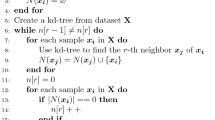Abstract
The Relief algorithm is a feature selection algorithm that uses the nearest neighbor to weight attributes. However, Relief only considers the correlation between features, which leads to a low classification accuracy on noisy datasets whose interaction effect is weak. To overcome the weaknesses of Relief, a novel feature selection algorithm, named Multidirectional Relief (MRelief), is proposed. The MRelief algorithm includes four improvements. First, the multidirectional neighbor search method, which finds all neighbors within a distance threshold from different orientations, is included to obtain regularly distributed neighbors. Therefore, the weights provided by MRelief are more accurate than those provided by Relief. Second, a novel objective function that incorporates the instances’ force coefficients is introduced to reduce the influence of noise. Thus, the new objective function improves the classification accuracy of MRelief. Third, subset generation is introduced to the MRelief algorithm and combined with the maximum Pearson maximum distance (MPMD) to generate a promising candidate subset for feature selection. Finally, a novel multiclass margin definition is proposed and introduced to the MRelief algorithm to handle multiclass data. As demonstrated by extensive experiments on eleven UCI datasets and eleven real-world gene expression benchmarking datasets, MRelief is significantly better than other algorithms including LPLIR, ReliefF, LLH-Relief, MultiSURF, MSLIR-NN, MRMR, MPMD and STIR in our study.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
An overwhelming amount of data is currently available [1, 2]. As the dimensionality of data increases, so does the number of features. Therefore, to select an optimal feature subset, feature selection is used [3, 4]. Feature selection [5, 6] is very effective in reducing dimensionality; and it is applied in numerous fields, such as pattern recognition [7], machine learning [8, 9], data mining [10] and some other fields [11]. Feature selection includes four processes, including subset generation, subset evaluation, stopping criterion and result validation.
Typical feature selection methods are divided into three classes: typical wrapper methods typical filter methods and embedded methods [12,13,14]. A typical wrapper method uses the performance of the classifier as an evaluation criterion while a typical filter method depends on the characteristics of the dataset to select a subset of features without evaluating any classifier [15]. Embedded methods have the advantage that they not only include the interaction with the classifier, but also take less computational time than wrapper methods [16]. Typical filter methods include the Maximum Relevance Minimum Redundancy (MRMR) [17] and ReliefF [18]. Typical wrapper methods include Particle Swarm Optimization (PSO) [19], the Genetic Algorithm (GA) [20], Bacterial Foraging Optimization (BFO) [21], Ant Colony Optimization (ACO) [22], Cuckoo Search (CS) [23], the Artificial Bee Colony (ABC) [24], the Whale Optimization Algorithm [25, 26] and the Dragonfly Algorithm (DA) [27]. Because the performance of a typical wrapper method heavily relies on the specified mining algorithm, typical filter methods are generally more popular than the wrapper methods.
Relief is an extensively studied filter method because it is simple and effective in high-dimensional feature space and nonparametric [28]. The improved Relief algorithms mainly address two aspects. First, the weight estimation function is improved. Second, different neighbor search methods are proposed.
In order to alleviate the deficiencies of Relief, improved algorithms with weight estimation functions, such as Relief-MM [29] and I-Relief [30], have been proposed. To improve the performance of the Relief algorithm, logistic local hyperplane-based Relief (LLH-Relief) has been proposed as a feature selection algorithm. LLH-Relief combines the advantages of logistic iterative-Relief (LI-Relief) and local hyperplane-Relief (LH-Relief). A multiclass semisupervised feature selection named the local preserving logistic I-Relief (LPLIR) algorithm that pays attention to the local characteristics of data has been proposed [31]. However, instance force coefficients are not considered in the above algorithms.
To avoid arbitrary selection of the nearest neighbors, recent research improves the number of selected nearest neighbors, such as using a fixed number of nearest neighbors in ReliefF, the constant neighborhood radius in SURF [32], the adaptive radius in MultiSURF [33] and a specific k for each feature in ReliefSeq [34]. However, none of the above algorithms solve the problem that neighborhoods of a center instance are irregularly distributed.
In summary, the above studies ignore four shortcomings. First, the neighborhoods of a center instance are irregularly distributed, thus, the neighborhoods that are far away from the center instance from different orientations are not involved in calculating the weights of Relief. Therefore, the weights of Relief are inaccurate for feature selection. Second, because the objective function assigns instances the same force coefficient based on their importance, noise and outliers affect the classification accuracy rate of Relief. Third, Relief ignores the correlation between features and classes. Furthermore, Relief does not consider the redundancy among features. Thus, Relief easily achieves the local optimal solution for classification. Finally, Relief was originally designed to solve two-class problems, and it is not applicable to multiclass data.
To address the above problems, a novel feature selection algorithm called MRelief is proposed. First, to solve the problem of irregularly distributed neighbors, MRelief finds all neighbors within a distance threshold in different orientations. The circle around the center instance within the distance threshold is split into specific uniform regions, and MRelief selects neighbors from all regions. Second, to improve the classification rate of MRelief, instance force coefficients are introduced to the margin-based objective function. This function integrates the Pearson correlation coefficient to represent the instance force coefficients. Third, a new subset generation process is proposed and combined with MPMD to obtain a promising candidate subset. Finally, to handle multiclass data, a novel multiclass margin definition is proposed.
Overall, we present four corresponding improvements to MRelief, as shown below.
-
1.
A multidirectional neighbor search method is proposed to select regularly distributed neighbors in different orientations.
-
2.
A novel objective function that incorporates instance force coefficients is formed to improve the classification accuracy rate.
-
3.
A subset generation method is proposed to obtain the optimal candidate subset.
-
4.
A multiclass margin definition is proposed to handle multiclass data.
The remainder of our research is organized as follows. In section 2, we introduce the theoretical background of our paper. In section 3, we propose a novel feature selection algorithm called MRelief. In section 4, we evaluate the performance of MRelief compared with ReliefF, LLH-Relief, LPLIR, MultiSURF and STIR. In section 5, the discussion and conclusion are given in detail.
2 Theoretical backgrounds
We introduce two types of filter algorithms. The filter algorithms are Relief-based algorithms and MPMD.
2.1 Relief-based algorithms
A number of Relief-based methods have been proposed. Relief-based algorithms output individual feature weights. In this section, some typical Relief-based methods, i.e., Relief, ReliefF, SURF, MultiSURF, and STIR, are briefly introduced.
2.1.1 Relief and ReliefF
Relief is a feature weighting algorithm [35]. Relief evaluates the weights of features consistent with the difference between instances that are similar. Relief randomly selects an instance X. According to a random sampling of instances, Relief finds its nearest hit H from the same class and its nearest miss M from the other different class. Then, Relief calculates the difference between two similar instances in order to influence the weights of attributes and updates the quality estimation W[Ai] depending on their values for X, M and H for all features A. In the same class, the feature has a negative influence if the difference between two similar instances is caused by the feature. In a different class, a feature has a positive influence if the difference between two similar instances is caused by the feature. The weight of feature Ai is calculated using the formula given in (1).
Function diff(Ai, X1, X2) calculates the differencebetween two instances X1 and X2 for featureAi. W[Ai] is the weight of feature Ai. T is number of the maximum iterative times.
The numerical attributes are calculated as follows.
where value(Ai, X1) is the value of feature Ai for instance X1.
The nominal attributes are calculated as follows.
The original Relief can handle both nominal and numerical attributes. However, it is limited to solving datasets that are two-class. In addition, the original Relief cannot handle incomplete data.
The ReliefF algorithm is able to handle problems with multiclass datasets that contain noisy data. Based on Relief,ReliefF finds its K nearest hits Hj from the same class and its K nearest misses Mj(Class) from a different class according to a randomsampling of instances. It updates the weight W[Ai] relying on , Mj(Class) and Hj for all features A.
whereP(C)is the prior probability of class C. T is the maximum number of iterations. Class(Xm) is the class of instance Xm.
2.1.2 SURF and MultiSURF
The SURF algorithm [32] is similar to the ReliefF algorithm. Different from ReliefF, SURF implements a new neighbor search method. Instead of using a fixed parameter k, SURF adopts a distance threshold T to determine which instances are selected. The radius T of a given target instance is calculated as the average of two instances.
MultiSURF defines a threshold Ti − σi/2 to determine which instances are considered neighbors [32]. Ti is the mean pairwise distance between the target instance and all others. σi is the standard deviation of the pairwise distances between the target instance and all others.
2.1.3 STIR
Combined with the pooled standard deviations, this novel Relief-based algorithm transforms the Relief-based score WSTIR into a pseudo t-statistic [36]. For feature A, the STIR weight is calculated as the difference of the means and the standard error.
where |M| and |H| are the total numbers of nearest misses and hits, respectively. The pooled standard deviation is calculated by the following formula. \( {\overline{M}}_A \) is the mean difference for feature A averaged over of all pairs of nearest misses. \( {\overline{H}}_A \) is the mean difference for feature A averaged over of all pairs of nearest hits.
where \( {S}_{M_A}^2 \) and \( {S}_{H_A}^2 \) are the pooled standard deviations.
2.2 MPMD
Zheng et al. proposed a new filter algorithm called the MPMD [37]. The relevance between classes and features is calculated using the Pearson correlation coefficient, and the redundancy among features is calculated using the correlation distance.
The value of the maximum Pearson correlation coefficient between the feature set A = {A1, A2, …Ai, AD} and the class set Class = {Class(X1), Class(X2), …Class(Xj), Class(XN)} is calculated by the maximum of Pearson correlation coefficient between the feature and the class using (7).
where PE(Ai, Class) is calculated using (8).
where cov(Ai, Class)is the covariance between feature Ai and class Class in (9), σ(Ai) is defined by (10) and σ(Class) is defined by (11).
where\( {A}_i^j \)is value of jth instance for feature Ai, \( \overline{A_i} \) is the average of feature \( {A}_i \),\( \overline{Class} \) is the average of class Class.
where N is the number of instances.
The maximum correlation distance between remaining feature Rm and selected feature Sk is calculated by the maximum correlation coefficient in (12).
where the remaining feature set is defined by R = {R1, R2, …Rm, RM}, the selected feature set is defined by S = {S1, S2, …Sk, SW} and D = M + W. D(Sk, Rj) is calculated using (13).
MPMD is generated by the maximum Pearson correlation coefficient and maximum correlation distance. Two factors, r1 and r2, are introduced to balance MP and MD.
where r1 and r2 are calculated using (15) and (16), respectively.
where t represents the current number of iterations, T represents the total number of iterations and a is a constant.
3 The proposed algorithm
In this part, we introduce the multidirectional neighbor search method, the new relief-feature weighting objective function, the subset generation and the multiclass extension.
3.1 Multidirectional neighbor search method
Because neighbors far away from the center instance are useless, MRelief finds all neighbors within a distance threshold r instead of selecting a fixed number of neighbors. However, the neighborhoods of a center instance are irregularly distributed, and we endeavor to select neighbors in different orientations. The circle around the center instance within the distance threshold r is split into 5 uniform regions. In Fig. 1, each region represents a direction, and region has an azimuth angle θ that is set to 72∘. We select m nearest neighbor instances from a region to represent the region. For example, when m = 1, 2, 3. .., K = 5, 10, 15. .. ., where K is the number of selected neighbors. To avoid insufficient instances inside a region, we use iterative farthest point sampling (FPS) to choose the neighbor set \( \left\{{X}_{i_1},{X}_{i_2},\dots, {X}_{i_m}\right\} \) of the center instance. Given input instances {X1, X2, …, XN}, N is the total number of instances. Thus, \( {X}_{i_j} \)is the most distant point from the set \( \left\{{X}_{i_1},{X}_{i_2},\dots, {X}_{i_{j-1}}\right\} \) with regard to the remaining instances.
In Fig. 1, the circle is drawn with the center instance as the circle center and the search radius as the circle radius. The circle is divided into 5 uniform directions. The azimuth θ, radius r and m selected instances are marked in one region. The existing methods use some neighbor search methods. ReliefF selects a fixed number of nearest neighbors. SURF uses a constant neighborhood radius to select neighbors. MultiSURF uses the adaptive radius for neighbor searches. ReliefSeq assigns a specific k to each feature. However, none of the existing methods solve the problem that the neighborhoods of a center instance are irregularly distributed.
In Fig. 2, Relief finds the nearest neighbor in the same class (green circle) and the nearest neighbor in the other class (orange rectangle). ReliefF finds a specified number of neighbors (five in this example) that are used to calculate the weights. MRelief finds all neighbors that are divided into 5 uniform regions within a distance threshold, which is expressed as a circle. The center instance that selects neighbors to be used for weighting is expressed as a filled red circle. The neighbors from the same class are highlighted in green and the neighbors from the other class are highlighted in orange for each algorithm. Parts A, B, and C represent Relief, ReliefF and MRelief, respectively.
3.2 New relief-feature weighting objective function
In Relief, each instance in a given dataset is equally important for constructing optimization objective functions. However, a reasonable objective function should assign instances different force coefficients based on their importance. Thus, two instance force coefficients are introduced to generate a new objective function to reduce the influence of noise.
As shown in (17), when the degree of coincidence is large between the instance and the farthest miss, the instance needs to be given a small instance force coefficient um for Xm
where M1(Class(Xm)) is the farthest miss of Xm from other different classes, coefficient r1 is used to adjust um. PE is the Pearson correlation coefficient calculated by (8). The constraints ‖um‖ = 1 prevent the maximization from increasing without bound, and um ≥ 0 ensures that the instance force coefficient induces a distance measure.
In (18), when the degree of coincidence between the instance and farthest hit is large, the instance needs to be given a large instance force coefficient.
where H1(Class(Xm)) is the farthest hit of instance Xm, coefficient r2 is used to adjust hm. PEis the Pearson correlation coefficient calculated by (8). The constraint ‖hm‖ = 1 prevents the maximization from increasing without bound, and hm ≥ 0 ensures that the instance force coefficient induces a distance measure.
When the instance force coefficient is introduced, the objective function for instance Xm is expressed as follows:
wherefunction diff(Ai, X1,X2) calculates thedifference for feature Ai between two instances X1 and X2. K is thenumber of nearest hits and misses. Mj(Class) is the selected nearest misses from other different classes. Hj is the selected nearest hits from the same class. N is the number of instances. Class(Xm)is the class of instance Xm.
J(w) is the objective function for a given training dataset computed with respect to w. (20) is constructed to optimize um and hm.
where w is the weight vector. w is defined by w = {w1, w2, …, wi, wD}. ρm is calculated by (19). N1 is the number of instances in a given training dataset.
3.3 Subset generation method
Better classification can be achieved if some features with small but important weights are added in the process of generating feature subsets. Relief uses a fixed threshold to select attributes directly according to the feature weight. However, some features whose weight is small but important cannot be selected. Therefore, MPMD is used to select features in MRelief. MPMD balances the maximum Pearson correlation coefficient and the maximum correlation distance in order to enrich candidate subsets. In (21), the probability of a feature Ai being selected is expressed by the result of combining the weights of MRelief and the weights of MPMD.
where β is a coefficient that is used to balance the degree of importance between MPMD and MRelief. When β is set to 1, it means that MPMD and MRelief are equally important. For example, when β is set to 0.5, it means that the force coefficient of MPMD is 0.25 while the force coefficient of MRelief is 1. Thus, MPMD is less important than MRelief. For example, when β is set to 1.5, it means that the force coefficient of MPMD is 1.25 while the force coefficient of MRelief is 1. Therefore, MPMD is more important than MRelief.
3.4 Multiclass extension
In order to handle multiclass data, the margin of an instance is calculated by MReliefF as follows:
where Y ∈ Class is the set of class labels, y(Xm) is the label of instance Xm, Mj(C) is the nearest miss of Xm from class C, Hj is the selected nearest hits from the same class, hm and um are the instance force coefficients, P(C) is the a priori probability of class C and Class(Xm) is the class of instance Xm.
As summarized by the pseudocode in Algorithm 1, MRelief randomly selects M instances in each cycle. For each selected instance, MRelief finds its K nearest hits Hj and its K nearest misses Mj(Class) according to the multidirectional neighbor search method. For each feature, MRelief uses a new Relief-Feature weighting objective function to update the weights. Moreover, the value of MPMD is calculated by (14). Finally, the probability of a feature is determined using (21) according to subset generation.

4 Experiment results
4.1 Benchmark data
We selected eleven UCI datasets and eleven microarray datasets in the following experiments. UCI datasets are downloaded from http:// archive.ics.uci.edu/ml. The microarray datasets are downloaded from http://datam.i2r.a-star.edu.sg/datasets/krbd/. The characteristics of the datasets, which contain numeric data, nominal data and mixed data, are introduced in Tables 1 and 2. As we can see from Tables 1 and 2, the number of instances varies from 72 to 10,992, the number of features varies from 13 to 16,063 and the number of classes varies from 2 to 26.
4.2 Parameter settings
The experiments include state-of-the-art Relief-based methods and mutual-information-based feature selection methods. The detailed parameter values of each algorithm are described in Table 3. For ReliefF, the number of iterations is set to 10, and the number of selected instances is set to 5. For LLH-Relief, the number of iterations is set to 10, the number of selected instances is set to 5, θ = 0.01, λr = 0.001,λ = 1 and η = 0.1. For LPLIR, the number of iterations is set to 10, the number of selected instances is set to 5, λ1 = 1 and λ2 = 0.01. For MultiSURF, the number of iterations is set to 10, and the number of selected instances is set to 5. For MSLIR-NN, the number of iterations is set to 10, and the number of selected instances is set to 5. For MRMR, the number of iterations is set to 10. For MPMD, the number of iterations is set to 10. For STIR, the number of iterations is set to 10, and the number of selected instances varies for each instance. For MRelief, the number of iterations is set to 10, the number of selected instances is set to 5, r1=1.1 and r2=0.9.
4.3 Experiments and analysis
The experiments are conducted by implementing ReliefF, LLH-Relief, LPLIR, MultiSURF, STIR, MSLIR-NN, MRMR, MPMD and MRelief using MATLAB R2014a. Each algorithm is run 10 times, and the average classification accuracy rate is calculated as the final result. The SVM classifier is used to evaluate the classification performance for each filter algorithm [38, 39]. The kernel function of the SVM classifier is the radial basis function (RBF). Penalty parameter C and RBF parameter gamma are obtained through the grid search method. Ten-fold cross validation is applied to test the algorithms mentioned above. Ten-fold cross validation consists of 10 cycles. The datasets are divided into 10 folds for every cycle. Then, 9 folds are used for training, and the remaining fold is used for testing. In our experiments, two criteria are considered including the classification accuracy rate Acc and the F1 value. Acc and the F1 value are calculated in every cycle. Therefore, we obtain the average classification accuracy rate of 10 cycles. The Acc criterion is computed by the following formula.
where TP (TN) represents the number of positive (negative) instances that are classified correctly and FP (FN) is the number of positive (negative) instances that are classified incorrectly.
The F1 value is calculated in every cycle. Therefore, we obtain the average F1 value of 10 cycles. The F1 criterion is computed by the following formula.
where TPrate and PPrate are calculated by the following formulas.
Table 4 shows the average classification accuracies of ReliefF, ReliefF with subset generation in (21) and ReliefF with new relief-feature weighting objective function in (22) applied to ten datasets. The results show the effectiveness of the formulas in (21) and (22) is able to work better than the traditional method. The subset generation in (21) generates a promising candidate subset for feature selection combined with MPMD. The novel objective function that incorporates the instances’ force coefficients in (22) improves the classification accuracy of MRelief.
Figure 3 indicates that MRelief performs the best on the general trend of the average classification accuracy. The result of Musk clearly shows that MRelief achieves the highest classification accuracy over the entire process. LLH-Relief obtains the local optimal classification accuracy other than that of MRelief. In addition, LLH-Relief achieves the highest classification accuracy in Wine and Vowel. As the Waveform results show, the classification accuracy of LPLIR is higher than those of the other algorithms except MRelief. The results of Sonar show that the classification accuracies of MRelief and STIR both achieve excellent performance. In Vowel and Pendigits, there are overlaps among curves. Furthermore, there are crosses in Parkinson’s and Heart. Although the nine algorithms sometimes obtain the same accuracy rate, we know that MRelief has excellent performance among the nine algorithms. The results of Fig. 3 show that the novel preprocessing method significantly promotes the classification accuracy of MRelief, and the improved objective function makes a huge contribution to jumping out of the local optimum for MRelief.
Table 5 and Table 6 show the mean accuracies and standard deviations (%) of the algorithms on the UCI and microarray datasets. The tables compare the average accuracies and standard deviations on the target sets among nine algorithms including ReliefF, LLH-Relief, LPLIR, MultiSURF, STIR, MSLIR-NN, MRMR, MPMD and MRelief. In Table 5, nine algorithms are tested on eleven UCI datasets. To compare the nine feature selection algorithms, 50-dimensional irrelevant data, which are obtained from a zero mean and unit variance Gaussian distribution, are added to the original UCI datasets. In Table 6, 200 top-ranked microarray features are selected for feature selection. Microarray datasets have redundant features, so we do not need to add extra noise to the original microarray datasets. The results show that MRelief achieves the highest average classification accuracy on most datasets except Vehicle Silhouettes, Wine and GCM. The results show that the instance force coefficient improves MRelief significantly, and subset generation greatly promotes MRelief. Moreover, the results show that MRelief obtains the lowest average standard deviation on six datasets. The multidirectional neighbor search method improves the stability of MRelief.
Friedman test are conducted with the corresponding post-hoc tests. The Bonferroni–Dunn test is selected as post-hoc tests, which is calculated by the differences between MRelief and other algorithms [42]. The performance of pairwise algorithms is significantly different if the corresponding differences between MRelief and other algorithms is higher than the critical difference (CD). The CD is calculated by (27).
where NA is the number of algorithms, ND is number of datasets. Generally, α is set to 0.1, q0.10 = 2.326. In this paper, NA = 9, ND = 11. Thus, CD0.1 = 2.72.
Table 7 and Table 8 show the Friedman test results. In UCI datasets, LPLIR has significant difference compared with ReliefF, LLH-Relief, LPLIR, MSLIR-NN and MRMR. In microarray datasets, LPLIR has significant difference compared with other eight algorithms.
Figure 4 shows that MRelief has a performance advantage over the compared Relief-based methods in average classification accuracy. According to the results of LUNG, Leukemia3, Pros1, Pros2, 11-Tumors, and 14-Tumors, it is clear that MRelief achieves the highest classification accuracy over the entire process. In Leukemia2, Lymphoma, DLBCL and Colon, there are crosses among curves. Furthermore, there are overlaps in GCM. In the case of microarray datasets, MRelief is able to achieve the best performance among the compared methods. This implies that MRelief is able to extract useful information from a large number of features. Thus, MRelief achieves better classification performance. The results of Fig. 4 show that the novel subset generation method significantly promotes the classification accuracy rate of MRelief.
Table 9 shows that MRelief achieves superior F1 results in ten out of eleven datasets. Table 10 shows that MRelief outperforms other algorithms over all datasets. In conclusion, MRelief is competitive among Relief-based methods, regardless of what criteria are adopted.
Tables 11 and 12 show the Friedman test results with the F1 values. In UCI datasets, MRelief has significant difference compared with ReliefF, LPLIR, MRMR and MPMD. In microarray datasets, LPLIR has significant difference compared with other eight algorithms.
The computational complexity of MRelief is O(T1N2D + T2N2D), which consists of two parts. In the first part, the computational complexity of MRelief is O(T1N2D). T1 represents the maximum number of iterations of MRelief without subset generation. D represents the number of features. N represents the number of instances. The computational complexity is the same as ReliefF because the multidirectional neighbor search method, new relief-feature weighting objective function and multiclass extension do not increase the computational complexity of MRelief. In the second part, the computational complexity of MRelief is O(T2N2D) because subset generation is combined with MPMD. T2 represents the maximum number of MPMD. The experiments include state-of-the-art relief-based methods and mutual-information-based methods. The representative relief-based method is ReliefF, and the representative mutual-information-based method is MRMR. Thus, ReliefF and MRMR are selected as comparison algorithms. The computational complexity of MRMR and ReliefF is O(N2D). Therefore, the computational complexity of MRelief is the highest compared to ReliefF and MRMR.
In Tables 13 and 14, without calculating the execution time of the classifier, the execution time consumption is calculated in one iteration. Table 13 shows the mean running times of nine algorithms on the top 200 features selected from eleven UCI datasets. Table 14 shows the mean running times of nine algorithms on the top 200 features selected from eleven microarray datasets. It can be observed that the mean running times of MRelief is higher than the other two algorithms in both UCI and microarray datasets. Since MRelief combines ReliefF and MRMR, the running time of MRelief is almost equal to the sum of the running time of ReliefF and MRMR.
Table 15 shows the results of eight pairs of Wilcoxon signed-rank tests performed on the Colon dataset. The results show that the performance of MRelief is statistically significantly better than those of ReliefF, LLH-Relief, MultiSURF, LPLIR, STIR, MSLIR-NN, MRMR and MPMD at a significance level of 0.05. MRelief has demonstrated its robustness in handling noisy datasets whose interaction effect is weak. MRelief is able to extract useful information from a large number of features. Therefore, the multidirectional neighbor search method, new relief-feature weighting objective function, subset generation and multiclass extension make contribution to MRelief.
5 Discussion
Tables 5 and 6 show that MRelief achieves the highest average classification accuracy rate on most datasets except Vehicle Silhouettes, Wine and GCM. Although MRelief does not obtain the best learning accuracies compared with other algorithms on all datasets, the learning accuracies of MRelief are better than those of other algorithms on ten microarray datasets except GCM. MRelief is more suitable for high-dimensional datasets. In addition, Tables 5 and 6 show that MRelief achieves the highest Rank compared with other algorithms on both UCI datasets and microarray datasets. Furthermore, Figs. 3 and 4 show that MRelief performs the best on the general trend with respect to the average classification accuracy. The results show that the instance force coefficient improves MRelief significantly, and subset generation greatly promotes MRelief. Moreover, Tables 5 and 6 show that MRelief obtains the lowest average standard deviation in most datasets. Therefore, the proposed multidirectional neighbor search method is able to select representative neighbors for feature selection.
6 Conclusion and further work
Relief is a feature selection method that reduces the number of features. Relief is the only individual evaluation filter algorithm that is capable of detecting feature dependencies. Due to the low classification accuracy rate of Relief, a novel filter feature selection algorithm named MRelief is proposed in this paper. First, the multidirectional neighbor search method with a distance threshold is designed. Second, to improve the classification accuracy, a reasonable objective function assigns instances different force coefficients based on instances’ contribution to classification. Third, subset generation is proposed to obtain the optimal candidate subset. Last, the MRelief algorithm implements a multiclass margin definition to handle multiclass data.
To demonstrate the effectiveness of MRelief, experiments were conducted on 11 UCI and 11 microarray datasets. MRelief is compared with LPLIR, ReliefF, LLH-Relief, MultiSURF, MSLIR-NN, MRMR, MPMD and STIR. The results show that the multidirectional neighbor search method, new relief-feature weighting objective function, subset generation and multiclass extension significantly improve MRelief.
In the future, we will improve running time of the MRelief in handling with largescale datasets and high dimensional datasets, which is important to their application to gene expression. Since MRelief combines ReliefF and MRMR, the running time of MRelief is consist of the running time of ReliefF and MRMR. To deal with this shortage, we will improve mRMR to save the execution time. Moreover, we will improve the classification accuracy rate of MRelief combined with wrapper method. Furthermore, we robust MRelief to deal with multiple types of datasets, such as missing datasets, unlabeled datasets and muti-label datasets.
References
Craven M, DiPasquoa D, Freitagb D, McCalluma A, Mitchella T, Nigama K, Slatterya S (2000) Learning to construct knowledge bases from the world wide web. Artif Intell 118(1–2):69–113
Roweis ST, Saul LK (2000) Nonlinear dimensionality reduction by locally linear embedding. Science 290(5500):2323–2326
Kushwaha N, Pant M (2018) Link based BPSO for feature selection in big data text clustering. Future Gener Comput Syst 82:190–199
Blum AL, Rivest RL (1992) Training a 3-node neural networks is NP-complete. Neural Netw 5(1):117–127
Men M, Zhong P, Wang Z, Lin Q (2020) Distributed learning for supervised multiview feature selection. Appl Intell 50(9):2749–2769
Xiang S, Shen XT, Ye JP (2015) Efficient nonconvex sparse group feature selection via continuous and discrete optimization. Artif Intell 224:28–50
Zhang P, Gao WF, Liu GX (2018) Feature selection considering weighted relevancy. Appl Intell 48(12):4615–4625
Cohen JP, Ding W, Kuhlman C, Chen AJ, Di LP (2016) Rapid building detection using machine learning. Appl Intell 45(2):443–457
Xiao J, Cao HW, Jiang X, Jiang XY, Gu X, Xie L (2017) GMDH-based semi-supervised feature selection for customer classification. Knowledge-Based Syst 132:236–246
Belkoura S, Zanin M, LaTorre A (2019) Fostering interpretability of data mining models through data perturbation. Expert Syst Appl 137:191–201
Ji CY, Li Y, Fan JH, Lan SM (2019) A novel simplification method for 3D geometric point cloud based on the importance of point. IEEE ACCESS 7:129029–129042
Zheng YF, Li Y, Wang G, Chen YP, Xu Q, Fan JH, Cui XT (2018) A novel hybrid algorithm for feature selection. Pers Ubiquit Comput 22(5–6):971–985
Chen YP, Li Y, Wang G, Zheng YP, Xu Q, Fan JH, Cui XT (2017) A novel bacterial foraging optimization algorithm for feature selection. Expert Syst Appl 83:1–17
Turabieh H, Mafarja M, Li XD (2019) An introduction to variable and feature selection. Expert Syst Appl 3:1157–1182
Cui XT, Li Y, Fan JH, Wang T, Zheng YF (2020) A hybrid improved dragonfly algorithm for feature selection. IEEE ACCESS 8:155619–155629
Guyon I, Gunn S, Nikravesh M, Zadeh LA (2006) Feature extraction, foundations and applications. Springer
Peng H, Long F, Ding C (2005) Feature selection based on mutual information criteria of max-dependency, max-relevance, and min-redundancy. IEEE Trans Pattern Anal Mach Intell 27(8):1226–1238
Urbanowicz RJ, Meeker M, Cava WL, Olson RS, Moore JH (2018) Relief-based feature selection: introduction and review. J Biomed Informat 85:189–203
Li Y, Wang G, Chen HL, Dong H, Zhu X, Wang S (2011) An improved particle swarm optimization for feature selection. J Bionic Eng 8(2):191–200
Nazemi A, Dehghan M (2015) A neural network method for solving support vector classification problems. Neurocomputing 152:369–376
Kim DH, Abraham A, Cho JH (2007) A hybrid genetic algorithm and bacterial foraging approach for global optimization. Inf Sci 177(18):3918–3937
Kashef S, Nezamabadi-pour H (2015) An advanced ACO algorithm for feature subset selection. Neurocomputing 147:271–279
Mohapatra P, Chakravarty S, Dash PK (2015) An improved cuckoo search based extreme learning machine for medical data classification. Swarm Evol Compu 24:25–49
Karaboga D, Gorkemli B, Ozturk C, Karaboga N (2014) A comprehensive survey: artificial bee colony (ABC) algorithm and applications. Artif Intell Rev 42(1):21–57
Chen H, Li WD, Yang X (2020) A whale optimization algorithm with chaos mechanism based on quasi-opposition for global optimization problems. Expert Syst Appl 158:113612
Abdel-Basset M, Mohamed R, Mirjalili S (2021) A novel whale optimization algorithm integrated with Nelder-Mead simplex for multi-objective optimization problems. Knowledge-Based Syst 212:106619
Mafarja M, Aljarah I, Heidari AA, Faris H, Fournier-Viger P, Li XD, Mirjalili S (2018) Binary dragonfly optimization for feature selection using time-varying transfer functions. Knowledge-Based Syst 161:185–204
Reyes O, Morell C (2015) Scalable extensions of the ReliefF algorithm for weighting and selecting features on the multi-label learning context. Neurocomputing 161:168–182
Yilmaz T, Yazici A, Kitsuregawa M (2014) RELIEF-MM: effective modality weighting for multimedia information retrieval. Multimedia Syst 20(4):389–413
Sun Y (2007) Iterative RELIEF for feature weighting: algorithms, theories, and applications. IEEE Trans Pattern Anal Mach Intell 29(6):1035–1051
Tan B, Zhang L (2020) Local preserving logistic I-relief for semi-supervised feature selection. Neurocomputing 399:48–64
Greene CS, Penrod NM, Kiralis J, Moore JH (2009) Spatially uniform relieff (SURF) for computationally-efficient filtering of gene-gene interactions. BioData Min 2(1):5
Urbanowicz RJ, Olson RS, Schmit P, Meeker M, Moore JH (2018) Benchmarking relief-based feature selection methods for bioinformatics data mining. J Biomed Informat 85:168–188
McKinney BA, White BC, Grill DE, Li PW, Kennedy RB, Poland GA, Oberg AL (2013) ReliefSeq: a gene-wise adaptive-K nearest-neighbor feature selection tool for finding gene-gene interactions and main effects in mRNA-Seq gene expression data. PLoS One 8:e81527
Robnik-Sikonja M, Kononenko I (2003) Theoretical and empirical analysis of ReliefF and RReliefF. Mach Learn 53(1–2):23–69
Le TT, Urbanowicz RJ, Moore JH, Mckinney BA (2018) Statistical inference relief (STIR) feature selection. Bioinformatics 35(8):1358–1365
Zheng YF, Li Y, Wang G, Chen YP, Xu Q, Fan JH, Cui XT (2019) A novel hybrid algorithm for feature selection based on whale optimization algorithm. IEEE ACCESS 7:14908–14923
Cortes C, Vapnik V (1995) Support-vector network. Mach Learn 20(3):273–297
Leopold E, Kindermann J (2001) Text categorization with support vector machines. How to represent texts in input space? Mach Learn 46(1–3):423–444
Tang BG, Zhang L (2019) Multi-class semi-supervised logistic i-relief feature selection based on nearest neighbor. In: Pacific-Asia conference on knowledge discovery and data mining, pp 281–292
Zhang L, Huang X, Zhou W (2019) Logistic local hyperplane-relief: a feature weighting method for classification. Knowledge-Based Syst 181:104741
Demšar J (2006) Statistical comparisons of classifiers over multiple data sets. J Mach Learn Res 7:1–30
Acknowledgments
This research was supported by the Department of Science and Technology of Jilin Province (No. 20190303135SF).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Cui, X., Li, Y., Fan, J. et al. A novel filter feature selection algorithm based on relief. Appl Intell 52, 5063–5081 (2022). https://doi.org/10.1007/s10489-021-02659-x
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10489-021-02659-x