Abstract
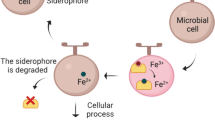
Siderophores are biosynthetically produced and secreted by many bacteria, yeasts, fungi and plants, to scavenge for ferric iron (Fe3+). They are selective iron-chelators that have an extremely high affinity for binding this trivalent metal ion. The ferric ion is poorly soluble but it is the form of iron that is predominantly found in oxygenated environments. Siderophore uptake in bacteria has been extensively studied and over the last decade, detailed structural information for many of the proteins that are involved in their transport has become available. Specifically, numerous crystal structures for outer membrane siderophore transporters, as well as for soluble periplasmic siderophore-binding proteins, have been reported. Moreover, unique siderophore-binding proteins have recently been serendipitously discovered in humans, and the structures of some of their siderophore-complexes have been characterized. The binding pockets for different ferric-siderophores in these proteins have been described in great molecular detail. In addition to highlighting this structural information, in this review paper we will also briefly discuss the relevant chemical properties of iron, and provide a perspective on our current understanding of the human and bacterial iron uptake pathways. Potential clinical uses of siderophores will also be discussed. The emerging overall picture is that iron metabolism plays an extremely important role during bacterial infections. Because levels of free ferric iron in biological systems are always extremely low, there is serious competition for iron and for ferric-siderophores between pathogenic bacteria and the human or animal host.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
The interplay between iron chemistry and its role in metabolism
Iron is one of the most abundant elements in the crust of the earth. It can exist in two oxidation states but in the presence of high concentrations of oxygen it is exclusively present in the ferric (Fe3+) form, which is not highly soluble. The ferrous (Fe2+) ion has a much better solubility, but it only exists under anoxic conditions or at very low pH. In aqueous solution iron can spontaneously become involved in Fenton chemistry, where toxic hydroxyl radicals are generated that would seriously harm a living organism. In spite of these two serious limitations all living organisms on earth, with the exception of a few specific bacterial strains, have an absolute requirement for iron to survive and to thrive. Several essential and highly conserved enzymes, such as the ribonucleotide reductases that produce deoxyribonucleic acids (DNA), require iron to carry out their function. Other well-known proteins such as hemoglobin and myoglobin utilize the same element to bind and transport oxygen in blood and muscle cells, respectively. As part of the cytochrome and cytochrome oxidase systems, iron also plays a major role in the generation of energy by mitochondria and bacterial cells (Crichton 2009). When it is bound to a protein the iron can be incorporated in prosthetic groups such as iron-sulfur clusters or the characteristic heme group, while other proteins such as transferrin can bind the metal ion directly. To fulfill their needs for iron all living organisms have their own dedicated highly specialized systems to solubilize, transport and store ferric or ferrous iron. Moreover, within a living organism as well as within a living cell, iron is always sequestered by proteins or other ligands in order to maintain solubility and to prevent the accidental formation of toxic radicals.
How do we acquire and control the levels of iron?
Humans and animals obtain iron directly from their diet. The metal ion is taken up in the ferrous form by the divalent metal ion transporter (DMT) proteins in the gut. However, after it enters the bloodstream, where oxygen is prevalent, it is converted to the ferric form. In serum the Fe3+ is normally bound tightly by the circulating transferrin protein, which can deliver the metal ion into cells that have a high demand for iron by binding to specific cell-surface transferrin receptors. Internalization of the transferrin receptor protein–protein complexes into endosomes allows for the intracellular release of iron and at the same time this process is extremely efficient as it also gives rise to the recycling of the receptor protein on the cell surface as well as the re-release of transferrin into the bloodstream (Crichton 2009). By far the largest amount of iron in our bodies is found in hemoglobin in circulating red blood cells. At the end of their lifespan the erythrocytes are consumed by macrophages and these carefully recycle the iron. As such, our daily requirements for iron are quite low and we only need to take up a few mg per day. The major storage for iron in our body is found in the liver, where the metal ion is stored in an insoluble form inside the multi-subunit ferritin protein (Harrison and Arosio 1996). The process of iron uptake and release from the hepatic or macrophage storage sites has to be carefully regulated. In some individuals, the iron uptake process is poorly controlled which can lead to iron overload diseases such as hereditary hemochromatosis. Conversely when there is not enough iron taken up from the diet some people can suffer from anemia. The recently discovered peptide hormone hepcidin plays an intricate role in orchestrating the regulation of iron uptake and release in our bodies. Hepcidin was originally purified as an antimicrobial peptide that is synthesized and secreted by the liver; later studies with knock-out mice revealed its essential role as the major control mediator for iron metabolism (Ganz 2003). Hepcidin has been found in all mammals, in amphibians and in some species of fish. This 25-residue peptide has an unusual highly disulfide crosslinked beta hairpin structure, which seems to be similar in human and fish hepcidin (Hunter et al. 2002; Lauth et al. 2005). There is still a debate about the exact positioning for two out of the four disulfide linkages (Jordan et al. 2009), but this only has a minor impact on the overall structure and this difference occurs in a region that seems to be removed from the ‘active site’ of hepcidin (Nemeth et al. 2005). Hepcidin acts by binding directly to ferroportin, the membrane transporter protein that is responsible for releasing iron from cells. In doing so hepcidin causes the internalization and degradation of the ferroportin and subsequently these cells, such as the macrophages, hepatocytes and enterocytes, are no longer able to release iron from intracellular stores (Nemeth et al. 2004). Clearly the uptake of iron is strictly controlled to maintain the level of iron in the body. The recent advances in understanding hepcidin action are likely to be translated into improved diagnostic and therapeutic modalities in the near future (Anderson et al. 2009).
Bacterial iron uptake and storage
The vast majority of the bacteria also need to acquire a substantial amount of iron to survive. In the environment bacteria are faced with the problem that the iron concentration in water is extremely low and that the iron in the soil is present in an insoluble form. To overcome these barriers many bacteria can synthesize ‘siderophores’ which are chelators that are specific for binding Fe3+ (van der Helm et al. 1987; Neilands 1995; Raymond et al. 2003). The siderophores, which are named after the Greek word for iron carriers, bind this relatively “hard metal ion” with great avidity; conversely they have a much lower affinity for Fe2+, which is from a chemical perspective, a much “softer metal ion”. Dissociation constants of better than 10−20 M have been reported for many of these iron-chelating agents. Siderophores are also produced and secreted by yeast, fungi and many plants; in fact numerous bacteria, including Escherichia coli, are quite opportunistic as they can utilize fungal siderophores, such as ferrichrome, in addition to the enterobactin and related compounds that they produce themselves. Most siderophores are biosynthetically produced by large multienzyme synthases that resemble the eukaryotic fatty acid synthases (Crosa and Walsh 2002; Barry and Challis 2009; Chan and Vogel 2010). They often have a peptidic backbone, with modified amino acid side chains creating the iron-coordinating ligands, such as the catecholate, hydroxamate and hydroxy-carboxylate groups (see Fig. 1). Each group provides two oxygen ligands in a fixed orientation, and a total of six ligands normally surrounds the ferric iron in an octahedral fashion.
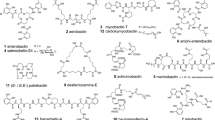
Crystal structures of three representative siderophores: enterobactin, ferrichrome and staphyloferrin A are depicted without (left side) and with Fe3+ bound (right side). Ferrichrome (a and b), a hydroxamate type siderophore, is produced by the fungus Ustilago sphaerogena. Enterobactin (c and d), a catecholate-type siderophore, is produced by numerous Gram-negative bacteria including E. coli. Staphyloferrin A (e and f), an α-hydroxy carboxylate type siderophore is produced by Staphylococcus aureus
Also pathogenic bacteria that need to proliferate within a host enter an environment where the free iron concentration is extremely low. Nevertheless, the high affinity of the secreted siderophores allows them to compete for the iron against host proteins such as transferrin and hemoglobin. Once a siderophore is transported inside the bacterial cell, the iron is released from the siderophores by the action of dedicated enzymes that carry out the reduction of siderophore-bound Fe3+ to Fe2+ (Matzanke et al. 2004); alternatively proteolysis of the backbone of some siderophores has also been reported. Once released, intracellular Fe2+ can be incorporated directly into metallo-enzymes, or if there is an excess, it is stored in bacterioferritins or in the related Dps proteins (Chiancone et al. 2004). After the bacteria have accumulated a sufficient amount of iron from their environment, further uptake is shut down through the action of the Fur repressor protein, which in its iron-saturated form, blocks the biosynthesis of the bacterial iron transport systems. The Fur protein has been extensively studied and it has a surprisingly broad regulatory action (for a recent review see Carpenter et al. 2009). Some pathogenic bacteria have adapted themselves even further towards the environment in the host and instead of siderophores they can actually use heme as an iron source, or they have receptors for transferrin that can extract the Fe3+ directly from this protein (see Fig. 2; Braun and Killmann 1999; Cescau et al. 2007; Krewulak and Vogel 2008).
Schematic diagram of iron, heme and siderophore uptake in Gram-negative bacteria. Transferrin and lactoferrin deliver iron, while hemoglobin, hemopexin and selected hemophore proteins deliver heme, to their specific OMT in the outer membrane. Interaction of the TonB protein with the OMT unplugs the barrel of the OMT, allowing free iron, heme or siderophores to enter the periplasmic space. A periplasmic binding protein carries each to their cognate ABC transporter complex that uses ATP hydrolysis to internalize the iron source
Iron uptake in Gram-negative and Gram-positive bacteria: TonB or not TonB?
Gram-negative bacteria are surrounded by two separate membranes; the space between the outer and the inner membranes is called the periplasm and this compartment can make up to 30% of the volume of the bacterial cell. The dual membrane protects Gram-negative bacteria from degradative enzymes released by the host. However, this arrangement of the bacterial cell envelope creates problems for the transport of nutrients into the cytoplasm of the cell. Nutrients such as glucose, amino acids or phosphate are small enough that they can pass directly through the small pores in the bacterial outer membranes that are created by the porin proteins. However, the ferric-siderophores and heme groups are too large to pass through the porins and hence, they require their own dedicated outer membrane transporters. All outer membrane transporters (OMTs) that are involved in the uptake of iron are made up of a 22-stranded beta barrel structure (see Fig. 3). The barrels are occluded by the presence of a ~160 residue independently folded ‘cork’ or ‘plug’ domain and the siderophore binds on top of this domain (Ferguson et al. 2002; for a recent review see Noinaj et al. 2010). In order to move the ferric–siderophore complex into the periplasmic space, the cork has to be at least partially dislodged from the interior of the beta-barrel OMT structure (see Fig. 4). The energy for this process is provided by the TonB protein, which can span the entire periplasm. Together with the ExbB and ExbD proteins that are anchored in the cytoplasmic membrane, it harvests the energy from the gradients across the inner membrane (Krewulak and Vogel 2008; Postle and Larsen 2007). Despite years of research, the molecular details of the action of TonB on the TonB-dependent OMTs are still unclear. Several solution and crystal structures have been reported for TonB, as well as for the periplasmic domain of ExbD, but these have not yet revealed the molecular details of the action of the TonB system (Peacock et al. 2005; Pawelek et al. 2006; Lopez et al. 2009; Garcia-Herrero et al. 2007). A bioinformatic analysis, surveying all sequenced bacterial genomes, revealed that all Gram-negative bacteria have one or more TonB proteins, that are characterized by a carboxy-terminal region with a conserved fold (Chu et al. 2007; see Fig. 5). Gram-positive bacteria do not have an outer membrane and, as expected, they do not appear to have the genes for OMTs or for the three proteins of the TonB system. Iron withholding has long been known to be a clinically useful treatment to combat bacterial infections (Weinberg 1984). Since the uptake of all ferric siderophores, heme or transferrin-derived iron are all mediated by specific TonB-dependent OMTs (see Fig. 2), the TonB system presents itself as a potential target for the development of new antibiotics aimed at Gram-negative bacteria.
FhuA outer membrane transporter (OMT) without (a; PDB 2FCP) and with (b–d; PDB 1FCP) bound ferrichrome. Residues 1–160 representing the cork domain are colored in green. b Side-view with ferrichrome sitting within the barrel of FhuA and on-top of the cork domain; c slab view revealing the cork domain; and d top–down view showing the structural ‘plugging’ of the FhuA barrel by the cork domain
Various TonB C-terminal domain (CTD) protein structures. a An overlay of the E. coli TonB-CTD NMR solution structure (blue; PDB 1XX3) and X-ray crystal structure (magenta; PDB 1U07) that makes minimal dimeric contacts through its extended β3-strand (RMSD1-233 of 1.153 Å). b An X-ray crystal structure (PDB 1IHR) of E. coli TonB-CTD in a domain swapped dimer conformation (PDB 1IHR) with chains A and B shown in light and dark blue, respectively. NMR solution structures of c E. coli TonB C-terminal domain (CTD) and d Vibrio anguillarum TonB2 CTD (PDB 2K9K). The primary differences between the two proteins are the lengthened loop 3 between α2-helix and β3-strand and the absence of the β4-strand in the V. anguillarum TonB2 CTD
Once the ferric-siderophores have been transported into the periplasmic space by the OMTs and TonB, they are ready to be transported into the cytoplasm. This process is initiated by periplasmic siderophore-binding proteins (Clarke et al. 2000, 2002). These proteins have a characteristic structure, where the ferric-siderophore is bound between two independently folded domains. The siderophore-binding proteins form a unique class of periplasmic-binding protein that have a rather distinct arrangement of the two lobes of the protein, when compared to the periplasmic proteins that are involved in the transport of amino acids or phosphate for example (Krewulak et al. 2004) (see Fig. 6). In their siderophore-bound state these proteins dock onto the periplasmic face of cognate ABC-transporters in the inner membrane, Subsequently, these helical proteins mediate the transport of the siderophores into the cytoplasm in an ATP-dependent process (see Fig. 3). Several crystal structures have been reported for the bacterial cytoplasmic membrane vitamin B12 uptake system, which is homologous in sequence, design and function to the siderophore transporters (Lewinson et al. 2010). It should also be noted that the uptake system for siderophores in Gram-positive bacteria closely resembles the ABC transport systems found in the inner membrane of Gram-negative bacteria; the only difference is that the siderophore-binding protein is anchored to the cell membrane, but its overall structure is very similar (see Fig. 6) (Grigg et al. 2010).
Structures of siderophore binding proteins from Gram-negative (in blue on the left) and Gram-positive (in green on the right) bacteria. Figures a and b are representative SBPs that bind catecholate-type siderophores; while c and d bind hydroxamate-type siderophores; and e and f have arginine-rich binding pockets which in the case of f binds an α-hydroxy carboxylate type siderophore. a CeuE (PDB 2CHU) complexed with MECAM (chain A and B in light and dark blue, respectively); b FeuA (PDB 2WHY) complexed with bacillibactin; c FhuD (PDB 1EFD) complexed with gallichrome; d BSU3320 (PDB 3G9Q) depicted in its apo-form is hypothesized to bind ferrichrome; e FitE (PDB 3BE6) in its apo dimeric form (chain A and B in light and dark blue, respectively); and f HtsA (3LI2) complexed with staphyloferrin
Iron, siderophores and the innate immune system of the host
How do we defend ourselves against an invading pathogen? Even though we are constantly surrounded by bacteria that land on our skin and that get into our lungs, it is rare that we become infected. Our bodies produce a large array of host-defense and antimicrobial proteins and peptides that are part of the innate immune system. Together these proteins and peptides usually manage to keep these bacteria at bay. Examples include the ubiquitous lysozyme, which was the first antibacterial discovered by Sir Alexander Fleming in 1922; we now know that this enzyme directly attacks the bacterial cell wall of Gram-positive bacteria (Fleming 1922). Also the defensins are a group of small amphipathic cationic proteins that can selectively disrupt bacterial membranes and stimulate the host immune system (Yang et al. 2007). Intriguingly, however, several of the host-defense proteins interfere with the ability of bacteria to acquire iron. In particular, the protein lactoferrin can be released by circulating neutrophils in our bodies at sites of infection (Legrand et al. 2008; Valenti and Antonini 2005). Lactoferrin, which was originally discovered as an iron-binding protein in milk, is quite similar to the serum transport protein transferrin (Baker et al. 2002), but it binds Fe3+ even more tightly, thereby making the metal ion locally unavailable for bacteria. It has also been shown that the extremely low iron concentrations that are created by the presence of lactoferrin, prevent bacteria from aggregating with each other and forming biofilms (Singh et al. 2002); by keeping the bacteria in a planktonic state they are more susceptible to endogenous and exogenous antimicrobial compounds. Likewise lipocalin proteins are also secreted by neutrophils in the host at sites of infection; these proteins were recently shown to avidly bind various ferric–siderophore complexes, thereby again directly preventing the bacteria from obtaining iron, by scavenging the bacterial iron-chelating entities (Goetz et al. 2002; Flo et al. 2004). It is interesting to note that this discovery was made, when ferric–siderophore complexes unintentionally co-purified with lipocalin proteins that were recombinantly expressed in E. coli. Also the distinct class of the tear lipocalins can bind various ferric-siderophores (Fluckinger et al. 2004). Because of this newly discovered function, where these proteins are now known to strongly bind ‘sidero’-phores rather than hydrophobic (‘lipo’) compounds, many researchers are now calling these proteins ‘siderocalins’ (see Fig. 7). In fact, several researchers have suggested that some bacteria may have recently escalated the fight by covalently attaching glucose to some of the bacterially-secreted siderophores; this modification does not alter their high affinity for Fe3+ or their uptake in bacteria, but it prevents them from binding to the host siderocalins (Hantke et al. 2003; Bister et al. 2004; Fischbach et al. 2006). Thus the bacteria that can chemically modify their siderophores in this manner, not only make them more soluble, but they cleverly evade the host-defense response. As such, these specific compounds are sometimes referred to as ‘stealth-siderophores’ (Hider and Kong 2010).
The structures of human lipocalins present a typical fold with an eight-stranded anti-parallel β-barrel that is open on one end and has an α-helix on its side. a and b show the structure of siderocalin (neutrophil gelatinase associated lipocalin; PDB 3CMP) crystallized bound with ferric-enterobactin (FeEnt). c and d are the structure of tear lipocalin (PDB 3EYC) bound with the artificial substrate 1,4-butanediol
Finally, it is important to realize, that bacterial infections also give rise to increased hepcidin biosynthesis in the liver. The increase in the serum concentration of the hepcidin peptide hormone directly blocks iron uptake by the gut and prevents the release of iron from iron stores in the liver and the macrophages into the circulatory system. This results in significantly decreased levels of iron in the bloodstream during a bacterial infection, an effect clinically known as “anemia of infection” (Ganz 2003, 2006). Obviously, to fend off invading bacteria, our bodies try to create a hostile environment, which includes a low iron concentration and release of ferric-siderophore scavenging proteins, to prevent the pathogens from growing and multiplying. This ‘battle for iron’ is now considered as an integral part of the innate immune response. The corollary to this is that if we can design new compounds that block bacterial iron uptake directly, for example by interfering in the TonB system in Gram-negative bacteria, we emulate this natural situation. Such a strategy could clearly be advantageous, as our immune systems are already well prepared to deal with this condition.
Clinical use of siderophores and related iron-chelators
Patients that suffer from the disease thalassemia major (also known as β-thalassemia) have a genetic defect in the biosynthesis of hemoglobin, which leads to the production of non-functional erythrocytes. These people suffer from severe anemia and to overcome this lethal deficiency they require additional healthy red blood cells, hence they are treated with frequent blood transfusions. As our bodies have a poor capacity for secreting excess iron, these patients quickly start to suffer from a significant iron overload, which, if left untreated, can cause serious damage to various organs. Consequently they have to be routinely treated with iron-chelation therapy to remove the excess iron. For more than 40 years the preferred treatment has been the administration of the siderophore desferrioxamine B (also known as deferoxamine, DFO or desferal). This compound binds Fe3+ tightly in serum and promotes its secretion into the urine. Desferrioxamine is produced by Streptomyces pilosus and like most siderophores, this molecule provides three bidentate ligands to preferentially bind ferric iron. While Fe3+ has a strong preference for a six-coordinate ‘octahedral’ surrounding, especially if it is made up mostly of oxygen atoms as the ligands, other metal ions such as Zn2+ and Fe2+, actually prefer a four-coordinate tetrahedral surrounding with several nitrogen and sulfur ligands. Thus siderophores such as desferrioxamine B, are designed to bind Fe3+ extremely tightly and they seem ideally suited to remove the ferric ion from the body. Unfortunately they are relatively large molecules and they are poorly absorbed if taken orally. To be effective desferioxamine needs to be injected intravenously, or infused subcutaneously through a little pump device. This procedure is cumbersome and results in poor patient compliance (Bring et al. 2008). Many researchers have been searching for alternative ferric-iron chelators that could be taken orally (see for example Dobbin et al. 1993). Currently two agents are available for clinical treatment that have met with considerable success: deferiprone (tradename Ferriprox, produced by Apotex) and deferasirox (marketed as Exjade by Novartis). The former provides two ligands and the latter three ligands and hence multiple molecules are required to fulfill the requirement of Fe3+ for hexadentate coordination (see Fig. 8). Consequently these compounds have a lower affinity for Fe3+ than desferrioxamine. However, in part because of their smaller size they appear to be well-tolerated and easily absorbed in the intestine and they are effective as oral agents. While deferiprone requires three daily doses and has some known side effects, early indications are that deferasirox is effective and may be relatively free of serious side effects. It requires only one daily dose and enjoys high patient compliance (Neufeld 2006; Cappellini and Pattoneri 2009). Be that as it may, this drug has only recently been approved by the FDA and its long-term effects still need to be carefully evaluated.
Crystal structures of the ferric complexes of a desferrioxamine B (desferal), b deferiprone and c deferasirox analogue Lz modified from Bernhardt (2007; H-atoms omitted, Fe—green, O—red, N—blue and C—grey). Reproduced by permission of The Royal Society of Chemistry
Individuals suffering from other serious iron-overload diseases such as hereditary hemochromatosis are usually not treated with iron-chelators. In most cases treatment by phlebotomy (‘blood-letting’) is very effective at reducing the overall iron load and this is currently considered the gold-standard for treatment (Bring et al. 2008). If the long-term safety of the new oral iron-chelator compounds can be demonstrated, these compounds could perhaps become part of a new treatment regimen for hereditary hemochromatosis; alternatively new drugs that can mimick the action of hepcidin could possibly be developed to help prevent the build-up of excess iron in these patients.
Another class of compounds that may have some clinical potential are naturally produced conjugates of siderophores and antibiotics or other bactericidal compounds. Because the siderophore uptake systems are used to actively recognize and transport these compounds into the bacteria, they are often referred to as ‘Trojan-horse’ antibiotics. For example, albomycin is an adduct of the hydroxamate ferrichrome siderophore that is chemically linked to a thioribosyl pyrimidine antibiotic that can inhibit the tRNA synthetase. This particular siderophore conjugate can be produced by various Streptomyces strains and the antibiotic portion can be enzymatically released once the albomycin has been transported into the bacterial cell. Albomycin was recently shown to be effective in curing bacterial infections in a mouse model (Pramanik et al. 2007). A closely related class of compounds is made up of covalently-linked complexes of antimicrobial peptides and siderophores. It was recently reported that microcin E422m, an antimicrobial peptide produced by Klebsiella pneumonia, contained several catecholate groups that allowed binding of Fe3+. This siderophore–peptide complex was shown to be actively taken up by Gram-negative bacteria through their siderophore outer membrane transporters (Thomas et al. 2004). It has been proposed to call this novel class of antibiotic compounds the ‘sideromycins’ (Pramanik et al. 2007). In addition to these naturally occurring compounds, numerous siderophore-antibiotic conjugates have already been produced through chemical synthesis, in an attempt to use the Trojan-horse strategy to develop novel bacteria specific antibiotics (e.g. Miller et al. 2009).
Conclusions
The first siderophores were discovered around 1950. Since then several hundred distinct biologically active iron-chelator compounds have been purified and characterized (Hider and Kong 2010). The three dimensional structures of many siderophores were uncovered, revealing the presence of unique chemistries specialized for binding Fe3+; ultimately this work has led to the development of novel oral iron-chelators that are now used clinically. As illustrated above, since 1998, numerous structures of siderophore transport proteins with or without bound siderophores have become available. The process of bacterial iron acquisition is now much better understood at a molecular level and this should ultimately lead to the rational design of novel classes of antibiotics that target this process. Moreover, hepcidin was discovered and its regulatory role as ‘the insulin of the iron world’ has been elucidated over the last 10 years. Intriguingly this peptide hormone also plays a major role during bacterial infections. Additionally the antibacterial function of the siderocalin/lipocalin proteins is now also coming into focus. Finally, the complex biosynthesis of siderophores is starting to be much better understood (Barry and Challis 2009) and perhaps modified iron-chelators with novel properties may be engineered in the future by altering the enzymes involved in this process. Obviously much has been learned and excellent progress has been made, yet there is always more to discover and lots more work to do. Undoubtedly, the clinical implications of the battle for iron will continue to stimulate research in this area for years to come.
References
Anderson GJ, Frazer DM, McLaren GD (2009) Iron absorption and metabolism. Curr Opin Gastroenterol 25:129–135
Baker EN, Baker HM, Kidd RD (2002) Lactoferrin and transferrin: functional variations on a common structural framework. Biochem Cell Biol 80:27–34
Barry SM, Challis GL (2009) Recent advances in siderophore biosynthesis. Curr Opin Chem Biol 13:205–215
Bernhardt PV (2007) Coordination chemistry and biology of chelators for the treatment of iron overload disorders. Dalton Trans 30:3214–3220
Bister B, Bischoff D, Nicholson GJ, Valdebenito M, Schneider K, Winkelmann G, Hantke K, Süssmuth RD (2004) The structure of salmochelins: C-glucosylated enterobactins of Salmonella enterica. Biometals 17:471–481
Braun V, Killmann H (1999) Bacterial solutions to the iron-supply problem. Trends Biochem Sci 24:104–109
Bring P, Partovi N, Ford JE, Yoshida EM (2008) Iron overload disorders: treatment options for patients refractory to or intolerant of phlebotomy. Pharmacotherapy 28:331–342
Cappellini MD, Pattoneri P (2009) Oral iron chelators. Annu Rev Med 60:25–38
Carpenter BM, Whitmire JM, Merrell DS (2009) This is not your mother’s repressor: the complex role of fur in pathogenesis. Infect Immun 77:2590–2601
Cescau S, Cwerman H, Létoffé S, Delepelaire P, Wandersman C, Biville F (2007) Heme acquisition by hemophores. Biometals 20:603–613
Chan DI, Vogel HJ (2010) Current understanding of fatty acid biosynthesis and the acyl carrier protein. Biochem J (in press)
Chiancone E, Ceci P, Ilari A, Ribacchi F, Stefanini S (2004) Iron and proteins for iron storage and detoxification. Biometals 17:197–202
Chu BC, Peacock RS, Vogel HJ (2007) Bioinformatic analysis of the TonB protein family. Biometals 20:467–483
Clarke TE, Ku SY, Dougan DR, Vogel HJ, Tari LW (2000) The structure of the ferric siderophore binding protein FhuD complexed with gallichrome. Nat Struct Biol 7:287–289
Clarke TE, Braun V, Winkelmann G, Tari LW, Vogel HJ (2002) X-ray crystallographic structures of the Escherichia coli periplasmic protein FhuD bound to hydroxamate-type siderophores and the antibiotic albomycin. J Biol Chem 277:13966–13972
Crichton R (2009) Iron metabolism, from molecular mechanisms to clinical consequences, 3rd edn. Wiley Interscience, Chichester, West Sussex
Crosa JH, Walsh CT (2002) Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiol Mol Biol Rev 66:223–249
Dobbin PS, Hider RC, Hall AD, Taylor PD, Sarpong P, Porter JB, Xiao G, van der Helm D (1993) Synthesis, physicochemical properties, and biological evaluation of N-substituted 2-alkyl-3-hydroxy-4(1H)-pyridinones: orally active iron chelators with clinical potential. J Med Chem 36:2448–2458
Ferguson AD, Chakraborty R, Smith BS, Esser L, van der Helm D, Deisenhofer J (2002) Structural basis of gating by the outer membrane transporter FecA. Science 295:1715–1719
Fischbach MA, Lin H, Liu DR, Walsh CT (2006) How pathogenic bacteria evade mammalian sabotage in the battle for iron. Nat Chem Biol 2:132–138
Fleming A (1922) On a remarkable bacteriolytic element found in tissues and secretions. Proc R Soc Lond B 93:306–317
Flo TH, Smith KD, Sato S, Rodriguez DJ, Holmes MA, Strong RK, Akira S, Aderem A (2004) Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature 432:917–921
Fluckinger M, Haas H, Merschak P, Glasgow BJ, Redl B (2004) Human tear lipocalin exhibits antimicrobial activity by scavenging microbial siderophores. Antimicrob Agents Chemother 48:3367–3372
Ganz T (2003) Hepcidin, a key regulator of iron metabolism and mediator of anemia of inflammation. Blood 102:783–788
Ganz T (2006) Hepcidin—a peptide hormone at the interface of innate immunity and iron metabolism. Curr Top Microbiol Immunol 306:183–198
Garcia-Herrero A, Peacock RS, Howard SP, Vogel HJ (2007) The solution structure of the periplasmic domain of the TonB system ExbD protein reveals an unexpected structural homology with siderophore-binding proteins. Mol Microbiol 66:872–889
Goetz DH, Holmes MA, Borregaard N, Bluhm ME, Raymond KN, Strong RK (2002) The neutrophil lipocalin NGAL is a bacteriostatic agent that interferes with siderophore-mediated iron acquisition. Mol Cell 10:1033–1043
Grigg JC, Cooper JD, Cheung J, Heinrichs DE, Murphy ME (2010) The Staphylococcus aureus siderophore receptor HtsA undergoes localized conformational changes to enclose staphyloferrin A in an arginine-rich binding pocket. J Biol Chem 285:11162–11171
Hantke K, Nicholson G, Rabsch W, Winkelmann G (2003) Salmochelins, siderophores of Salmonella enterica and uropathogenic Escherichia coli strains, are recognized by the outer membrane receptor IroN. Proc Natl Acad Sci USA 100:3677–3682
Harrison PM, Arosio P (1996) The ferritins: molecular properties, iron storage function and cellular regulation. Biochim Biophys Acta 1275:161–203
Hider RC, Kong X (2010) Chemistry and biology of siderophores. Nat Prod Rep 27:637–657
Hunter HN, Fulton DB, Ganz T, Vogel HJ (2002) The solution structure of human hepcidin, a peptide hormone with antimicrobial activity that is involved in iron uptake and hereditary hemochromatosis. J Biol Chem 277:37597–37603
Jordan JB, Poppe L, Haniu M, Arvedson T, Syed R, Li V, Kohno H, Kim H, Schnier PD, Harvey TS, Miranda LP, Cheetham J, Sasu BJ (2009) Hepcidin revisited, disulfide connectivity, dynamics, and structure. J Biol Chem 284:24155–24167
Krewulak KD, Vogel HJ (2008) Structural biology of bacterial iron uptake. Biochim Biophys Acta 1778:1781–1804
Krewulak KD, Peacock RS, Vogel HJ (2004) Periplasmic binding proteins involved in bacterial iron uptake. In: Iron transport in bacteria. ASM Press, Washington, DC
Lauth X, Babon JJ, Stannard JA, Singh S, Nizet V, Carlberg JM, Ostland VE, Pennington MW, Norton RS, Westerman ME (2005) Bass hepcidin synthesis, solution structure, antimicrobial activities and synergism, and in vivo hepatic response to bacterial infections. J Biol Chem 280:9272–9282
Legrand D, Pierce A, Elass E, Carpentier M, Mariller C, Mazurier J (2008) Lactoferrin structure and functions. Adv Exp Med Biol 606:163–194
Lewinson O, Lee AT, Locher KP, Rees DC (2010) A distinct mechanism for the ABC transporter BtuCD-BtuF revealed by the dynamics of complex formation. Nat Struct Mol Biol 17:332–338
Lopez CS, Peacock RS, Crosa JH, Vogel HJ (2009) Molecular characterization of the TonB2 protein from the fish pathogen Vibrio anguillarum. Biochem J 418:49–59
Matzanke BF, Anemüller S, Schünemann V, Trautwein AX, Hantke K (2004) FhuF, part of a siderophore-reductase system. Biochemistry 43:1386–1392
Miller MJ, Zhu H, Xu Y, Wu C, Walz AJ, Vergne A, Roosenberg JM, Moraski G, Minnick AA, McKee-Dolence J, Hu J, Fennell K, Kurt Dolence E, Dong L, Franzblau S, Malouin F, Möllmann U (2009) Utilization of microbial iron assimilation processes for the development of new antibiotics and inspiration for the design of new anticancer agents. Biometals 22:61–75
Neilands JB (1995) Siderophores: structure and function of microbial iron transport compounds. J Biol Chem 270:26723–26726
Nemeth E, Tuttle MS, Powelson J, Vaughn MB, Donovan A, Ward DM, Ganz T, Kaplan J (2004) Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science 306:2090–2093
Nemeth E, Preza GC, Jung CL, Kaplan J, Waring AJ, Ganz T (2005) The N-terminus of hepcidin is essential for its interaction with ferroportin: structure-function study. Blood 107:328–333
Neufeld EJ (2006) Oral chelators deferasirox and deferiprone for transfusional iron overload in thalassemia major: new data, new questions. Blood 107:3436–3441
Noinaj N, Guillier M, Barnard TJ, Buchanan SK (2010) TonB-dependent transporters: regulation, structure, and function. Annu Rev Microbiol 64 (in press). doi:10.1146/annurev.micro.112408.134247
Pawelek PD, Croteau N, Ng-Thow-Hing C, Khursigara CM, Moiseeva N, Allaire M, Coulton JW (2006) Structure of TonB in complex with FhuA, E. coli outer membrane receptor. Science 312:1399–1402
Peacock RS, Weljie AM, Howard PS, Price FD, Vogel HJ (2005) The solution structure of the C-terminal domain of TonB and interaction studies with TonB box peptides. J Mol Biol 345:1185–1197
Postle K, Larsen RA (2007) TonB-dependent energy transduction between outer and cytoplasmic membranes. Biometals 20:453–465
Pramanik A, Stroeher UH, Krejci J, Standish AJ, Bohn E, Paton JC, Autenrieth IB, Braun V (2007) Albomycin is an effective antibiotic, as exemplified with Yersinia enterocolitica and Streptococcus pneumoniae. Int J Med Microbiol 297:459–469
Raymond KN, Dertz EA, Kim SS (2003) Enterobactin: an archetype for microbial iron transport. Proc Natl Acad Sci USA 100:3584–3588
Singh PK, Parsak MR, Greenberg EP, Welsh MJ (2002) A component of innate immunity prevents bacterial biofilm development. Nature 417:552–555
Thomas X, Destoumieux-Garzón D, Peduzzi J, Afonso C, Blond A, Birlirakis N, Goulard C, Dubost L, Thai R, Tabet JC, Rebuffat S (2004) Siderophore peptide, a new type of post-translationally modified antibacterial peptide with potent activity. J Biol Chem 279:28233–28242
Valenti P, Antonini G (2005) Lactoferrin: an important host defence against microbial and viral attack. Cell Mol Life Sci 62:2576–2587
van der Helm D, Jalal MAF, Hossain MB (1987) The crystal structures, conformations and configurations of siderophores. In: Iron transport in bacteria, plant and animals. VCH Publishers, New York
Weinberg ED (1984) Iron withholding: a defense against infection and neoplasia. Physiol Rev 64:65–102
Yang D, Liu ZH, Tewary P, Chen Q, de la Rosa G, Oppenheim JJ (2007) Defensin participation in innate and adaptive immunity. Curr Pharm Des 13:3131–3139
Acknowledgments
Research in the author’s laboratory has been supported by operating grants from the Canadian Institutes for Health Research. HJV is the recipient of a Scientist award from the Alberta Heritage Foundation for Medical Research.
Author information
Authors and Affiliations
Corresponding author
Additional information
This article is dedicated to the memory of Dr. Dick van der Helm who passed away in April this year. Dick committed much of his research career to structural studies of siderophores and the bacterial proteins that mediate their transport. His insights and encouragement will be missed.
Rights and permissions
About this article
Cite this article
Chu, B.C., Garcia-Herrero, A., Johanson, T.H. et al. Siderophore uptake in bacteria and the battle for iron with the host; a bird’s eye view. Biometals 23, 601–611 (2010). https://doi.org/10.1007/s10534-010-9361-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10534-010-9361-x