Abstract
A series of complexes of type [ML(CH3COO)(OH2)2] (M: Co, Ni; HL: 2-[(E)-1H-1,2,4-triazol-3-ylimino)methyl]phenol)) and [M2L2(CH3COO)2(OH2)n] (M: Cu, n = 2; M: Zn, n = 0) were synthesised by template condensation. The compounds were characterised with microanalytical, ESI–MS, IR, electronic, EPR spectra and magnetic data at room temperature. Based on the IR and ESI–MS spectra, a dinuclear structure with the acetate as bridge was proposed for Cu(II) and Zn(II) complexes. The dinuclear structure of Cu(II) complex is also consistent with both magnetic behaviour and EPR spectrum. The thermal analyses have evidenced processes as water elimination, acetate decomposition, as well as oxidative degradation of the Schiff base. The final decomposition product was the most stable metal oxide as indicated by powder X-ray diffraction. The cobalt and copper compounds exhibited a broad spectrum of antibacterial activity towards both planktonic and biofilm-embedded cells. The complexes exhibit a low cytotoxicity except for Cu(II) species that induces the early apoptosis for the HEp 2 cells.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Schiff bases represent an interesting class of ligands widely used in the coordination chemistry of transition, inner transition and main group metals. The azomethine linkage was found to be responsible for a broad spectrum of biological activities, such as antimicrobial [1], antiviral [2, 3] and anti-inflammatory [4]. The polyfunctional species based on nitrogen heterocyclic derivatives as 1,2,4-triazole are important having in view their anti-inflammatory [5] or cytotoxic [6] activity. As a result, triazole ring is widely used in the synthesis of Schiff bases, and some complexes with such ligands were evaluated and demonstrated the ability to catalyze superoxide decomposition [7] or a nuclease-like activity [8].
On the other hand, some transition metal ions play a vital role in different biological processes, and their activity can be enhanced by coordination to a Schiff base. Thus, complexes of Schiff bases derived from heterocyclic compounds containing nitrogen as donor atoms were studied as structural, spectral or functional models of complicated biological systems as hydrolytic enzymes [9, 10]. This type of complexes has been also investigated having in view useful properties as reversible binding of oxygen [11, 12], oxidation or binding and cleavage of DNA [13–17]. Moreover, the antimicrobial [18–22], antiviral [23] or cytotoxic [24] activity was reported for some of these species.
In order to overcome the challenging problem of microbial resistance, there is an increasing need for the development of novel antimicrobials, acting both in suspension and to adherent microbial cells, with acceptable toxicity. In the last time, the attention in the field was directed to inorganic compounds, some complexes proving a good activity mostly on assays on planktonic strains [25].
In the present work, we report the synthesis and thermal characterization of Co(II), Ni(II), Cu(II) and Zn(II) complexes containing Schiff base derived from 3-amino-4H-1,2,4-triazole and salicylaldehyde. The complexes were characterised by elemental analyses, IR and UV–Vis-NIR spectra as well as thermogravimetric analysis. Having in view that complexes are sparingly soluble in water, a proper drug delivery systems developing requires the thermal analysis, amongst others. As a result, the thermal analysis (TG, DTG and DTA) of the complexes was performed in order to elucidate the composition, the number and nature of the water molecules and also the behaviour at heating. The bioevaluation of antimicrobial and cytotoxicity activity of the obtained compounds was also performed.
Experimental
Materials
The high purity reagents were purchased from Sigma-Aldrich Co(CH3COO)2·4H2O, Ni(CH3COO)2·4H2O, Cu(CH3COO)2·H2O, Zn(CH3COO)2·2H2O), Loba (salicylaldehyde) and Fluka (3-amino-4H-1,2,4-triazole) and were used as received without further purification.
Instruments
Chemical analysis of carbon, nitrogen and hydrogen has been performed using a Perkin Elmer PE 2400 analyzer.
The molar conductance was determined for 10−3 M solutions of complexes in DMSO with a Multi parameter analyser CONSORT C861.
Mass spectra were recorded by electrospray ionization tandem mass spectrometry (ESI–MS) technique operating both in the positive and in negative ion mode. The sample was dissolved in acetonitrile:water (1:1) containing 0.2 % formic acid, and the solution was injected directly in Agilent triple quadrupole mass spectrometer with an electrospray interface.
IR spectra were recorded in KBr pellets with a Bruker Tensor 37 spectrometer in the range 400–4,000 cm−1.
1H NMR spectra were recorded on a Bruker DPX 200 spectrometer (working frequency 200 MHz) at 25 °C. Chemical shifts were measured in parts per million from internal standard TMS.
Electronic spectra by diffuse reflectance technique, with spectralon as standard, were recorded in the range 200–1,500 nm, on a Jasco V670 spectrophotometer.
Magnetic measurements were done by Faraday’s method, at room temperature, using Hg[Co(NCS)4] as a standard. The molar magnetic susceptibilities were calculated and corrected for the atomic diamagnetism.
The X-band EPR measurements were recorded at 20 °C for solid samples and at −173 °C for DMSO solutions, on a JEOL FA100 spectrometer. The general settings used were as follows: sweep field 1,000 G, frequency 100 kHz, gain in the range 100–200, sweep time 1,800 s, time constant 1 s, modulation width 2 G and microwave power 1 mW. The magnetic field calibration was performed with a DPPH (diphenylpicrylhydrazyl) standard marker, exhibiting a narrow EPR line at g = 2.0036.
The heating curves (TG, DTG and DTA) were recorded using a Labsys 1200 SETARAM instrument with a sample mass of 6–17 mg over the temperature range of 20–900 °C, using a heating rate of 10 °C min−1. The measurements were carried out in synthetic air atmosphere (flow rate 16.66 cm3 min−1) by alumina crucibles.
The X-ray powder diffraction patterns were collected on a DRON-3 diffractometer with a nickel filtered Cu Kα radiation (λ = 1.5418 Å) in 2θ 5–70° range a step width of 0.05° and an acquisition time of 2 s per step.
Antimicrobial assays
The antimicrobial activity of the obtained compounds was assayed on a large spectrum of microbial strains, i.e. Gram-negative (Escherichia coli ATCC 25922, E. coli 832, Klebsiella pneumoniae ATCC 134202, K. pneumoniae 806, Pseudomonas aeruginosa ATCC 27853) and Gram-positive (Staphylococcus aureus MRSA 1263, S. aureus ATCC 25923, Bacillus subtilis ATCC 6633) bacterial strains, as well as Candida albicans 22 fungal strain. Microbial suspensions of 1.5 × 108 CFU mL−1 (0.5 McFarland density) obtained from 15 to 18 h bacterial cultures developed on solid media were used in our experiments. The antimicrobial activity was tested on Mueller–Hinton Agar (MHA) medium, while yeast peptone glucose (YPG) medium was used in case of C. albicans. The compounds (ligand and complexes) were solubilised in DMSO, and the starting stock solution was of 1,000 μg mL−1 concentration. The qualitative screening was performed by an adapted dis diffusion method as previously reported [25].
The quantitative assay of the antimicrobial activity was performed by the liquid medium microdilution method, in 96 multi-well plates, in order to establish the minimal inhibitory concentration (MIC). In this purpose, serial twofold dilutions of the compounds ranging between 1,000 and 1.95 µg mL−1) were performed in a 200 μL volume of broth, and each well was seeded with 50 μL microbial inoculum. Sterility control (wells containing only culture medium) and culture controls (wells containing culture medium seeded with the microbial inoculum) were used. The influence of the DMSO solvent was also quantified in a series of wells containing DMSO, diluted accordingly with the dilution scheme used for the complexes. The plates were incubated for 24 h at 37 °C, and MIC values were considered as the lowest concentration of the tested compound that inhibited the visible growth of the microbial overnight cultures.
The assessment of the complexes influence on the microbial ability to colonize an inert substratum was performed by the microtiter method, following previously described protocols [25]. The absorbance at 490 nm was measured with an ELISA reader Apollo LB 911. All biological experiments were performed in triplicates.
Cytotoxicity assay
The cytotoxicity evaluation of ligand and complexes solution in DMSO was performed on human tumor cell line HEp 2 (human laryngeal carcinoma). A number of 5x105 HEp 2 cells were seeded in 3.5 cm diameter Petri dishes in DMEM/F12 (Gibco, NY, SUA) supplemented with 10 % heat-inactivated bovine serum and 100 µg mL−1 DMSO solution of the compound. After 24 h, the effect of the compound was evaluated using Annexin V-FITC Apoptosis Detection Kit I (BD Bioscience Pharmingen, USA), according to manufacturer protocol. Briefly, the cells were suspended in 100 µL of binding buffer and stained with 5 µL Annexin V-FITC and 5 µL propidium iodide (PI) for 10 min in dark. The analysis was performed by a Beckman Coulter flow cytometer, and at least 10,000 events from each sample were acquired and analysed using FlowJo software [26, 27].
Cell cycle analysis
A number of 5 × 105 HEp 2 cells were plated in 3.5 cm diameter Petri dishes for 24 h and treated with 100 µg mL−1 DMSO solution of the compound. After 24 h, the HEp 2 cells were taken from the substratum, fixed in 70 % cold ethanol for at least 30 min at −20 °C, washed twice in phosphate-buffered saline (PBS), and then incubated for 15 min, at 37 °C, with RNAse A (1 µg mL−1), and 1 h with propidium iodide (100 µg mL−1). After cells staining, the acquisition was done using an Epics Beckman Coulter flow cytometer. Data were analysed using the FlowJo software and expressed as fractions of cells found in different cell cycle phases [26, 27].
Synthesis, analytical and spectral data for ligand and complexes
To a solution containing 5 mmol metal (II) acetate hydrated (M: Co, Ni, Cu and Zn) in 50 mL ethanol, there were added 5 mmol salicylaldehyde and 5 mmol 3-amino-4H-1,2,4-triazole. The reaction mixture pH was adjusted by adding few drops of ammonia hydroxide, except for Co(II) system, and then was magnetically stirred at 50 °C for 4–5 h, until a sparingly soluble species was formed. The precipitate was filtered off, washed with ethanol and air-dried. The colour of complexes is dark red for (1), light green for (2), olive for (3) and light yellow for (4).
[CoL(CH3COO)(OH2)2] (1): Analysis found: C, 38.84; H, 4.05; N, 16.43, CoC11H14N4O5 requires: C, 38.72; H, 4.14; N, 16.42; ΛM, 13.5 Ω−1 cm2 mol−1.
[NiL(CH3COO)(OH2)2] (2): Analysis found: C, 38.82; H, 4.06; N, 16.47, NiC11H14N4O5 requires: C, 38.75; H, 4.14; N, 16.43; ΛM, 13.0 Ω−1 cm2 mol−1.
[Cu2L2(CH3COO)2(OH2)2] (3): Analysis found: C, 40.59; H, 3.63; N, 17.15, Cu2C22H24N8O8 requires: C, 40.31; H, 3.69; N, 17.09; ΛM, 14.5 Ω−1 cm2 mol−1.
[Zn2L2(CH3COO)2] (4): Analysis found: C, 42.43; H, 3.04; N, 18.03, Zn2C22H20N8O6 requires: C, 42.40; H, 3.23; N, 17.98; ΛM, 12.8 Ω−1 cm2 mol−1.
The ligand was synthesised by a literature procedure [28] and identified through chemical analysis, IR (Table 1) and 1H NMR spectra. 1H NMR (DMSO) δ (ppm): 5.77 (s, NH), 7.04 (dd, J = 6.40, 3.00 Hz, Ar–H), 7.49 (dd, J = 6.78; 2.50 Hz, Ar–H), 7.81 (d, J = 6.10 Hz, Ar–H), 8.46 (s, CH=N), 9.46 (s, CH), 12.40 (s, OH).
Results and discussion
The condensation of 3-amino-4H-1,2,4-triazole and salicylaldehyde in cobalt(II), nickel (II), copper(II) or zinc (II) acetate presence produced the species [ML(CH3COO)(OH2)2] (M: Co, Ni; HL: 2-[(E)-(1H-1,2,4-triazol-3-ylimino)methyl]phenol)) and [M2L2(CH3COO)2(OH2) n ] (M: Cu, n = 2; M: Zn, n = 0) (Scheme 1) as chemical analyses indicates. The ligand was synthesised by [1 + 1] condensation of 3-amino-4H-1,2,4-triazole and salicylaldehyde by a method reported in literature [28]. The complexes were characterised based on IR, UV–Vis-NIR and EPR spectroscopy, magnetic and thermal analysis data, some of them indicating the self deprotonation of Schiff base upon coordination.
The complexes behave as non-electrolyte as their molar conductance values (in DMSO) are in the range 12.8–14.5 Ω−1 cm2 mol−1 [29].
Infrared spectra
The most relevant IR bands and assignments for the complexes are summarised in Table 1. The formation of Schiff base was confirmed by the absence of characteristic bands of the aldehyde and primary amine groups stretching vibrations in spectra of both ligand and complexes. Furthermore, the appearance of a strong band in the range 1,615–1,625 cm−1 assigned to azomethine ν(HC=N) vibration is observed. The band assigned to ν(C=N) vibration for the triazole unit appear at lower wave numbers in the range 1,577–1,582 cm−1 as a result of withdrawing character of this ring [28] and is slightly shifted in comparison with ligand. In complexes, the band corresponding to azomethine linkage is shifted towards higher wavenumbers by 5–10 cm−1, indicating the ligand coordination through the nitrogen of this group [30–32].
The medium band at 3,375 cm−1 in the ligand spectrum disappears from the complexes spectra as an indicative of phenolic group deprotonation. Instead, the band at 1,277 cm−1 assigned to ν(C–O) vibration is shifted also by 50–88 cm−1 in comparison with the ligand, as a result of both deprotonation and coordination of phenolic oxygen [33].
In the absence of the single-crystal X-ray studies, the difference (Δ) between ν as(COO) and ν s(COO) stretching vibrations is often used as a criterion for establishing the nature of acetate in complexes, respectively, free ion, unidentate or bidentate (bridge or chelate) ligand. A Δ value lower than 170 cm−1 indicates a bidentate mode (chelate or bridge), but usually Δ(chelate) < Δ(bridge) [34]. This value can be moreover influenced by the hydrogen bonds [35] or other non-covalent interactions in solid state.
As a result, a Δ value in the range 101–142 cm−1 can be considered an indicative of the acetate coordination as bidentate for all complexes, as observed for similar species characterised by single-crystal X-ray diffraction. A second criterion used to differentiate the chelate for bridge coordination takes into consideration the shifting of the band assigned to ν as(COO) in comparison with CH3COONa. For complexes (1) and (2), a difference about 100 cm−1 correlated with the shifting of the band assigned to ν as(COO) to lower wave numbers, suggest the acetate presence as chelate [36, 37]. Instead, a difference of 142 and 126 cm−1, respectively, indicates for complexes (3) and (4) rather bridge coordination [38, 39].
In the characteristic ranges for water, a broad band in the 3,400–3,450 cm−1 range can be assigned to ν(OH) stretching vibrations, except for complex (4) [38]. Moreover, the new band in the range 580–610 cm−1 assigned to ρ w(H2O) supports the coordination of some water molecules [40].
NMR and ESI–MS spectra
In the 1H NMR spectrum of ligand, the peak for proton of phenolic group appears as a singlet at 12.40 ppm, whilst the singlet at 8.46 ppm is characteristic for azomethine group proton [41].
The 1H NMR spectrum of complex (4) was not possible to register, having in view the low solubility of this compound in all deuterated solvents.
The m/z values identified in the MS spectra confirm the structures proposed in Scheme 1 for complexes. Thus, in the positive mode, the pseudomolecular ions were found as [CoL(CH3COO) + CH3CN + 4H]+ (m/z: 350.10), [NiL(CH3COO)(OH2)2–2H]+ (m/z 338.40), [Cu2L2(CH3COO)2 + 2CH3–H]+ (m/z 648.40) and [Zn2L2(CH3COO) + H]+ (m/z 565.00), respectively. Moreover, in the negative mode, the pseudomolecular ions [Cu2L2(CH3COO)2 + 4H + OH]− (m/z 640.30) and [Zn2L2(CH3COO)2 + OH]+ (m/z 640.40) represent a proof for the dimeric structure of complexes (3) and (4), respectively. The fragmentation pattern is complex for all species, and as a result, the other peaks were difficult be assigned.
Electronic, EPR spectra and magnetic moments
The electronic spectrum of the cobalt (II) complex exhibits a pattern characteristic for octahedral distorted species with band ν1 splitted in two components as a result of different nature of donor atoms [42] as is presented in Table 2. The splitting parameter value of 10,190 cm−1 is close to an average value for a [Co(II)NO5] chromophore [43], while the nephelauxetic parameter of 0.61 is an indicative of a higher degree of covalency. The mean value for ν 1 band was calculated taking into consideration the energy barycentre of their components, while the crystal field parameters (10Dq, B and β) have been calculated with König formulas [44]. The Co(II) complex exhibits a magnetic moment of 4.54 B.M. at room temperature, value in the range accepted for octahedral species (4.3–5.2 B.M.) with three unpaired electrons and orbital contribution [43].
The Ni(II) complex exhibits octahedral geometry as indicated by the two bands at 630 and 1,065 nm that can be assigned to 3A2g → 3T1g(F) and 3A2g → 3T2g spin allowed transitions, respectively [42]. The splitting parameter value of 9,390 cm−1 is according with an [Ni(II)NO5] chromophore having a high number of donor atoms that generate a low field, such oxygen ones [43], while the nephelauxetic parameter of 0.85 is an indicative of a lower degree of covalency. This stereochemistry is further sustained by the room temperature magnetic moment of 3.26 B.M., which is close to the spin-only magnetic moments (3.3 B.M.) for two unpaired electrons, as usual for complexes with an A ground term [43].
The electronic spectrum of the Cu(II) complex shows the characteristic broad band with maximum at 680 nm tentatively assigned to 2B1 → 2E transition in square pyramidal stereochemistry. The value of 1.66 B.M. for the magnetic moment indicates for Cu(II) complex a weak interaction between paramagnetic ions with S = 1/2 at room temperature that may arises from an antiferromagnetic interaction mediated by acetato bridge [43].
A tetrahedral stereochemistry was tentatively proposed for the Zn(II) complex, considering that both ligands are negatively charged and as result, such a stereochemistry minimizes their repulsions [43].
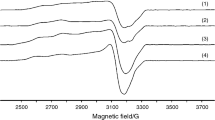
EPR spectra of Cu(II) complexes usually reveal information concerning both local surrounding of metal ion and monomeric or dimeric structure. The X-band EPR spectrum both in powder and frozen DMSO solution of complex (2) exhibits a broad and low intensity signal with g av = 2.1483 characteristic to local symmetry of copper ion lower than an octahedral such as the square pyramidal one. This aspect of the spectrum can be related to a dimeric structure for a ion pair system with magnetically isolated centres [45].
Thermal behaviour
Thermal analysis techniques represent a useful tool for determining the composition and stability of complexes [46, 47]. Furthermore, the understanding of the thermal behaviour of biological active species represents an important step in order to develop new metallotherapeutics.
In order to obtain such information, the thermal behaviour of ligand and complexes was investigated by simultaneous TG/DTA analysis, while the final residue was examined by powder X-ray diffraction. The species isolated after the water elimination were also isolated and characterised in order to establish its presence as ligand or as hydration species.
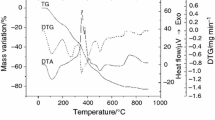
The simultaneous TG/DTA curves registered for the ligand are shown in Fig. 1, and indicate that this species undergoes a three steps thermal decomposition. The data collected from these curves are summarised in Table 3.
The first step of thermal degradation of ligand consists in an endothermic elimination of water up to 155 °C followed by its melting at 172 °C. The melting point was considered from the maximum of DTA peak having in view that this process is partially overlapped by the water elimination. The thermal decomposition of anhydrous species begins at 195 °C with eliminations of 14 % from Schiff base, process accompanied by several exothermic effects according with DTA curve. The next step is not a single one, but an overlapping of several processes, according to DTA curve profile. The oxidative degradation of the rest of Schiff base proceeds with elimination of about 12 % from organic species. The small exothermic peaks observed in the 345–580 °C temperature range are probably due to both endothermic and exothermic reactions that occur simultaneously, such as cleavage and rearrangement of the bonds, as well as some moieties oxidative degradation. According with the mass loss, 44 % from the Schiff base is eliminated in this step, and the rest of organic part oxidative degradation occurs in the final step. As a result, several broad exothermic processes can be noticed on DTA curve.
The TG and DTA curves corresponding to the complex (1) indicate that decomposition follows four steps (Fig. 2). The first two endothermic steps consist in the stepwise release of water molecules up to 240 °C (Table 3), but the spectral data indicate the water presence as ligand. This assumption is furthermore supported by the electronic spectra of intermediates isolated at about 150 and 240 °C that preserves the pattern of parent spectrum, but the splitting parameters are different. Thus, their variation from 10,190 to 10,635 cm−1 and 10,815 cm−1, respectively, indicates an increase of crystalline field for intermediates. This behaviour can arise by the gradual replacement of coordinated water molecules through nitrogen atoms from the triazole moiety. Based on these observations, one may consider that the water molecules are involved in different interactions, one being more stronger attached through hydrogen bonds or π-OH2 interaction besides coordinative ones [48].
The anhydrous species undergoes then an exothermic transformation in the third step that consists in the transformation of acetate into carbonate together with the oxidative degradation of about 50 % from Schiff base, according with DTA curve. The thermal studies on mixed ligand complexes evidenced the fact that the coordinated acetate decomposes before other organic derivatives, such as Schiff bases [49, 50].
The final step is not a single one being an overlapping of several exothermic processes, according to both DTG and DTA curves profile. The oxidative degradation of the rest of Schiff base and carbonate decomposition occur as mass loss indicates. All these transformations lead to Co3O4 as final product (found/calc. overall mass loss: 76.4/76.5 %) (ASTM 78-1970).
For complex (2), the decomposition proceeds in three steps (Fig. 3). In the first endothermic step, the coordination water is eliminated up to 190 °C as was observed for other similar complexes [51]. The transformation of acetate into carbonate occurs in the second step. These two processes are not well delimitated each other, but the DTG curve sustains this assumption. The mass loss up to 848 °C corresponds to the oxidative degradation of the Schiff base and carbonate decomposition. Several overlapped processes occur until the NiO is formed, according with both DTA and DTG curves (ASTM 78-0429) (found/calc. overall mass loss: 78.0/78.1 %).
The first step for complex (3) is an endothermic one and according to the mass loss corresponds to water elimination up to 180 °C (Fig. 4) as an indicator of its presence as ligand [51]. The acetate decomposition together with the Schiff base oxidative degradation starts immediately, being accompanied by a strong exothermic effect. The small exothermic peaks observed in the 278–443 °C temperature range are probably due to both endothermic and exothermic reactions that occur simultaneously such as cleavage and rearrangement of the bonds as well as some moieties oxidative degradation. In the final step, both the rest of organic part oxidative degradation and the carbonate decomposition occur. As a result of these reactions, several exothermic processes can be noticed on DTA curve. All these transformations finally lead to copper (II) oxide (ASTM 5-661) (found/calc. overall mass loss: 75.6/75.7 %).
Complex (4) is an anhydrous species and as result, their thermal degradation starts at 286 °C and the decomposition pattern is different from that of other species from this series (Fig. 5). The first step corresponds to the elimination of about 14 % from the Schiff base and acetate transformation into carbonate. The bonds cleavage occurs as main processes in this step considering the strong endothermic effect noticed on DTA curve. The second step is not a single one, but an overlap of two processes, one endo- and one exothermic, that may correspond to bond cleavage and oxidative degradation of 68 % from the Schiff base ligand. The final step comprises several exothermic processes, as indicated by the DTA curve profile that corresponds to the organic component pyrolysis, carbonate decomposition and zinc (II) oxide generation (ASTM 036-1451) (found/calc. overall mass loss: 74.0/73.9 %).
Having in view all these data, a dimeric structure was proposed for complexes (3) and (4) in order to correlate all physico-chemical data, as shown in Scheme 1.
Antimicrobial activity assay
The emergence of multidrug-resistant Gram-positive and Gram-negative bacteria, including methicillin-resistant Staphylococcus aureus (MRSA), vancomycin-resistant S. aureus (VRSA), vancomycin-resistant enterococci (VRE), extended-spectrum β-lactamase (ESBL)-producing E. coli and K. pneumoniae, carbapenemase-producing Enterobacteriaceae or multidrug-resistant P. aeruginosa, has limited therapeutic options, in certain cases, the infections being untreatable with the existing antimicrobial agents [52, 53]. Fungal infections, mainly caused by Candida and Apergillus species, are also emerging in severely immunocompromised patients, being often produced by resistant strains [54, 55].
As a result, the antimicrobial activity of the obtained compounds was assayed using a wide range of bacterial and fungal strains, both reference and of clinical origin, belonging to the major pathogenic genera that produce difficult to treat human infections. It is to be pointed that the E. coli 832 and K. pneumoniae 806 strains were isolated from urine culture and wound secretion, respectively, and exhibited a multidrug resistance phenotype. It was observed that the used DMSO solved does not influence the antimicrobial activity of the tested compounds at the working concentrations. The assays revealed a very good antimicrobial activity for the complex (3) with a MIC value of 31.25 μg mL−1 in the case of E. coli ATCC 25922, K. pneumoniae ATCC 134202 and B. subtilis ATCC 6633 and a MIC of 62.50 μg mL−1 in case of C. albicans 22 fungal strain. The complex (2) had a MIC of 62.50 μg mL−1 had on the C. albicans 22 fungal strain. A good activity was also obtained for the complex (1) with a MIC value of 62.50 μg mL−1 in the case of E. coli ATCC 25922, K. pneumoniae ATCC 134202 and B. subtilis ATCC 6633 and of 31.25 μg mL−1 in the case of C. albicans 22 fungal strain (Table 4). The antimicrobial activity displayed by complex combinations was generally lower against the Gram-positive cocci, with MIC of 125 μg mL−1.
Moreover, medical microbial biofilms defined as sessile microbial communities composed of cells irreversibly attached to a substratum and between them, embedded in a self-produced matrix of extracellular polymeric substances are amplifying the genetic resistance [56] due to their phenotypical, behavioural and, more recently, called tolerance [57], mediated by different and incompletely elucidated mechanisms, such as (1) failure or restricted penetration of antimicrobial agents into biofilms; (2) biofilm cells entry in a latent metabolic state, in which they are no more susceptible to antibiotics; (3) selection of persistent cells and (4) gene expression changes leading to the occurrence of more tolerant phenotypes [58].
Having in view the differences in the physiology and susceptibility to antibiotics of biofilm-embedded microorganisms, the obtained complexes were investigated concerning their efficiency against the adherent cells grown in biofilms developed in plastic wells. From this point of view, it was observed that the complexes interact differently with the biofilm formed by different microbial strains. The complexes (1) and (3) exhibited the most evident inhibitory effect upon de adherence ability of K. pneumoniae ATCC 134202, S. aureus ATCC 25923, B. subtilis ATCC 6633 and C. albicans 22, up to the concentration of 15.62 μg mL−1. The complexes (2) and (4) also exhibited an important inhibitory effect upon the adherence ability of the C. albicans 22 fungal strain with a minimal biofilm eradication concentration (MBEC) of 15.62 μg mL−1 (Table 5). This aspect is important considering that the genetic resistance of different microbial strains to antimicrobials is amplified when pathogenic microorganisms are developing in biofilm.
The complexes activity may be related to several factors such as the stereochemistry, thermodynamic and kinetic stability, solubility, diffusion ability, and a combined effect of the metal and the ligands for the inactivation of specific microbial targets.
In order to reach the intracellular targets, the compounds can be either liposoluble in order to diffuse through the bilipidic layer of the microbial membrane or hydrophilic in order to be actively internalized by porins. The complexes (1)–(4) may have a lipophilic character having in view their non-electrolytic behaviour as well as the presence of hydrophobic periphery. As a result, after diffusion through the microbial membrane, these species may interfere with the cytoplasmatic components and thus affecting both the microbial cell viability and pathogenicity. The good activity of complexes (1) and (3) can be related to their ability to change easily the oxidation state as well as their stereochemical versatility.
Cytotoxicity assay
The treatment of HEp 2 cells with 100 µg mL−1 solution of the compound (2) induced a low percentage of apoptotic cells, 2.12 % for early apoptosis and 2.82 % for late apoptosis, respectively. The early apoptotic effect was more evident for the compound (3), for which 26.00 % of cells were positive for PI, and 8.89 % for both Annexin V and PI. The PI is impermeant for live cells and early apoptotic cells, but it stains dead cells with red fluorescence, binding tightly to the nucleic acids in the cell (Table 6). Apoptosis is a stringently regulated process of cellular death, that could be distinguished from necrosis based on phosphatidylserine (PS), that are normally located on the cytoplasmic surface of the cellular membrane. This protein is translocated from the inner to the outer leaflet of the plasmatic membrane in apoptotic cells, thus exposing PS to the external cellular environment and interaction with Annexin V/FTIC system.
The enhanced activity of compound (3) could arise from the known toxicity of copper compounds that generate reactive oxygen species (ROS) via Fenton or Haber–Weiss reactions. Another possible mechanism underlying copper-induced toxicity originating from the high affinity for thiol groups of cysteine residues of this ion that leads to the modification of the biological functions of many proteins [59].
On the other hand, the increase of cellular death induced by the compound (3) was also observed from the analysis of the cellular cycle, where for this complex only few cells reach in the cell growth and replication phases (Table 7).
The very good activity of complex (3) concerning both antimicrobial and cytotoxicity activity might be related to its lower stability that allows an easy ligand substitution and rapid interaction with target biomolecules. A low thermodynamic and kinetic stability of this complex is in accordance with its relatively lower thermal stability, having in view that its decomposition as anhydrous species starts at 180 °C.
Conclusions
The cobalt(II), nickel(II), copper(II) and zinc(II) complexes with Schiff base resulted in the condensation of salicylaldehyde and 3-amino-4H-1,2,4-triazole have been synthesised by template condensation.
The ligand nature and stereochemistry were characterised by means of IR and UV–Vis-NIR spectroscopy. The modifications in the IR spectra of complexes are in accordance with the condensation process, coordination of Schiff base through azomethinic nitrogen and phenolic oxygen and of acetate as bidentate ligand. Electronic spectra of Co(II) and Ni(II) complexes display the pattern of octahedral stereochemistry that is furthermore supported by magnetic moments at room temperature. The Cu(II) complex exhibits a square pyramidal stereochemistry according with electronic spectrum. The dimeric nature of Cu(II) and Zn(II) complexes is sustained by ESI–MS spectra.
Thermal decomposition of complexes allowed establishing the number and nature of water molecules, the composition and also the intervals of thermal stability. The thermal degradation occurs in three or four steps, respectively, and comprises water elimination, acetate into carbonate transformation, Schiff base decomposition and oxidative degradation. These results are in good concordance with the complexes composition.
The temperature corresponding to the decomposition of the anhydrous complexes increases in order (3) < (2) < (1) < (4) indicating that Cu(II) compound is the most covalent from this series.
The antimicrobial assays revealed a very good antimicrobial activity for the obtained complexes, especially towards the Gram-negative bacterial strains (E. coli and K. pneumoniae), followed by B. subtilis and C. albicans strains. The complexes (1) and (3) exhibited the most evident anti-biofilm effect, inhibiting the biofilm development in the inert substratum, in case of the majority of the tested strains, being promising agents for the development of new antimicrobial agents and surfaces.
The copper compound (3) exhibits the highest cytotoxicity that may result from either ROS generation or by cooper ion binding to proteins rich in cysteine residues.
References
Sumangala V, Poojary B, Chidananda N, Arulmoli T, Shenoy S. Synthesis and biological evaluation of some Schiff bases of 4-amino-5-(4-methylsulfonyl)benzyl-2,4-dihydro-3H-[1, 2, 4]-triazole-3-thione. Med Chem Res. 2013;22:2921–8.
Abbas S, Farag AA, Ammar YA, Trees AA, Mohamed F, El-Henawy AA. Synthesis, characterization, and antiviral activity of novel fluorinated isatin derivatives. Monatsh Chem. 2013;144:1725–33.
Jarrahpour A, Sheikh J, El Mounsi I, Juneja H, Hadda TB. Computational evaluation and experimental in vitro antibacterial, antifungal and antiviral activity of bis-Schiff bases of isatin and its derivatives. Med Chem Res. 2013;22:1203–11.
Chandramouli, Shivanand MR, Nayanbhai TB, Bheemachari, Udupi RH (2012) Synthesis and biological screening of certain new triazole Schiff bases and their derivatives bearing substituted benzothiazole moiety. J Chem Pharm Res 4: 1151–9.
Abdel-Megeed AM, Abdel-Rahman HM, Alkaramany G-ES, Mahmoud A, El-Gendy MA. Design, synthesis and molecular modeling study of acylated 1,2,4-triazole-3-acetates with potential anti-inflammatory activity. Eur J Med Chem. 2009;44:117–23.
Li X, Li X-Q, Liu H-M, Zhou X-Z, Shao Z-H. Synthesis and evaluation of antitumor activities of novel chiral 1,2,4-triazole Schiff bases bearing γ-butenolide moiety. Org Med Chem Lett. 2012;2:2–26.
Serbest K, Ozen A, Unver Y, Er M, Değirmencioğlu I, Sancak K. Spectroscopic and theoretical study of 1,2,4-triazole-3-one based salicylaldimine complexes and evaluation of superoxide-scavenging properties. J Mol Struct. 2009;922:1–10.
Patil SA, Kamble UV, Badami PS. Antimicrobial and DNA-cleavage studies of 22-membered N4 tetraaza macrocyclic triazoles: template synthesis and physicochemical characterization. Nucleosides Nucleotides Nucleic Acids. 2010;29:658–75.
Halcrow MA, Christou G. Biomimetic chemistry of nickel. Chem Rev. 1994;94:2421–81.
Banu KS, Chattopadhyay T, Banerjee A, Bhattacharya S, Zangrando E, Das D. Catechol oxidase activity of dinuclear copper(II) complexes of Robson type macrocyclic ligands: syntheses, x-ray crystal structure, spectroscopic characterization of the adducts and kinetic studies. J Mol Catal A Chem. 2009;310:34–41.
Basolo F, Hoffman BM, Ibers JA. Synthetic oxygen carriers of biological interest. Acc Chem Res. 1975;8:384–92.
Wei XY, Li JZ, Du Y, Qing SY. Dioxygen affinities and biomimetic catalytic oxidation performance of transition-metal complexes with unsymmetrical bis-Schiff bases. Chin Chem Lett. 2004;15:529–31.
Liu C, Wang M, Zhang T, Sun H. DNA hydrolysis promoted by di- and multi-nuclear metal complexes. Coord Chem Rev. 2004;248:147–68.
Patil SA, Naik VH, Kulkarni AD, Badami PS. DNA cleavage, antimicrobial, spectroscopic and fluorescence studies of Co(II), Ni(II) and Cu(II) complexes with SNO donor coumarin Schiff bases. Spectrochim Acta Part A. 2010;75:347–54.
Y-c Liu, Z-y Yang. Crystal structures, antioxidation and DNA binding properties of Dy(III) complexes with Schiff-base ligands derived from 8 hydroxyquinoline-2-carboxaldehyde and four aroylhydrazines. Eur J Med Chem. 2009;44:5080–9.
Khan TA, Naseem S, Khanb SN, Khan AU, Shakir M. Synthesis and spectral characterization of 14- and 16-membered tetraazamacrocyclic Schiff base ligands and their transition metal complexes and a comparative study of interaction of calf thymus DNA with copper(II) complexes. Spectrochim Acta Part A. 2009;73:622–9.
Yuan H, Wang X, Zhou H, Pan Z, Li J, Huang Q. DNA cleavage activities and mechanism of a novel heterodinuclear macrocyclic complex. Inorg Chem Commun. 2010;133:314–8.
Chohan ZH, Arif M, Shafiq Z, Yaqub M, Supuran CT. In vitro antibacterial, antifungal and cytotoxic activity of some isonicotinoylhydrazide Schiff’s bases and their cobalt (II), copper (II), nickel (II) and zinc (II) complexes. J Enzym Inhib Med Chem. 2006;21:95–103.
Chohan ZH, Pervez H, Rauf A, Khan KM, Supuran CT. Antibacterial cobalt (II), copper (II), nickel (II) and zinc (II) complexes of mercaptothiadiazole-derived furanyl, thienyl, pyrrolyl, salicylyl and pyridinyl Schiff bases. J Enzym Inhib Med Chem. 2006;21:193–201.
Aslantaş M, Kendi E, Demir N, Şabik AE, Tumer M, Kertmen M. Synthesis, spectroscopic, structural characterization, electrochemical and antimicrobial activity studies of the Schiff base ligand and its transition metal complexes. Spectrochim Acta Part A. 2009;74:617–24.
Mohamed GG, Omar MM, Ibrahim AA. Biological activity studies on metal complexes of novel tridentate Schiff base ligand. Spectroscopic and thermal characterization. Eur J Med Chem. 2009;44:4801–12.
Rosu T, Pahontu E, Maxim C, Georgescu R, Stanica N, Almajan GL, Gulea A. Synthesis, characterization and antibacterial activity of some new complexes of Cu(II), Ni(II), VO(II), Mn(II) with Schiff base derived from 4-amino-2,3-dimethyl-1-phenyl-3-pyrazolin-5-one. Polyhedron. 2010;29:757–66.
Al-Masoudi NA, Aziz NM, Mohammed AT. Synthesis and in vitro anti-HIV activity of some new Schiff base ligands derived from 5-Amino-4-phenyl-4H-1,2,4-triazole-3-thiol and their metal complexes. Phosphorus Sulfur Silicon Relat Elem. 2009;184:2891–901.
Chaviara AT, Kioseoglou EE, Pantazaki AA, Tsipis AC, Karipidis PA, Kyriakidis DA, Bolos CA. DNA interaction studies and evaluation of biological activity of homo- and hetero-trihalide mononuclear Cu(II) Schiff base complexes. Quantitative structure–activity relationships. J Inorg Biochem. 2008;102:1749–64.
Calu L, Badea M, Falcescu D, Duca D, Marinescu D, Olar R. Thermal study on complexes with Schiff base derived from 1,2,4-triazole as potential antimicrobial agents. J Therm Anal Calorim. 2013;111:1725–30.
Awtar K. Rapid flow cytofluorometric analysis of mammalian cell cycle by propidium iodide stainin. J Cell Biol. 1975;66:188–93.
Hawley BRG, Hawley TS. Flow cytometry protocols. 2nd ed. New York: Humana Press; 2004.
Hassan FSM, Mohamed AE, Gabr AA, Gad AA. The preparation and characterisation of divalent copper, nickel, cobalt and manganese complexes of some Schiff base ligands. Science. 1994;6:125–34.
Geary WJ. The use of conductivity measurements in organic solvents for the characterisation of coordination compounds. Coord Chem Rev. 1971;7:81–122.
Chohan ZH, Shad HA, Youssoufi MH, Hadda TB. Some new biologically active metal-based sulfonamide. Eur J Med Chem. 2010;45:2893–901.
Chohan ZH, Sumrra SH, Youssoufi MH, Hadda TB. Metal based biologically active compounds: design, synthesis, and antibacterial/antifungal/cytotoxic properties of triazole-derived Schiff bases and their oxovanadium(IV) complexes. Eur J Med Chem. 2010;45:2739–47.
Reddy V, Patil N, Reddy T, Angadi SD. Synthesis, characterization and biological activities of Cu(II), Co(II), Ni(II), Mn(II) and Fe(III) complexes with Schiff base derived from 3-(4-chlorophenoxymethyl)-4-amino-5-mercapto-1,2,4-triazole. J Chem. 2008;5:529–38.
Abdel-Latif SA, Hassib HB, Issa YM. Studies on some salicylaldehyde Schiff base derivatives and their complexes with Cr(III), Mn(II), Fe(III), Ni(II) and Cu(II). Spectrochim Acta Part A. 2007;67:950–7.
Deacon GB, Philips JR. Relationships between the carbon-oxygen stretching frequencies of carboxylato complexes and the type of carboxylate coordination. Coord Chem Rev. 1980;33:227–50.
Zeleńák V, Vargová Z, Györyová K, Večerníková E, Balek V. Cooper(II) acetates with aliphatic/heterocycles amines coupled TG-DTA-EGA study, IR characterization and structure correlation. J Therm Anal Calorim. 2005;82:747–54.
Dojer B, Golobič A, Jagličić Z, Krist M, Drofenik M. Two new nickel(II) carboxylates with 3- and 4-aminopyridine: syntheses, structures, and magnetic properties. Monatsh Chem. 2012;143:73–8.
Karmakar T, Kuang Y, Neamati N, Baruah JB. Cadmium complexes and cocrystals of indium complexes of benzothiazole derivatives and anticancer activities of the cadmium complexes. Polyhedron. 2013;54:285–93.
Hernández-Gil J, Ferrer S, Cabedo N, López-Gresa MP, Castiñeiras A, Lloret F. Two copper complexes from two novel naphthalene-sulfonyl-triazole ligands: different nuclearity and different DNA binding and cleavage capabilities. J Inorg Biochem. 2013;125:50–63.
Menzel R, Weiß D, Täuscher E, Beckert R, Görls H. Formation, X-ray structure and magnetic properties of two carboxy-bridged copper(II) complexes with tridentate bipyridine- and phenanthroline-based 1,3-thiazoles ligands. Inorg Chem Com. 2012;18:65–8.
Nakamoto K. Infrared and raman spectra of inorganic and coordination compounds. New York: Wiley; 1986.
Chohan ZH, Hanif M. Syhthesis and characterization of biologically active new Schiff bases containing 3-functionalized 1,2,4-triazoles and their zinc(II) complexes: crystal structure of 4-bromo-2-[(E)-(1H-1,2,4-triazol-3-ylimino)-methyl]phenol. Appl Organomet Chem. 2011;25:753–60.
Lever ABP. Inorganic electronic spectroscopy. Amsterdam: Elsevier; 1986.
Gispert JR. Coordination chemistry. Weinheim: Wiley-VCH; 2008.
König E. The nephelauxetic effect. Calculation and accuracy of the interelectronic repulsion parameters I. Cubic high-spin d 2, d 3, d 7 and d 8 systems. Struct Bound. 1972;9:175–372.
Smtih TD, Pilbrow JR. The determination of structural properties of dimeric transition metal ion complexes from EPR spectra. Coord Chem Rev. 1974;13:173–278.
Tatucu M, Rotaru P, Rau I, Spinu C, Kriza A. Thermal behaviour and spectroscopic investigation of some methyl 2-pyridyl ketone complexes. J Therm Anal Calorim. 2010;100:1107–14.
Zianna A, Vecchio S, Gdaniec M, Czapik A, Hatzidimitriou A, Lalia-Kantouri M. Synthesis, thermal analysis, and spectroscopic and structural characterizations of zinc(II) complexes with salicylaldehydes. J Therm Anal Calorim. 2013;112:455–64.
Meyer EA, Castellano RK, Diederich F. Interactions with aromatic rings in chemical and biological recognition. Angew Chem Int Ed. 2003;42:1210–50.
Singh K, Kumar Y, Pari P, Kumar M, Sharma C. Cobalt, nickel, copper and zinc complexes with 1,3-diphenyl-1H-pyrazol-4-carboxyaldehyde Schiff bases: antimicrobial, spectroscopic, thermal and fluorescence studies. Eur J Med Chem. 2012;52:313–21.
Dong W-K, Sun Y-X, He X-N, Tong J-F, Wu J-C. Trinuclear and mononuclear copper(II) complexes incorporating tetradentate 2,2′-[1,1′(ethylenedioxydinitrilo)diethylidyne]diphenol ligand: syntheses, crystal structures, spectral and thermal behaviors. Spectrochim Acta Part A. 2010;76:476–83.
Patil SA, Unki SN, Badami PS. Synthesis, characterization, biological and thermal behaviour of Co(II), Ni(II) and Cu(II) complexes with Schiff bases having coumarin moieties. J Therm Anal Calorim. 2013;111:1281–9.
Coates A, Hu Y, Bax R, Page C. The future challenges facing the development of new antimicrobial drugs. Nat Rev Drug Discov. 2002;1:895–910.
Da Re S, Garnier F, Guerin E, Campoy S, Denis F, Ploy MC. The SOS response promotes qnrB quinolone-resistance determinant expression. EMBO Rep. 2009;10:929–33.
Kriengkauykiat J, Ito JI, Dadwal SS. Epidemiology and treatment approaches in management of invasive fungal infections. Clin Epidemiol. 2011;3:175–91.
Chapeland-Leclerc F, Hennequin C, Papon N, Noël T, Girard A, Socié G, Ribaud P, Lacroix C. Acquisition of flucytosine, azole, and caspofungin resistance in Candida glabrata bloodstream isolates serially obtained from a hematopoietic stem cell transplant recipient. Antimicrob Ag Chemother. 2010;54:1360–2.
Donlan RM, Costerton JW. Biofilms: survival mechanisms of clinically relevant microorganisms. Clinical Microbiol Rev. 2002;15:167–93.
Lazăr V, Chifiriuc MC. Architecture and physiology of microbial biofilms. Roum Arch Microbiol Immunol. 2010;69:92–8.
Lazăr V. Microbial adherence. Bucharest: Romanian Academy Publications House; 2003.
Letelier ME, Lepe AM, Faúndez M, Salazar J, Marín R, Aracena P, Speisky H. Possible mechanisms underlying copper-induced damage in biological membranes leading to cellular toxicity. Chem Biol Interact. 2005;151:71–82.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Badea, M., Calu, L., Chifiriuc, M.C. et al. Thermal behaviour of some novel antimicrobials based on complexes with a Schiff base bearing 1,2,4-triazole pharmacophore. J Therm Anal Calorim 118, 1145–1157 (2014). https://doi.org/10.1007/s10973-014-3821-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10973-014-3821-4