Abstract
The richness and composition of herbivore communities can be influenced by the genetic variation of host plants. Hybrid plant populations are ideal to test these effects because they usually harbor high genetic variation and display a mosaic of phenotypic characters. The goal of this study was to examine the effect of hybridization between two Mexican white oaks, Q. magnoliifolia and Q. resinosa, on the composition and diversity of the associated cynipid gall wasp community. We used eight nuclear microsatellite markers to genotype 150 oak individuals sampled at three different altitudes at the Tequila volcano and conducted monthly samplings of galls in each individual over the course of 2 years. A Bayesian assignment analysis indicated genetic admixture between the two oak species at the study site and allowed classifying individuals as Q. magnoliifolia, Q. resinosa or hybrids. Gall morphospecies richness was significantly higher in the hybrids, intermediate in Q. magnoliifolia and lower in Q. resinosa. Overall, 48 different gall morphospecies were found, with 21 of them being shared among the three groups of plants, 13 between two groups of plants, and 14 were unique to one group of plants, with eight of these being found in hybrids. Several of the shared galls showed differences in abundance among plant groups. Therefore, genetic structure in this oak complex significantly influences the diversity and composition of the associated gall wasp community, and hybrid individuals are probably acting as potential sinks and bridges for the colonization of plant hosts by these highly specialized insect species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The diversity and structure of herbivorous insect communities usually have been explained by abiotic factors such as temperature, humidity and soil fertility, as well as by biotic factors with bottom-up effects, like plant diversity, plant architecture, chemical defense and nutritional quality of hosts (Larson and Whitham 1997; Tscharntke et al. 2002; Cuevas-Reyes et al. 2004a; Whitham et al. 2006). In addition, the top-down effects of natural enemies have been considered important (Cooper and Rieske 2010; Maldonado-López et al. 2015a). Only recently, the richness and composition of the herbivore community have been associated with the intra- and interspecific genetic variation of plants (Booth and Grime 2003; Morin 2003; Wade 2003). For example, Wimp et al. (2004) found that almost 60 % of the variation in arthropod diversity could be explained by differences in the genetic diversity of Populus host trees. In another study, Johnson and Agrawal (2005) showed experimentally that the genotypic variation of Oenothera biennis was the main factor that explained variation in the diversity and structure of the arthropod community associated with this plant. However, a significant effect of the genotype-by-habitat interaction was also observed.
Hybrid zones are ideal systems to study the effects of host-plant genetic variation on the community of associated herbivores. Hybrid plant populations usually harbor high genetic variation and display a mosaic of phenotypic characters that can be intermediate between parental populations, identical to one of the parental populations, extreme or novel (Rieseberg and Ellstrand 1993; Arnold 1997). As a result of this variation, herbivore communities associated with hybrid plant populations could hypothetically be a combination of the herbivore communities of both parental populations, be more similar to the community of one of the parental populations than to the other, or be extreme in terms of species abundance and composition (Whitham et al. 1994, 1999; Johnson and Agrawal 2005; Johnson et al. 2006). In the latter case, hybrid populations could show a lower abundance or diversity of associated herbivores than both parental populations, as Boecklen and Spellenberg (1990) found in hybrids between oak species in Mexico. Alternatively, higher herbivore abundance or even novel interactions with herbivore species not associated with either of the parental populations could exist (Whitham et al. 1994, 1999; Dungey et al. 2000; Hochwender and Fritz 2004; Wimp et al. 2005; Nakamura et al. 2010). Therefore, plant hybridization can significantly influence the ecology and evolution of plant-herbivore interactions (Fritz et al. 1994).
The genus Quercus (Fagaceae) is a highly diverse group of woody plants with temperate origin that is also known for a high frequency of interspecific hybridization (Boecklen and Spellenberg 1990; González-Rodríguez et al. 2004; Tovar-Sánchez and Oyama 2004). The genus includes between 300 and 600 species distributed throughout the northern hemisphere (Jones 1986). Mexico is considered a main center of diversification of Quercus (Rzedowski 1978; Nixon 1993), with a total number of species of about 161, and 86 endemics (Valencia 2004).
Oaks also host a great diversity of herbivorous insects (Stone and Schönrogge 2003). Particularly notorious are the highly specialized cynipid gall wasps (Hymenoptera: Cynipidae) (Ronquist and Liljeblad 2001). More than 80 % of the approximately 1000 species of cynipids is associated with oak species (Stone et al. 2002). This interaction is highly specific in terms of the oak species used and the particular organ of induction. Galls are more frequent on leaves and twigs, but also form on flowers, fruits branches, stems and roots (Abrahamson et al. 2003; Stone and Schönrogge 2003; Maldonado-López et al. 2015a). In addition, each wasp species induces a particular gall with a distinct morphology and structure, which is mainly controlled by the insect (Crespi et al. 1997; Nyman et al. 2000; Stone and Schönrogge 2003). Related to their high specificity, cynipids are very sensitive to variation in their host-plants and it has been reported that they can discriminate among closely related host species (Boecklen and Spellenberg 1990; Aguilar and Boecklen 1992; Fritz et al. 1994; Floate and Whitham 1995; Evans et al. 2012). In general, galls induced by cynipid wasps on oak species are considerably more complex and diverse than galls induced on other host-plant groups (Dreger-Jauffret and Shorthouse 1992).
In Mexico, the frequent formation of hybrid zones among Quercus species with different degrees of relatedness (González-Rodríguez et al. 2004; Tovar-Sánchez and Oyama 2004; Albarrán-Lara et al. 2010; Peñaloza-Ramírez et al. 2010; Valencia-Cuevas et al. 2014) offers an excellent opportunity to analyze the effects of host-plant genetic variation on the composition and structure of highly diverse and complex communities of gall wasps. This information is relevant to understand the ecology and evolution of ecological interactions, as well as the processes and mechanisms that shape and maintain biological diversity. Therefore, the goal of this study was to examine the effect of hybridization in a complex of two Mexican white oaks, Q. magnoliifolia and Q. resinosa, on the composition and diversity of the cynipid gall wasp community. Hybridization between these two species has been previously analyzed at the Tequila volcano using morphometry and genetic markers (Albarrán-Lara et al. 2010). According to this previous study, Q. magnoliifolia individuals predominate at altitudes between 1400 and 1800 m, while Q. resinosa individuals mostly occur at 1700–2100 m. A proportion of individuals with mixed phenotypes are found between 1600 and 1800 m. Even though the two species are clearly distinguishable on the basis of morphological traits and also differ in ecological traits such as leaf phenology (Hernández-Calderon et al. 2013), genetic differentiation between them is low and a considerable proportion of individuals show evidence of introgression at all altitudes, independently of their phenotype (Albarrán-Lara et al. 2010).
In this study, we genotyped 150 individuals of the Q. magnoliifolia-Q. resinosa complex sampled at three different altitudes at the Tequila volcano and conducted monthly surveys over the course of 2 years to determine the richness and abundance of gall morphospecies associated with each tree. Our specific questions were the following: (1) What is the richness and structure of the community of gall-inducing insects associated to Q. magnoliifolia and Q. resinosa at the Tequila volcano? (2) How do parental species and hybrid plants compare in terms of richness and composition of their gall wasp communities?
Materials and methods
Study system
This study was carried out at the Tequila volcano, Jalisco state, Mexico (20°50′N, 103°5′W). At this site, Quercus magnoliifolia Née and Q. resinosa Liebm. are distributed along an altitudinal gradient from about 1400 to 2100 m. Q. magnoliifolia occurs mostly from 1400 to 1800 m. It is a tree 5–25 m in height, characterized by large leaves 7.5–23 cm long and 3.5–13 cm wide, obovate in shape, lustrous and almost glabrous on the adaxial surface, tomentose on the abaxial surface and with glabrescent petioles (Arizaga et al. 2009). Quercus resinosa is distributed from 1700 to 2100 m. It is a tree 7–10 m in height with very large leaves 15–36 cm long and 5–26 cm wide, obovate in shape, rugous on the adaxial surface and pale-green or yellowish, tomentose on the abaxial surface and with densely tomentose petioles (Arizaga et al. 2009). Staminate flowers of both species are produced in March and April (González Villarreal 1986).
Sampling and classification of galls
Based on previous studies in this site (Albarrán-Lara et al. 2010; Hernández-Calderon et al. 2013), we sampled trees of the Q. magnoliifolia-Q. resinosa complex at three different altitudes: 1400–1500, 1600–1800 and 1900–2100 m. At each altitude, 50 trees were randomly chosen and marked with aluminum tags. Leaf and branch galls were sampled for each tree using a systematic-stratified design, collecting three branches of similar length (e.g. 70–80 cm) from the lower, medium and upper parts of the crown with a pruning pole. Sampling was performed monthly from July to February during 2 years (2011–2012 and 2012–2013). Inflorescences were not collected since these are produced in spring during a short period. All collected galls were classified as morphospecies according to their morphology and counted (Cuevas-Reyes et al. 2004a, b). The use of morphospecies assumes that each gall wasp species induces a particular gall with a unique morphology, and this seems to be the case in most cases (Cuevas-Reyes et al. 2004a, b; Araújo et al. 2013).
Genetic analysis
Five young intact leaves were collected from each tree for genetic analysis. Leaves were placed in plastic bags on ice in the field and transferred to a −80 °C ultrafreezer in the laboratory. DNA extraction was performed using the DNeasy Plant Mini Kit (QIAGEN) following the manufacturer’s instructions. Purified DNA was stored in deionized water at −20 °C. DNA concentration was determined for each sample with a Qubit 2.0 (Life Technologies) fluorometer.
Genetic analysis was carried out using eight nuclear microsatellite loci previously employed to characterize this hybrid zone (Albarrán-Lara et al. 2010): QpZAG36, QpZAG110 (Steinkellner et al. 1997), QrZAG39 (Kampfer et al. 1998), GA-0C19, quru-GA-0C11, quru-GA-0M07, quru-GA-0I01 and quru-Ga-01C08 (Aldrich et al. 2003). Multiplex reactions were prepared with primers organized into two groups according to allele size range and fluorescent label. The first group was constituted with primers QpZAG36, QpZAG110, QrZAG39 and quru-GA-0C19, and the second group with primers quru-GA-0C11, quru-GA-0M07, quru-GA-0I01 and quru-Ga-01C08 (Albarrán-Lara et al. 2010). Reactions were prepared using the Multiplex PCR Kit (QIAGEN) in a final volume of 5 µl containing 2.5 µl Multiplex PCR Master Mix, 0.5 µl of primer mix, 1.5 µl deionized water and 0.5 µl of template DNA. The thermal cycling was conducted using an applied biosystems thermal cycler. The program consisted of one cycle at 95 °C for 15 min and then 35 cycles, each with a denaturation step at 94 °C for 30 s, alignment at 50 °C for 1.5 min and extension at 72 °C for 1 min. A final extension at 60 °C for 30 min was included. One micro litre of PCR product was mixed with HI-DI formamide and 0.3 µl GeneScan-500 LIZ as size standard and analyzed in an ABI-PRISM 3100-Avant capillary sequencer. Results were analyzed with the Peak Scanner Software v1.0 (Applied Biosystems) for final sizing of the amplified fragments.
To assign individuals to a genotypic class (i.e. pure parental or hybrid), data were subjected to a Bayesian assignment analysis in STRUCTURE ver. 2.1 (Pritchard et al. 2000). We used the admixture model with correlated allele frequencies without prior population information. The number of potential genetic groups (k) was set to vary from one to five with ten replicate runs for each value of k. Each run consisted of a burn-in period of 105 steps followed by 106 iterations. To select the most probable value of k for these data we followed the method of Evanno et al. (2005) based on the calculation of Δk, which represents the second-order rate of change of the likelihood function with respect to k. The Structure Harvester Web v. 0.6.93 software (Earl and von Holdt 2011) was used to obtain Δk. After this analysis, individuals were assigned to a genotypic class depending on their inferred admixture coefficient (q value). Trees with q values equal or higher than 0.8 were considered to belong to a single genetic group (i.e. the two parental species), and trees with q values between 0.19 and 0.79 were considered hybrids. Additionally, we estimated individual observed heterozygosity (i.e. the proportion of heterozygous loci out of the total number of loci analyzed) as an estimator of genetic variation for each tree. The program CERNICALIN was used for this purpose (Aparicio et al. 2006).
Statistical analysis
Individual observed heterozygosity values were compared among the three groups of plants (i.e. Q. magnoliifolia, Q. resinosa and hybrid) with an analysis of variance (ANOVA) after all individuals had been assigned to one group as explained above. The richness of the gall morphospecies community present in each of the three groups of plants was obtained using the program EstimateSWin820 (Colwell et al. 2012). Since the number of plants in the three categories differed, the richness value was standardized and compared using a rarefaction approach (Gotelli and Entsminger 2001).
For each individual tree, we also calculated the Shannon–Wiener diversity index on the basis of the gall morphospecies present and their abundance. Comparisons among the three groups of plants were performed for diversity values as well as total abundance of galls (independently of morphospecies) using non-parametric Wilcoxon tests (Sokal and Rohlf 1995). These tests were also used to compare the abundance of each gall morphospecies among plant categories. To determine if the proportions in the number of leaf/branch gall morphospecies differed among the plant groups, a Chi square test was used (Sokal and Rohlf 1995).
Associations between heterozygosity and the richness and total abundance of galls at the level of individual trees were evaluated with non-parametric Spearman correlation tests.
Finally, we used a logistic regression analysis with the CATMOD procedure (SAS 2000) (a general procedure for modelling categorical data), to determine the differences in the incidence of gall morphospecies according to morphological categories (i.e. discoidal, irregular, ellipsoidal, cylindrical, ornamented, bulbous, cottony, globular and pubescent) on Q. magnoliifolia, Q. resinosa and hybrids. The model used had two conditions as categorical independent variables (plant species and morphological categories) and the frequency of gall morphospecies as the dependent variable (Stokes et al. 2000).
Results
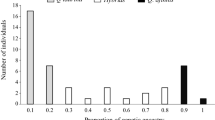
The results obtained with STRUCTURE followed by calculation of Δk with STRUCTURE HARVESTER clearly indicated that k = 2 is the most probable number of genetic groups (Fig. 1a). Considering these two main genetic groups, 30 individuals (24.6 %) were assigned as Q. resinosa, 35 (28.7 %) as Q. magnoliifolia and 57 (46.7 %) as genetically intermediate (Fig. 1b). The remaining 28 individuals could not be assigned due to failure with the amplification of some microsatellite loci. At the lower altitude (1400–1500 m), 29 of 43 analyzed individuals (67.4 %) were assigned as Q. magnoliifolia, 13 (30.2 %) as hybrids and one (2.3 %) as Q. resinosa. At the intermediate altitude (1600–1800 m) out of 42 individuals, 6 (14.3 %) were Q. magnoliifolia, 24 (57.1 %) hybrids and 12 (28.6 %) were Q. resinosa. Finally, at the higher altitude (1900–2100) out of 37 individuals 20 (54 %) were hybrids and 17 (46 %) were Q. resinosa. Individuals assigned to Q. magnoliifolia were not observed at this altitude.
In total, we found 33 gall morphospecies on the Q. magnoliifolia group, 28 morphospecies on the Q. resinosa group and 42 morphospecies on the hybrids. The rarefaction analysis indicated that with a standard sample size of 30 for each of the three groups, gall morphospecies richness is significantly higher in the hybrids, intermediate in Q. magnoliifolia and lower in Q. resinosa (Fig. 2). In contrast, mean gall abundance per tree did not differ among the three groups of individuals (χ 2 = 0.42; P = 0.8). However, there was a significant correlation between the number of gall morphospecies and the total gall abundance per individual (Spearman’s r = 0.79; P < 0.0001) (Fig. 3).
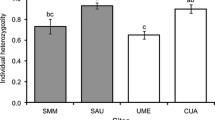
There was a significant difference in genetic diversity levels (F = 5.13; P = 0.007), measured as individual heterozygosity, between the three groups of plants, with Q. magnoliifolia individuals having, on average, a higher individual heterozygosity (0.82 ± 0.023) than Q. resinosa (0.72 ± 0.025) and the hybrid group (0.74 ± 0.018), which did not differ among themselves. The number of gall morphospecies per individual was weakly correlated negatively with individual heterozygosity (Spearman’s r = −0.18; P = 0.04) (Fig. 4), but not gall abundance per individual (Spearman’s r = 0.04; P = 0.6).
Most of the gall morphospecies were foliar galls (75 % in Q. magnoliifolia, 79 % in hybrids and 93 % in Q. resinosa), while the rest of the morphospecies were branch galls (25 % in Q. magnoliifolia, 21 % in the hybrids and 7 % in Q. resinosa). These differences among plant groups in the proportion of morphospecies induced on each organ were significant according to a Chi square test (χ 2 = 6.28; P < 0.04).
Of the total gall morphospecies registered, twenty-one (44 %) gall morphospecies were found to be shared among the three groups of plants, while six (12 %) were shared between Q. resinosa and the hybrids and seven (15 %) between Q. magnoliifolia and the hybrids (Table 1). No morphospecies were shared only between Q. magnoliifolia and Q. resinosa. Finally, five morphospecies (10 %) were exclusively found on Q. magnoliifolia (M18, M32, M33, M72, M80), one (2 %) on Q. resinosa (M76) and eight morphospecies (17 %) were unique to the group of hybrid individuals (M21, M23, M70, M75, M81, M82, M84, M85) (see Fig. 6 in Appendix).
Some of the gall morphospecies that were shared among the three groups of plants or between two groups differed in their abundance in each group. Out of the 21 morphospecies shared among the three plant groups, four morphospecies (M2, M16, M24 and M27) showed significant differences in their mean abundance per individual (Table 1). M2 and M27 were more abundant on Q. magnoliifolia trees, had intermediate abundance on hybrids and had a lower abundance on Q. resinosa individuals, while M16 had higher, intermediate and lower abundance on Q. resinosa, hybrid and Q. magnoliifolia individuals, respectively. Interestingly, M16 and M27 were the two morphospecies with the highest overall abundance. Finally, M24 had a higher abundance on the hybrid trees (Table 1). One of the gall morphospecies shared between Q. magnoliifolia and the hybrids (M31) was significantly more abundant on Q. magnoliifolia, while morphospecies M26, shared between Q. resinosa and the hybrids, was significantly more abundant on Q. resinosa (Table 1).
Regarding the morphological classification of gall morphospecies, in total nine categories were identified: discoidal, irregular, ellipsoidal, cylindrical, ornamented, bulbous, cottony, globular and pubescent (Fig. 6 in Appendix). We found differences among Q. magnoliifolia, Q. resinosa and hybrids in the incidence of these gall morphospecies (Plant species: χ 2 = 49.8.4; d.f. = 2; P < 0.0001) and between morphological categories (χ 2 = 77.1; d.f. = 8; P < 0.0001) (Fig. 5). Seven of the morphological categories were found on the three groups of plants (irregular, pubescent, globular, cottony, bulbous, ornamented, and ellipsoidal). Cylindrical galls were found on Q. magnoliifolia and hybrids but not on Q. resinosa. Discoidal galls were only found on the group of hybrid individuals, which presented the nine morphological categories of galls (Fig. 5; Fig. 6 in Appendix).
Discussion
As previously reported by Albarrán-Lara et al. (2010), there is clear evidence for genetic admixture between Q. magnoliifolia and Q. resinosa at the Tequila Volcano. The hybrid zone has a clear structure with each parental species predominating at opposite ends of the altitudinal gradient. However, since hybrid individuals are present with high frequency across the whole gradient, the classification of the hybrid zone into a specific category (i.e. tension zone, mosaic hybrid zone, bounded hybrid superiority zone) is difficult (Abbott and Brennan 2014).
In terms of genetic diversity, the group formed by Q. magnoliifolia individuals had a higher average heterozygosity than Q. resinosa and the hybrid individuals. Inter-specific gene flow is usually regarded as a mechanism that can result in an increased genetic variation of hybrid populations (Rieseberg et al. 2003; Hedge et al. 2006). However, this may depend on many other factors, such as the formation or not of different genealogical classes (e.g. advanced generation hybrids, backcrosses) within the hybrid population, the relative fitness of parental and hybrid individuals, etc. (Arnold 1997). Other studies of hybrid tree populations have reported either higher genetic variation on the hybrid group or equivalent levels of diversity in the hybrids and one or both of the parental populations (Robertson et al. 2004; Zalapa et al. 2009; Brunet et al. 2013).
Ecological studies have shown controversial results of herbivore richness in plant hybrid zones. For example, Boecklen and Spellenberg (1990) found that oak hybrids support lower abundance and species richness of gall-inducing insects and leaf miners in comparison with their parental species, indicating that hybrid plants are more resistant than either parent is. Other studies, such as Aguilar and Boecklen (1992) showed intermediate densities of gall-inducing insects and leaf miners on hybrid individuals in the Q. grisea × Q. gambellii complex. Similarly, Pearse and Baty (2012) found intermediate herbivory levels by chewing and leaf miner insects on hybrid oaks, and Tovar-Sanchez and Oyama (2006a) reported intermediate density and diversity of both endophagous and free-feeding insects in a Mexican oak hybrid complex. Finally, higher herbivore densities or higher herbivore performance on hybrid plants compared to parental taxa have been reported (Whitham 1989; Floate et al. 1993). Differences in geographic host ranges, plant architecture and phenology have been suggested as possible causes for these differences. Another important factor could be the structure of the hybrid zone and the abundance of the different genealogical classes. For example, in Populus hybrid zones the direction of introgression is unidirectional (i.e. F1′s backcross with only one of the parental species), while in most of the studied Quercus hybrid zones (including this one), introgression is bidirectional (Whitham 1989; González-Rodríguez et al. 2004; Wimp et al. 2005; Peñaloza-Ramírez et al. 2010).
In our study, we found that the richness of gall morphospecies was higher in the group of hybrid individuals, intermediate in Q. magnoliifolia and lower in Q. resinosa. This pattern results from the fact that some gall morphospecies are shared between one of the parental species and the hybrids but not between the two parental species (six morphs in the case of Q. resinosa and seven in the case of Q. magnoliifolia). Our results are consistent with other studies in hybrid complexes showing that hybrid plants have a higher incidence of herbivore insects in comparison with parental species (Fritz et al. 1994; Whitham et al. 1994). Since plant hybridization may affect directly plant traits such as phenology, physiology, morphology and chemical defense, in turn, these traits can influence the preference and performance of insect herbivores (Dungey et al. 2000; Rehill et al. 2005; Carmona et al. 2011; Evans et al. 2012). Considering the high degree of host plant specificity of gall wasps, small changes in plant traits of hybrid hosts can affect the incidence and performance of this insect guild (Valencia-Cuevas and Tovar-Sánchez 2015). Overall, our results are congruent with the hybrid sink hypothesis (Whitham 1989), that proposes that hybrid plants have a higher load of herbivores in comparison with their parental plant species because co-adapted gene complexes for resistance in the parental species can be disrupted in the hybrids. In consequence, hybrid plants may represent potential niches to be colonized by insect herbivores (Tovar-Sanchez and Oyama 2006a, b). Our results imply that gall-inducing insects, indeed, are able to discriminate between closely related plant species, hybrid and parental plants (Aguilar and Boecklen 1992; Floate and Whitham 1995; Fritz et al. 1996; Donaldson and Lindroth 2007).
Additionally, eight gall morphs were exclusively found in the hybrid group, in comparison with five morphs exclusively found in Q. magnoliifolia and one in Q. resinosa. These results suggest that the presence of hybrids in our study system can potentially influence race formation and promote host shifts among plant species in the community of gall-inducing insects (Floate and Whitham 1993). In addition, some studies suggest that herbivore adaptation to alternative hosts may lead to reproductive isolation and genetic differentiation (Moran and Whitham 1988; Evans et al. 2012).
Several studies have indicated that genetic diversity of host plants is related with the incidence of arthropod species suggesting two general not mutually exclusive patterns. The first pattern indicates that genetic diversity is positively related with arthropod diversity (Wimp et al. 2004; Tovar-Sanchez and Oyama 2006a, b; Tovar-Sánchez et al. 2013). Because an increment of genetic diversity in host plants results in morphological, phenological and chemical variation (Hunter et al. 1997; González-Rodríguez et al. 2004; Cheng et al. 2011), hybrid plants could represent to arthropods potential sites to be colonized. The second pattern shows that genetically more similar hosts harbor similar arthropod communities (Bangert and Whitham 2007; Kiers et al. 2010; Maldonado-López et al. 2015b). In this case, similar plant attributes such as phenology and chemical defense in genetically related individuals may promote the presence of similar arthropod communities (Valencia-Cuevas and Tovar-Sánchez 2015). Surprisingly, in our study, the number of gall morphospecies per individual was negatively (but weakly) correlated with individual heterozygosity. Considering the high specificity of gall-inducing insects (Nieves-Aldrey 2001; Stone et al. 2009), a possible explanation of our results is that more heterozygous individuals are more resistant to gall induction than plants with lower levels of genetic variation. Several studies conducted on different plant and animal species have shown that more heterozygous individuals are generally more resistant to parasites and infectious diseases (Stevens et al. 1997; Luikart et al. 2008; Leimu et al. 2008).
Most studies that have evaluated gall-plant interactions have assumed that gall morphology is unique to each gall-inducing species and that each gall species is specific to a single plant species (Nieves-Aldrey 2001; Cuevas-Reyes et al. 2004a; Stone et al. 2009). Particularly, each gall wasp is specific to a single oak species or species group, inducing particularly morphologically complex galls on their host plants (Stone and Schönrogge 2003; Oyama et al. 2003). Our data show that the oak complex formed by Q. magnoliifolia, Q. resinosa and hybrids supports a highly diverse and complex community of gall wasp species. Therefore, these plants can be considered as “super hosts” for gall wasps (Araújo et al. 2013; Maldonado-López et al. 2015a). Also, as we have shown, a large proportion of the gall morphospecies have been able to colonize the three plant groups, while a smaller proportion are restricted to two or a single plant group. However, differences among the three plant groups in traits relevant to the gall wasps should exist, since the overlap among the three communities was high but not complete. Particularly interesting was the differential incidence of morphological categories among the plant groups. For example, cylindrical galls were found on Q. magnoliifolia and hybrids but not on Q. resinosa and discoidal galls were only found on the group of hybrid individuals. This result suggests that there could be differences in genes controlling gall morphogenesis among the three plant groups, or at least that there is a differential genetic response to the insect chemical signals that elicit gall development (Bangert and Whitham 2007; Kiers et al. 2010).
In conclusion, we have shown that the genetic structure of the hybrid zone between the Mexican white oaks, Q. magnoliifolia and Q. resinosa, has important consequences for the richness and structure of a highly diverse associated community of gall wasps. This hybrid complex represents an example of “super host” situation and emphasizes the role of hybrid individuals as potential sinks or bridges for the colonization of plant hosts by highly specialized insects. Hybrid zones represent ideal study models incorporating habitat conservation, genetic diversity, diversity hotspots and zones of transition. Therefore, our study provides important information to establish a feasible conservation strategy by preserving the genetic diversity of plants as a way of conserving dependent animal species.
References
Abbott RJ, Brennan AC (2014) Altitudinal gradients, plant hybrid zones and evolutionary novelty. Philos Trans R Soc Lond B Biol Sci B 369:20130346. doi:10.1098/rstb.2013.0346
Abrahamson WG, Hunter MD, Melika G, Price PW (2003) Cynipid gall-wasp communities correlate with oak chemistry. J Chem Ecol 29:209–223. doi:10.1023/A:1021993017237
Aguilar JM, Boecklen WJ (1992) Patterns of herbivory in the Quercus grisea × Quercus gambelii species complex. Oikos 64:498–504. doi:10.2307/3545167
Albarrán-Lara AL, Mendoza-Cuenca L, Valencia-Avalos S, González-Rodríguez A, Oyama K (2010) Leaf fluctuating asymmetry increases with hybridization and introgression between Quercus magnoliifolia and Quercus resinosa (Fagaceae) through an altitudinal gradient in Mexico. Int J Plant Sci 171:310–322. doi:10.1086/650317
Aldrich PR, Jagtap M, Michler CH, Romero-Severson J (2003) Amplification of north american red oak microsatellite markers in european white oaks and chinese chestnut. Silvae Genet 52:176–179
Aparicio JM, Ortego J, Cordero PJ (2006) What should we weigh to estimate heterozygosity, alleles or loci? Mol Ecol 15:4659–4665. doi:10.1111/j.1365-294X.2006.03111.x
Araújo SW, Scareli-Santos C, Guimaraes FAG, Cuevas-Reyes P (2013) Comparing galling insect richness among neotropical savannas: effects of plant richness, vegetation structure and super-host presence. Biodivers Conserv 22:1083–1094. doi:10.1007/s10531-013-0474-8
Arizaga S, Martínez-Cruz J, Salcedo-Cabrales M, Bello-González MA (2009) Aspectos generales de los encinos. In: Arizaga S, Cruz JM, Cabrales MS, González MAB (eds) Manual de la biodiversidad de encinos michoacanos. Secretaría de Medio Ambiente y Recursos Naturales (Semarnat), Instituto Nacional de Ecología (INESemarnat), México, pp 12–141
Arnold ML (1997) Natural hybridization and evolution. Oxford University Press, New York
Bangert RK, Whitham TG (2007) Genetic assembly rules and community phenotypes. Evol Ecol 21:549–560. doi:10.1007/s10682-006-9135-7
Boecklen WJ, Spellenberg R (1990) Structure of herbivore communities in two oak. Oecologia 85:92–100. doi:10.1007/BF00317348
Booth RE, Grime JP (2003) Effects of genetic impoverishment on plant community diversity. J Ecol 91:721–730. doi:10.1046/j.1365-2745.2003.00804.x
Brunet J, Zalapa JE, Pecori F, Santini A (2013) Hybridization and introgression between the exotic Siberian elm, Ulmus pumila, and the native field elm, U. minor, in Italy. Biol Invasion 15:2717–2730. doi:10.1007/s10530-013-0486-z
Carmona D, Lajeunesse MJ, Johnson MTJ (2011) Plant traits that predict resistance to herbivores. Funct Ecol 25:358–367. doi:10.1111/j.1365-2435.2010.01794.x
Cheng D, Vrieling K, Klinkhammer PGL (2011) The effect of hybridization on secondary metabolites and herbivore resistance: implications for the evolution of chemical diversity in plants. Phytochem Rev 10:107–117. doi:10.1007/s11101-010-9194-9
Colwell RK, Chao A, Gotelli NJ, Lin SY, Mao CX, Chazdon RL, Longino JT (2012) Models and estimators linking individual-based and sample-based rarefaction, extrapolation, and comparison of assemblages. J Plant Ecol 5:3–21. doi:10.1093/jpe/rtr044
Cooper WR, Rieske LK (2010) Gall structure affects ecological associations of Dryocosmus kuriphilus Yasumatsu (Hymenoptera: Cynipidae). Environ Entomol 39:787–797. doi:10.1603/EN09382
Crespi BJ, Carmean DA, Chapman TW (1997) Ecology and evolution of galling thrips and their allies. Ann Rev Entomol 42:51–71. doi:10.1146/annurev.ento.42.1.51
Cuevas-Reyes P, Quesada M, Hanson P, Dirzo R, Oyama K (2004a) Diversity of gall-forming insects in a Mexican tropical dry forest: the importance of plant species richness, life forms, host plant age and plant density. J Ecol 92:707–716. doi:10.1111/j.0022-0477.2004.00896.x
Cuevas-Reyes P, Quesada M, Siebe C, Oyama K (2004b) Spatial patterns of herbivory by gall-forming insects: a test to the soil fertility hypothesis in a Mexican tropical dry forest. Oikos 107:181–189. doi:10.1111/j.0030-1299.2004.13263.x
Donaldson JR, Lindroth RL (2007) Genetics, environment, and their interaction determine efficacy of chemical defense in trembling aspen. Ecology 88:729–739. doi:10.1890/06-0064
Dreger-Jauffret F, Shorthouse JD (1992) Diversity of gall-inducing insects and their galls. In: Shorthouse JD, Rohfritsch O (eds) Biology of insect-induced galls. Oxford University Press, New York, pp 8–33
Dungey H, Potts BM, Whitham TG, Li H (2000) Plant genetics affects arthropod community richness and composition: evidence from a synthetic eucalypt hybrid population. Evolution 54:1938–1946. doi:10.1554/0014-3820(2000)054[1938:PGAACR]2.0.CO;2
Earl DA, von Holdt BM (2011) Structure harvester: a website and program for visualizing structure output and implementing the Evanno method. Conserv Genet Resour 4:359–361. doi:10.1007/s12686-011-9548-7
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620. doi:10.1111/j.1365-294X.2005.02553.x
Evans LM, Allan GJ, Whitham TG (2012) Populus hybrid hosts drive divergence in the herbivorous mite, Aceria parapopuli: implications for conservation of plant hybrid zones as essential habitat. Conserv Genet 13:1601–1609. doi:10.1007/s10592-012-0409-z
Floate KD, Whitham TG (1993) The “hybrid bridge” hypothesis: host shifting via plant hybrid swarms. Am Nat 141:651–662. doi:10.1086/285497
Floate KD, Whitham TG (1995) Insects as traits in plant systematics: their use in discriminating between hybrid cottonwoods. Can J Bot 73:1–13. doi:10.1139/b95-001
Floate K, Kearsley MJC, Whitham TG (1993) Elevated herbivory in plant hybrid zones: Chrysomela confluens, Populus and phenological sinks. Ecology 74:2056–2065
Fritz RS, Nichols-Orians CM, Brunsfeld SJ (1994) Interspecific hybridization of plants and resistance to herbivores: hypotheses, genetics, and variable responses in a diverse herbivore community. Oecologia 97:106–117. doi:10.1007/BF00317914
Fritz RS, Roche BM, Brunsfeld SJ, Orians CM (1996) Interspecific and temporal variation in herbivore responses. Oikos 108:121–129. doi:10.1007/BF00333223
González Villarreal LM (1986) Contribuciones al conocimiento del género Quercus en el estado de Jalisco. Colección Flora de Jalisco. Instituto de Botánica, Universidad de Guadalajara, Zapopan
González-Rodríguez A, Arias DM, Valencia S, Oyama K (2004) Morphological and RAPD analysis of hybridization between Quercus affinis and Q. laurina (Fagaceae), two Mexican red oaks. Am J Bot 91:401–409. doi:10.3732/ajb.91.3.401
Gotelli NJ, Entsminger GL (2001). Ecosim: null models software for ecology, version 6.0. Acquired Intelligence Inc, & Kesey-Bear. http://homepages.together.net/gentsmin/ecosim.htm
Hedge SG, Nason JD, Clegg JM, Ellstrand NC (2006) The evolution of California’s wild radish has resulted in the extinction of its progenitors. Evolution 60:1187–1197. doi:10.1554/05-634.1
Hernández-Calderon E, González-Rodríguez A, Méndez-Alonzo R, Vega-Peña E, Oyama K (2013) Contrasting leaf phenology in two white oaks, Quercus magnoliifolia and Quercus resinosa, along an altitudinal gradient in Mexico. Can J For Res 43:208–2013. doi:10.1139/cjfr-2012-0406
Hochwender CG, Fritz RS (2004) Plant genetic differences influence herbivore community structure: evidence from a hybrid willow system. Oecologia 138:547–557. doi:10.1007/s00442-003-1472-4
Hunter MD, Varley GC, Gradwell GR (1997) Estimating the relative roles of top-down and bottom-up forces on insect herbivore populations: a classic study revisited. Proc Natl Acad Sci 94:9176–9181. doi:10.1007/s00442-010-1802-2
Johnson MTJ, Agrawal AA (2005) Plant genotype and environment interact to shape a diverse arthropod community on evening primrose (Oenothera biennis). Ecology 86:874–885. doi:10.1890/04-1068
Johnson MTJ, Lajeunesse MJ, Agrawal AA (2006) Additive and interactive effects of plant genotypic diversity on arthropod communities and plant fitness. Ecol Lett 9:24–34. doi:10.1111/j.1461-0248.2005.00833.x
Jones JH (1986) Evolution of the Fagaceae: implications of defoliation. Bot Gard 73:228–275
Kampfer S, Lexer C, GloÈssl J, Steinkellner H (1998) Characterization of (GA)n microsatellite loci from Quercus robur. Hereditas 129:183–186. doi:10.1111/j.1601-5223.1998.00183.x
Kiers ET, Palmer TM, Ives AR, Bruno JF, Bronstain JL (2010) Mutualism in a changing world: an evolutionary perspective. Ecol Lett 13:1459–1474. doi:10.1111/j.1461-0248.2010.01538.x
Larson KC, Whitham TG (1997) Competition between gall aphids and natural plant sinks: plant architecture affects resistance to galling. Oecologia 109:575–582. doi:10.1007/s004420050119
Leimu R, Kloss L, Fischer M (2008) Effects of experimental inbreeding on herbivore resistance and plant fitness: the role of history of inbreeding, herbivory and abiotic factors. Ecol Lett 11:1001–1110. doi:10.1111/j.1461-0248.2008.01222.x
Luikart G, Pilgrim K, Visty J, Ezenwa VO, Schwartz MK (2008) Candidate gene microsatellite variation is associated with parasitism in wild bighorn sheep. Biol Lett 4:228–231. doi:10.1098/rsbl.2007.0633
Maldonado-López Y, Cuevas-Reyes P, Stone GN, Nieves-Aldrey JL, Oyama K (2015a) Gall wasp community response to fragmentation of oak tree species: importance of fragment size and isolated trees. Ecosphere 6:1–15. doi:10.1890/ES14-00355.1
Maldonado-López S, Cuevas-Reyes P, González-Rodríguez A, Pérez-López G, Acosta-Gómez C, Oyama K (2015b) Relationships among plant genetics, phytochemistry and herbivory patterns in Quercus castanea across a fragmented landscape. Ecol Res 30:133–143. doi:10.1007/s11284-014-1218-2
Moran NA, Whitham TG (1988) Evolutionary reduction of complex life cycles: loss of host alternation in Pemphigus (Homoptera: Aphididae). Evolution 42:717–728. doi:10.2307/2408863
Morin PJ (2003) Community ecology and the genetics of interacting species. Ecology 84:577–580. doi:10.1890/0012-9658(2003)084[0577:CEATGO]2.0.CO;2
Nakamura M, Asanuma M, Hiura T (2010) Differential effects of host plant hybridization on herbivore community structure and grazing pressure on forest canopies. Oikos 119:1445–1452. doi:10.1111/j.1600-0706.2010.18255.x
Nieves-Aldrey JL (2001) Hymenoptera, Cynipidae. In: Ramos MA, Nieves-Aldrey JL (eds) Fauna Ibérica, vol 16. Museo Nacional de Ciencias Naturales. CSIC, Madrid
Nixon KC (1993) The genus Quercus in Mexico. In: Nixon KC (ed) Biological diversity of Mexico: origins and distributions. Oxford University Press, New York, pp 447–458
Nyman T, Widmer A, Roininen H (2000) Evolution of gall morphology and host-plant relationships in willow-feeding sawflies (Hymenoptera: Tenthredinidae). Evolution 54:526–533. doi:10.1111/j.0014-3820.2000.tb00055.x
Oyama K, Pérez-Pérez M, Cuevas-Reyes P, Luna R (2003) Regional and local species richness of gall-forming insects in two tropical rain forest in Mexico. J Trop Ecol 19:595–598
Pearse IS, Baty JH (2012) The predictability of traits and ecological interactions on 17 different crosses of hybrid oaks. Oecologia 169:489–497. doi:10.1007/s00442-011-2216-5
Peñaloza-Ramírez JM, González-Rodríguez A, Mendoza-Cuenca L, Caron H, Kremer A, Oyama K (2010) Interspecific gene flow in a multispecies oak hybrid zone in the sierra Tarahumara of Mexico. Ann Bot 105:389–399. doi:10.1093/aob/mcp301
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Rehill B, Clauss A, Wieczorek L, Whitham T, Lindroth R (2005) Foliar phenolic glycosides from Populus fremontii, Populus angustifolia, and their hybrids. Biochem Syst Ecol 33:125–131. doi:10.1016/j.bse.2004.06.004
Rieseberg LH, Ellstrand NC (1993) What can morphological and molecular markers tell us about plant hybridization. Crit Rev Plant Sci 12:213–241. doi:10.1080/07352689309701902
Rieseberg LH, Raymond O, Rosenthal DM, Lai Z, Livingstone K, Nakazato T, Durphy JL, Schwarzbach AE, Donovan LA, Lexer C (2003) Major ecological transitions in wild sunflowers facilitated by hybridization. Science 301:1211–1216. doi:10.1126/science.1086949
Robertson A, Newton AC, Liljeblad J (2004) Multiple hybrid origins, genetic diversity and population genetic structure of two endemic Sorbus taxa on the Isle of Arran, Scotland. Mol Ecol 13:123–134. doi:10.1046/j.1365-294X.2003.02025.x
Ronquist FJ, Liljeblad J (2001) Evolution of the gall waps-host plant association. Evolution 55:2503–2522. doi:10.1126/science.1086949
Rzedowski J (1978) Vegetación de México. Limusa, México
SAS (2000) Categorical data analysis using the SAS system. SAS Institute, Cary
Sokal RR, Rohlf FJ (1995) Biometry: the principles of statistics in biological research, 3rd edn. W.H. Freeman, New York
Steinkellner H, Lexer C, Turetschek E, Glössl J (1997) Conservation of (GA)n loci between Quercus species. Mol Ecol 6:1189–1194. doi:10.1046/j.1365-294X.1997.00288.x
Stevens L, Guiyun Y, Pray LA (1997) Consequences of inbreeding on invertebrate host susceptibility to parasitic infection. Evolution 51:2032–2039. doi:10.2307/2411025
Stokes ME, Davis CS, Koch GG (2000) Categorical data analysis using the SAS system, 2nd edn. SAS, Cary
Stone GN, Schönrogge K (2003) The adaptive significance of insect gall morphology. Tree 18:512–522. doi:10.1016/S0169-5347(03)00247-7
Stone GN, Schönrogge K, Atkinson R, Bellido D, Pujade-Villar J (2002) The population biology of oak gall wasps (Hymenoptera: Cynipidae). Ann Rev Entomol 47:633–668. doi:10.1146/annurev.ento.47.091201.145247
Stone GN, Hernández-López A, Nicholls JA, di Pierro E, Pujade-Villar J, Melika G, Cook JM (2009) Extreme host plant conservatism during at least 20 million years of host plant pursuit by oak gallwasps. Evolution 63:854–869. doi:10.1111/j.1558-5646.2008.00604.x
Tovar-Sanchez E, Oyama K (2006a) Effect of hybridization of the Quercus crassifolia × Quercus crassipes complex on the community structure of endophagous insects. Oecologia 147:702–713. doi:10.1007/s00442-005-0328-5
Tovar-Sanchez E, Oyama K (2006b) Community structure of canopy arthropods associated to Quercus crassifolia × Quercus crassipes complex. Oikos 112:370–381
Tovar-Sánchez E, Oyama K (2004) Natural hybridization and hybrid zones between Quercus crassifolia and Q. crassipes in Mexico. Morphological and molecular evidence. Am J Bot 91:1352–1363. doi:10.3732/ajb.91.9.1352
Tovar-Sánchez E, Valencia-Cuevas L, Castillo-Mendoza E, Mussali-Galante P, Pérez-Ruíz RV, Mendoza A (2013) Association between individual genetic diversity of two oak host species and canopy arthropod community structure. Eur J For Res 132:165–179. doi:10.1007/s10342-012-0665-y
Tscharntke T, Stefan-Dewenter I, Kruess A, Thies C (2002) Contribution of small habitat fragments to conservation of insect communities of grassland cropland landscapes. Ecol Appl 12:354–363. doi:10.2307/3060947
Valencia S (2004) Diversidad del género Quercus (Fagaceae) en México. Sociedad Botánica de México, México
Valencia-Cuevas L, Tovar-Sánchez E (2015) Oak canopy arthropod communities: which factors shape its structure? Rev Chil Hist Nat 88:1–22. doi:10.1186/s40693-015-0045-3
Valencia-Cuevas L, Piñero D, Mussali-Galante P, Valencia-Ávalos S, Tovar-Sánchez E (2014) Effect of a red oak species gradient on genetic structure and diversity of Quercus castanea (Fagaceae) in Mexico. Tree Genet Genomes 10:641–652. doi:10.1007/s11295-014-0710-8
Wade MJ (2003) Community genetics and species interactions. Ecology 84:583–585. doi:10.1890/0012-9658(2003)084[0583:CGASI]2.0.CO;2
Whitham TG (1989) Plant hybrid zones as sinks for pests. Science 244:1490–1493. doi:10.1126/science.244.4911.1490
Whitham TG, Morrow PA, Potts BM (1994) Plant hybrid zones as centers of biodiversity: the herbivore community of two endemic Tasmanian eucalypts. Oecologia 97:481–490. doi:10.1007/BF00325886
Whitham TG, Martinsen GD, Keim P, Floate KD, Dungey HS, Potts BM (1999) Plant hybrid zones affect biodiversity: tools for a genetic-based understanding of community structure. Ecology 80:416–428. doi:10.2307/176622
Whitham TG, Bailey JK, Schweitzer JA, Shuster SM, Bangert RK, LeRoy CJ, Lonsdorf EV, Gery Allan J, DiFazio SP, Potts BM, Fischer DG, Gehring CA, Lindroth RL, Marks JC, Hart SC, Wimp GM, Wooley SC (2006) A framework for community and ecosystem genetics:from genes to ecosystems. Nat Rev Genet 7:510–523. doi:10.1038/nrg1877
Wimp GM, Young WP, Woolbright SA, Martinsen PK, Whitham TG (2004) Conserving plant genetic diversity for dependent animal communities. Ecol Lett 7:776–780. doi:10.1111/j.1461-0248.2004.00635.x
Wimp GM, Martinsen GD, Floate KD, Bangert RK, Whitham TG (2005) Plant genetic determinants of arthropod community structure and diversity. Evolution 59:61–69. doi:10.1111/j.0014-3820.2005.tb00894.x
Zalapa JE, Brunet J, Guries RP (2009) Patterns of hybridization and introgression between invasive Ulmus pumila (Ulmaceae) and native U. rubra. Am J Bot 96:1116–1128. doi:10.3732/ajb.0800334
Acknowledgments
This project was supported by CONACYT project CB105755 and DGAPA-PAPIIT-UNAM project RV201015. Cuevas-Reyes P thanks Coordinación de la Investigación Científica UMSNH for their generous support. We also thank V. Rocha for technical assistance with microsatellite amplification. Finally, we acknowledge two anonymous reviewers for their constructive comments.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Raphael K. Didham.
Appendix
Appendix
See Fig. 6.
Rights and permissions
About this article
Cite this article
Pérez-López, G., González-Rodríguez, A., Oyama, K. et al. Effects of plant hybridization on the structure and composition of a highly rich community of cynipid gall wasps: the case of the oak hybrid complex Quercus magnoliifolia x Quercus resinosa in Mexico. Biodivers Conserv 25, 633–651 (2016). https://doi.org/10.1007/s10531-016-1074-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10531-016-1074-1