gallic acid (SD3) was utilized as the reference agent. The results revealed weak to moderate activity of synthesized compounds (given in Fig. 8). Whereas the gallic acid has shown the maximum activity (78%). Among the synthesized compounds, Fe(II) complex (2) exhibited highest (72%) antioxidant activity, Cu(II) and Zn(II) complexes possessed moderate activity whereas the Ni(II) complex has shown least activity (Table S11). The order of increasing percentage inhibition was; Ni(II) < H2L < Co(II) < VO(IV) < Zn(II) < Cu(II) < Fe(II) < Gallic acid.
Reducing power assay
Reducing power assay was executed for all the synthesized compounds by measuring their absorbance at 700 nm wavelength to evaluate the extent of test compounds for converting the ferric ions (Fe3+) to ferrous ions (Fe2+). The antioxidant activity was estimated as the total iron reducing power (%) of all the as-synthesized compounds. All the compounds exhibited significant to excellent values when compared with gallic acid (standard). Gallic acid (SD3) has shown the maximum activity (86%). The complexes (4) and (6) had the higher value of 83 and 81%, respectively whereas the ligand (H2L) possessed the least value of 47% (given in Fig. 8). The decreasing order of total iron reducing power for all of the samples was visualized as; Gallic acid > Ni(II) > Zn(II) > Fe(II) > Co(II) > Cu(II) > VO(IV) > H2L. The results of this assay (Table S11) displayed that the triazole ligand (H2L) exhibited the least reducing ability while the Ni(II) complex (4) demonstrated the highest reducing ability.
Total phenolic content assay
The total phenolic content of triazole ligand (H2L) and its transition complexes (1)–(6) was calculated by measuring their absorbance at 760 nm. For each sample, the measurements were made in triplicates and then their average results were expressed, according to the standard calibration curve (given in Fig. 9) plotted by using the results of various gallic acid standard solutions (Table S11). The total phenolic contents of all the tested samples have been deduced as milligram equivalent of gallic acid for the triazole ligand (H2L) and its corresponding complexes (1)–(6). All the tested compounds have exhibited weak to good activity in the range of 25–83% (as shown in Fig. 8). Standard (SD3) has shown the maximum activity with a value of 83%. Among the synthesized compounds, the highest (79%) value was recorded for the Fe(II) complex (2), and a minimum value of 25% was obtained for the Cu(II) complex (5). Overall the descending order of total phenolic content was obtained as; Gallic acid > Fe(II) > H2L > Co(II) > Ni(II) > Zn(II) > VO(IV) > Cu(II).
Conclusion
In this study, we have synthesized a triazole ligand (2-methoxy-4-[(1H-1,2,4-triazol-3-ylimino)methyl]phenol) along with its VO(IV), Fe(II), Co(II), Ni(II), Cu(II) and Zn(II) complexes. Synthesized ligand and its derived complexes were characterized by physical, spectroscopic and elemental analysis. The DFT study based quantum chemical calculations were employed to attain the optimized geometries of triazole ligand and its derived complexes at B3LYP/6-31G(d,p) level of theory. The computational data was in accordance with experimental data, thus validating the investigated compounds. All the complexes (2)–(6) exhibited octahedral geometry with the exception of vanadium complex (1) which showed square pyramidal geometry. The FMO energies, molecular descriptors, Mulliken atomic charges, natural bond orbitals and electrostatic potential capacities indicated that the studied compounds are effective bioactive compounds. The ligand and complexes were further analyzed for antibacterial, antifungal and antioxidant activity. The results have shown that the bioactivity of ligand was increased significantly upon chelation. To conclude, these promising results are supportive for different biological applications in the future endeavors.
References
Abu-Dief AM, Abdel-Rahman LH, Abdel-Mawgoud AAH (2020) A robust in vitro anticancer, antioxidant and antimicrobial agents based on new metal-azomethine chelates incorporating Ag(I), Pd(II) and VO(II) cations: probing the aspects of DNA interaction. Appl Organomet Chem 34(2):1–20. https://doi.org/10.1002/aoc.5373
Akin S, Demir EA, Colak A, Kolcuoglu Y, Yildirim N, Bekircan O (2019) Synthesis, biological activities and molecular docking studies of some novel 2,4,5-trisubstituted-1,2,4-triazole-3-one derivatives as potent tyrosinase inhibitors. J Mol Struct 1175:280–286. https://doi.org/10.1016/j.molstruc.2018.07.065
Ali A, Al-Hassani R, Hussain D, Jabir M, Meteab H (2020) Anti-proliferative activity and tubulin targeting of novel micro and nanoparticles complexes of 4-amino-3-thion-1,2,4-triazole derivatives. Nano Biomed Eng 12(1):75–89. https://doi.org/10.5101/nbe.v12i1.p75-89
Alphonsa AT, Loganathan C, Anand, SAA, Kabilan S (2016) Molecular structure, NMR, UV-visible, vibrational spectroscopic and HOMO, LUMO analysis of (E)-1-(2,6-bis(4-methoxyphenyl)-3,3-dimethylpiperidine-4-ylidene)-2-(3-(3,5-dimethyl-1H-pyrazol-1-yl)pyrazin-2-yl)hydrazine by DFT method. J Mol Struct 1106:277–285. https://doi.org/10.1016/j.molstruc.2015.11.005
Aly HM, Moustafa ME, Nassar MY, Abdelrahman EA (2015) Synthesis and characterization of novel Cu(II) complexes with 3-substituted-4-amino-5-mercapto-1,2,4-triazole Schiff bases: a new route to CuO nanoparticles. J Mol Struct 1086:223–231. https://doi.org/10.1016/j.molstruc.2015.01.017
Badea M, Calu L, Chifiriuc MC, Bleotu C, Marin A, Ion S, Ioniţă G, Stanică N, Măruţescu L, Lazăr V (2014) Thermal behaviour of some novel antimicrobials based on complexes with a Schiff base bearing 1,2,4-triazole pharmacophore. J Therm Anal Calorim 118(2):1145–1157. https://doi.org/10.1007/s10973-014-3821-4
Bheemarasetti M, Palakuri K, Raj S, Saudagar P, Gandamalla D, Yellu NR, Kotha LR (2018) Novel Schiff base metal complexes: synthesis, characterization, DNA binding, DNA cleavage and molecular docking studies. J Iran Chem Soc 15(6):1377–1389. https://doi.org/10.1007/s13738-018-1338-7
Bouhidel Z, Cherouana A, Durand P, Doudouh A, Morini F, Guillot B, Dahaoui S (2018) Synthesis, spectroscopic characterization, crystal structure, Hirshfeld surface analysis and antimicrobial activities of two triazole Schiff bases and their silver complexes. Inorg Chim Acta 482:34–47. https://doi.org/10.1016/j.ica.2018.05.028
Bulut N, Kocyigit UM, Gecibesler IH, Dastan T, Karci H, Taslimi P, Dastan SD, Gulcin I, Cetin A (2018) Synthesis of some novel pyridine compounds containing bis-1,2,4-triazole/thiosemicarbazide moiety and investigation of their antioxidant properties, carbonic anhydrase, and acetylcholinesterase enzymes inhibition profiles. J Biochem Mol Toxic 32(1):1–10. https://doi.org/10.1002/jbt.22006
Chaurasia M, Tomar D, Chandra S (2019) Synthesis, spectral characterization, and DNA binding studies of Co(II), Ni(II), Cu(II) and Zn(II) complexes of Schiff base 2-((1H–1, 2, 4-triazol-3-ylimino)methyl)-5-methoxyphenol. J Mol Struct 1179:431–442. https://doi.org/10.1016/j.molstruc.2018.11.027
Chohan ZH, Hanif M (2013) Antibacterial and antifungal metal based triazole Schiff bases. J Enzyme Inhib Med Chem 28(5):944–953. https://doi.org/10.3109/14756366.2012.696246
Emam SM, Tolan DA, El-Nahas AM (2020) Synthesis, structural, spectroscopic, and thermal studies of some transition-metal complexes of a ligand containing the amino mercapto triazole moiety. Appl Organomet Chem 34(5):1–26. https://doi.org/10.1002/aoc.5591
Eze FI, Ajali U, Ukoha PO (2014) Synthesis, physicochemical properties, and antimicrobial studies of Iron(III) complexes of ciprofloxacin, cloxacillin, and amoxicillin. Int J Med Chem 2014:1–7. https://doi.org/10.1155/2014/735602
Frisch E, Hratchian HP, Dennington II RD, Keith TA, Millam J, Nielsen B, Holder AJ Hiscocks J (2009) GaussView Version 5.0.8. Gaussian, Inc. Wallinford
Gaber M, El-Ghamry HA, Fathalla SK, Mansour MA (2018) Synthesis, spectroscopic, thermal and molecular modeling studies of Zn2+, Cd2+ and UO22+ complexes of Schiff bases containing triazole moiety. Antimicrobial, anticancer, antioxidant and DNA binding studies. Mater Sci Eng C 83:78–89. https://doi.org/10.1016/j.msec.2017.11.004
Ghanaat J, Khalilzadeh MA, Zareyee D (2020) Molecular docking studies, biological evaluation and synthesis of novel 3-mercapto-1,2,4-triazole derivatives. Mol Divers 25:223–232. https://doi.org/10.1134/S1068162020010069
Hanif M, Chohan ZH (2013a) Design, spectral characterization and biological studies of transition metal(II) complexes with triazole Schiff bases. Spectrochim Acta Part A Mol Biomol Spectrosc 104:468–476. https://doi.org/10.1016/j.saa.2012.11.077
Hanif M, Chohan ZH (2013b) Synthesis, spectral characterization and biological studies of transition metal(II) complexes with triazole Schiff bases. Appl Organomet Chem 27(1):36–44. https://doi.org/10.1002/aoc.2936
Hassan AU, Sumrra SH, Zafar MN, Nazar MF, Mughal EU, Zafar MN, Iqbal M (2021) New organosulfur metallic compounds as potent drugs: synthesis, molecular modeling, spectral, antimicrobial, drug likeness and DFT analysis. Mol Divers. https://doi.org/10.1007/s11030-020-10157-4
Jamil W, Kumari D, Taha M, Khan MN, Baharudin MS, Ali M, Kanwal M, Lashari MS, Khan KM (2018) Synthesis, β-glucuronidase inhibition, and molecular docking studies of 1,2,4-triazole hydrazones. J Iran Chem Soc 15(11):2441–2454. https://doi.org/10.1007/s13738-018-1433-9
Jin R, Wang Y, Guo H, Long X, Li J, Yue S, Zhang S, Zhang G, Meng Q, Wang C, Yan H, Tang Y, Zhou S (2020) Design, synthesis, biological activity, crystal structure and theoretical calculations of novel 1,2,4-triazole derivatives. J Mol Struct 1202:127234. https://doi.org/10.1016/j.molstruc.2019.127234
Joshi R, Kumari A, Singh K, Mishra H, Pokharia S (2019) New diorganotin(IV) complexes of Schiff base derived from 4-amino-3-hydrazino-5-mercapto-4H-1,2,4-triazole: synthesis, structural characterization, density functional theory studies, atoms-in-molecules analysis and antifungal activity. Appl Organomet Chem 33(5):1–22. https://doi.org/10.1002/aoc.4894
Kaproń B, Łuszczki JJ, Płazińska A, Siwek A, Karcz T, Gryboś A, Nowak G, Kocka AM, Walczak KK, Langner E, Szalast K, Marciniak S, Paczkowska M, Cielecka-Piontek J, Ciesla LM, Plech T (2019) Development of the 1,2,4-triazole-based anticonvulsant drug candidates acting on the voltage-gated sodium channels. Insights from in-vivo, in-vitro, and in-silico studies. Eur J Pharm Sci 129:42–57. https://doi.org/10.1016/j.ejps.2018.12.018
Kaushal R, Thakur S (2013) Synthesis and biological screening of Schiff base of titanium(IV). Chem Eng Trans 32:1801–1806. https://doi.org/10.3303/CET1332301
Khan B, Naiyer A, Athar F, Ali S, Thakur SC (2021) Synthesis, characterization and anti-inflammatory activity evaluation of 1,2,4-triazole and its derivatives as a potential scaffold for the synthesis of drugs against prostaglandin-endoperoxide synthase. J Biomol Struct Dyn 39(2):457–475. https://doi.org/10.1080/07391102.2019.1711193
Machado PM, Cassaro RAA, de Assis VM, Machado SDP, Horn A Jr, Lachter ER (2019) Synthesis, characterization and DFT studies of a new unsymmetrical dinuclear vanadium(IV) complex with a bipodal N2O-donor ligand. J Mol Struct 1193:110–117. https://doi.org/10.1016/j.molstruc.2019.04.098
Maharramov AM, Mahmudov KT, Kopylovich MN, Pombeiro AJL (2016) Non-covalent interactions in the synthesis and design of new compounds. Wiley, London
Matshwele JT, Nareetsile F, Mapolelo D, Matshamek P, Leteane M, Nkwe DO, Odisitse S (2020) Synthesis of mixed ligand ruthenium(II/III) complexes and their antibacterial evaluation on drug-resistant bacterial organisms. J Chem 2020:1–10. https://doi.org/10.1155/2020/2150419
Munawar K, Ali S, Tahir M, Khalid N, Abbas Q, Qureshi I, Shahzadi S (2015) Investigation of derivatized Schiff base ligands of 1,2,4-triazole amine and their oxovanadium(IV) complexes: synthesis, structure, DNA binding, alkaline phosphatase inhibition, biological screening, and insulin mimetic properties. Russ J Gen Chem 85(9):2183–2197. https://doi.org/10.1134/S1070363215090248
Naik S, Naik PP, Krishnamurthy G, Venugopal N, Naik N, Naik TR (2020) Synthesis, characterization, DFT studies and biological activity of Ru(III), La(III) and Ce(III) triphenylphosphine complexes containing 2-aminothiazole and 2-aminotriazole. J Inorg Organomet Polym 30:3332–3356. https://doi.org/10.1007/s10904-020-01492-y
Nasli-Esfahani E, Mohammadi-Khanaposhtani M, Rezaei S, Sarrafi Y, Sharafi Z, Samadi N, Faramarzi MA, Bandarian F, Hamedifar H, Larijani B (2019) A new series of Schiff base derivatives bearing 1,2,3-triazole: design, synthesis, molecular docking, and α-glucosidase inhibition. Arch Pharm 352(8):1–10. https://doi.org/10.1002/ardp.201900034
Nastasa C, Vodnar D, Ionut I, Stana A, Benedec D, Tamaian R, Oniga O, Tiperciuc B (2018) Antibacterial evaluation and virtual screening of new thiazolyl-triazole Schiff bases as potential DNA-gyrase inhibitors. Int J Mol Sci 19(1):222–240. https://doi.org/10.3390/ijms19010222
Nayak SG, Poojary B (2020) Design, synthesis, in silico docking studies, and antibacterial activity of some thiadiazines and 1,2,4-triazole-3-thiones bearing pyrazole moiety. Russ J Bioorg Chem 46(1):97–106. https://doi.org/10.1134/S1068162020010069
Ngo TC, Dao DQ, Nguyen MT, Nam PC (2017) A DFT analysis on the radical scavenging activity of oxygenated terpenoids present in the extract of the buds of Cleistocalyx operculatus. RSC Adv 7(63):39686–39698. https://doi.org/10.1039/C7RA04798C
Ommenya F, Nyawade E, Andala D, Kinyua J (2020) Synthesis, characte rization and antibacterial activity of Schiff base, 4-chloro-2-{(E)-[(4-fluorophenyl)imino]methyl}phenol metal(II) complexes. J Chem 2020:1–8. https://doi.org/10.1155/2020/1745236
Patil SA, Unki SN, Kulkarni AD, Naik VH, Badami PS (2011) Co (II), Ni (II) and Cu (II) complexes with coumarin-8-yl Schiff-bases: spectroscopic, in vitro antimicrobial, DNA cleavage and fluorescence studies. Spectrochim Acta Part A Mol Biomol Spectrosc 79:1128–1136. https://doi.org/10.1016/j.saa.2011.04.032
Qin J, Lei N, Zhu HL (2014) Synthesis, structural characterization, molecular docking, and urease inhibition studies of dinuclear cobalt(II) complexes derived from 3,5-bis(pyridin-2-yl)-4-amino-1,2,4-triazole. Coord Chem 67(7):1279–1289. https://doi.org/10.1080/00958972.2014.909591
Rambabu A, Ganji N, Daravath S, Venkateswarlu K, Rangan K (2020) Mononuclear Co(II), Ni(II) and Cu(II) complexes of the Schiff base, 2-(((4-trifluoromethoxy) phenylimino)methyl)-6-tert-butylphenol: synthesis, spectroscopic characterization, X-ray study and biological evaluation. J Mol Struct 1199:127006. https://doi.org/10.1016/j.molstruc.2019.127006
Sert Y, Doğan OE, Gökce H, Ağar T, Ucun F, Dege N (2020) Synthesis, crystal structure, Hirshfeld surface analysis, spectral characterization, and quantum computational evaluation of (E)-2-(((4-bromophenyl)imino)methyl)-6-methylphenol. J Phys Chem Solids 144:109478. https://doi.org/10.1016/j.jpcs.2020.109478
Shafi S, Alam MM, Mulakayala N, Mulakayala C, Vanaja G, Kalle AM, Pallu R, Alam MS (2012) Synthesis of novel 2-mercapto benzothiazole and 1,2,3-triazole based bisheterocycles: their anti-inflammatory and anti-nociceptive activities. Eur J Med Chem 49:324–333. https://doi.org/10.1016/j.ejmech.2012.01.032
Shen QK, Gong GH, Li G, Jin M, Cao LH, Quan ZS (2020) Discovery and evaluation of novel synthetic 5-alkyl-4-oxo-4,5-dihydro-[1,2,4]triazolo[4,3-a]quinoxaline-1-carbox-amide derivatives as anti-inflammatory agents. J Enzyme Inhib Med Chem 35(1):85–95. https://doi.org/10.1080/14756366.2019.1680658
Subashchandrabose S, Krishnan AR, Saleem H, Parameswari R, Sundaraganesan N, Thanikachalam V, Manikandan G (2010) Vibrational spectroscopic study and NBO analysis on bis (4-amino-5-mercapto-1,2, 4-triazol-3-yl) methane using DFT method. Spectrochim Acta Part A Mol Biomol Spectrosc 77(4):877–884. https://doi.org/10.1016/j.saa.2010.08.023
Sumrra SH, Chohan ZH (2012) Metal based new triazoles: their synthesis, characterization and antibacterial/antifungal activities. Spectrochim Acta Part A Mol Biomol Spectrosc 98:53–61. https://doi.org/10.1016/j.saa.2012.08.026
Sumrra SH, Chohan ZH (2013) Antibacterial and antifungal oxovanadium(IV) complexes of triazole-derived Schiff bases. Med Chem Res 22(8):3934–3942. https://doi.org/10.1007/s00044-012-0388-0
Sumrra SH, Hanif M, Chohan ZH (2015) Design, synthesis and in vitro bactericidal/fungicidal screening of some vanadyl(IV) complexes with mono-and di-substituted ONS donor triazoles. J Enzyme Inhib Med Chem 30(5):800–888. https://doi.org/10.3109/14756366.2014.976565
Sumrra SH, Hanif M, Chohan ZH, Akram MS, Akhtar J, Al-Shehri SM (2016) Metal based drugs: design, synthesis and in-vitro antimicrobial screening of Co(II), Ni(II), Cu(II) and Zn(II) complexes with some new carboxamide derived compounds: crystal structures of N-[ethyl (propan-2-yl) carbamothioyl] thiophene-2-carboxamide and its copper(II) complex. J Enzyme Inhib Med Chem 31(4):590–598. https://doi.org/10.3109/14756366.2015.1050011
Tadavi SK, Rajput JD, Bagul SD, Hosamani AA, Sangshetti JN, Bendre RS (2017) Synthesis, crystal structures, biological screening and electrochemical analysis of some salen-based transition metal complexes. Res Chem Intermed 43(8):4863–4879. https://doi.org/10.1007/s11164-017-2917-4
Tarawneh AH, Leon F, Jain SK, Gadetskaya AV, Abu-Orabi ST, Tekwani BL, Cutler SJ (2018) Evaluation of triazole and isoxazole derivatives as potential anti-infective agents. Med Chem Res 27(4):1269–1275. https://doi.org/10.1007/s00044-018-2146-4
Utthra PP, Raman N (2018) Probing the potency of triazole tethered Schiff base complexes and the effect of substituents on their biological attributes. Int J Biol Macromol 116:194–207. https://doi.org/10.1016/j.ijbiomac.2018.05.009
Vinusha H, Kollur SP, Revanasiddappa H, Ramu R, Shirahatti PS, Prasad MN, Chandrashekar S, Begum M (2019) Preparation, spectral characterization and biological applications of Schiff base ligand and its transition metal complexes. Results Chem 1:100012. https://doi.org/10.33263/LIANBS93.13721388
Wang X, Ling N, Zhang Y, Zeng D, Yang H (2019) Synthesis, crystal structure and biological properties of two Cu(II) complexes based on 1-(benzotriazole-1-methyl)-1-(2-ethylimidazole). J Mol Struct 1193:348–356. https://doi.org/10.1016/j.molstruc.2019.05.036
Xavier RJ, Gobinath E (2012) FT-IR, FT-Raman, ab initio and DFT studies, HOMO–LUMO and NBO analysis of 3-amino-5-mercapto-1,2,4-triazole. Spectrochim Acta Part A Mol Biomol Spectrosc 86:242–251. https://doi.org/10.1016/j.saa.2011.10.031
Yakan H, Bakır TK, Çavuş MS, Muğlu H (2020) New β-isatin aldehyde-N,N′-thiocarbohydrazones: preparation, spectroscopic studies and DFT approach to antioxidant characteristics. Res Chem Intermed 46(12):5417–5440. https://doi.org/10.1007/s11164-020-04270-0
Yasmeen S, Sumrra SH, Akram MS, Chohan ZH (2017) Antimicrobial metal-based thiophene derived compounds. J Enzyme Inhib Med Chem 32(1):106–112. https://doi.org/10.1080/14756366.2016.1238363
Yousef TA, El-Reash GA, AL-Zahab MA, Safaan MAA (2019) Physicochemical investigations, biological studies of the Cr(III), Mn(II), Fe(III), Co(II), Ni(II), Cu(II), Zn(II), Cd(II), Hg(II) and UO2(VI) complexes of picolinic acid hydrazide derivative: a combined experimental and computational approach. J Mol Struct 1197:564–575. https://doi.org/10.1016/j.molstruc.2019.07.088
Zafar W, Sumrra SH, Chohan ZH (2021) A review: Pharmacological aspects of metal based 1,2,4-triazole derived Schiff bases. Eur J Med Chem 222:113602. https://doi.org/10.1016/j.ejmech.2021.113602
Acknowledgements
The authors are thankful to the Higher Education Commission (HEC) of Pakistan for providing financial support through the NRPU Project # 7800. The authors also express their appreciation to the Deanship of Scientific Research at King Khalid University Saudi Arabia for funding through research groups program under Grant Number R.G.P. 2/3/42.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest:
The authors report no declarations of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sumrra, S.H., Zafar, W., Javed, H. et al. Facile synthesis, spectroscopic evaluation and antimicrobial screening of metal endowed triazole compounds. Biometals 34, 1329–1351 (2021). https://doi.org/10.1007/s10534-021-00345-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10534-021-00345-6