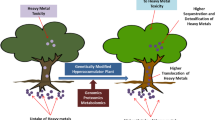
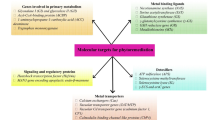
Abstract
Contamination by heavy metals including As, Cd, Co, Cu, Fe, Hg, Mn, Ni and Zn in agricultural fields is a global safety issue. Indeed, excessive accumulations of metals have detrimental effects on life by altering cell components such as lipids, proteins, enzymes and DNA. Phytoremediation appears as a solution to remove metals from contaminated sites, yet metal uptake is usually low in most common plants. Therefore, genetically engineered plants have been designed for higher efficiency of metal accumulation. Here, we review metal phytoremediation by genetically engineered plants with focus on metal uptake and transport, mechanisms involving phytochelatin and metallothionein proteins, toxicity, plant species, methods of gene transfer and gene editing.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Pollution, especially since the beginning of industrial revolution (Akguc et al. 2008), is causing negative impacts on the biosphere, and altering the hydrological cycle and ecological balance (Gomes et al. 2016; Nanasato et al. 2016; Ozturk et al. 2017; Xia et al. 2018). The common anthropogenic sources causing widespread pollution are mining activities, consumption of fossil fuel, fertilizer and pesticide utilization in agriculture, discharges of various types of urban wastes, potential emerging pollutants (Dharupaneedi et al. 2019) and productions of petroleum-based materials including petroleum hydrocarbons, polycyclic aromatic hydrocarbons, polychlorinated biphenyls, halogenated hydrocarbons, solvents (trichloroethylene) and explosives (Gerhardt et al. 2009; Osma et al. 2012; Gerhardt et al. 2017). The strategies that comprise membrane-based filtration techniques including microfiltration, ultrafiltration, nanofiltration, reverse osmosis and forward osmosis could be applied particularly to waste/surface water or groundwater for removing potential emerging pollutants (reviewed by Dharupaneedi et al. (2019). To remove potential emerging pollutants from wastewater, the methods involving using of nanomaterial-based filtration including Fe-nanoparticle (Reddy et al. 2020), Zr-nanoparticle (Reddy et al. 2019), Bi-nanoproduct (Koutavarapu et al. 2020), Ce-nanoproduct (Tavangar et al. 2020) photocatalytic activities have successful been applied. Many researchers meet on a common ground point that the above-mentioned contaminants adversely affect living organisms including human (Mudgal et al. 2010; Sarwar et al. 2010; Ali et al. 2013; Kelishadi et al. 2014; Ramasahayam et al. 2017). These contaminants are subdivided into two groups: organic contaminants, including pesticides, pharmaceuticals, petroleum compounds such as polycyclic aromatic hydrocarbons, polychlorinated biphenyls and inorganic contaminants, including mostly heavy metals such as Cd, Co, Cr, Cu, Fe, Hg, Mn, Mo, Ni, Pb and Zn (Mudgal et al. 2010; Fasani et al. 2018; Ozyigit et al. 2018). As and U are also toxic elements causing pollution in environment. Some elements known as essential microelements including Cu, Fe, Mn, Mo, Ni and Zn required for optimum growth and development could cause toxicity because of disturbed biological processes in organisms if posed to excessive amounts via primarily soil and water-borne transportation. Each of these essential elements has unique and critical role-play in plants life cycle and therefore should be well supplied for regular growth and development. Accumulation of toxic and nonessential contaminants in different environmental layers (soil, air and groundwater) is become continually growing major concern of modern world nowadays (Kang 2014; Tue et al. 2014; Pandey et al. 2015; Ozyigit et al. 2017). Recent estimation showed that over 5 million hectares is contaminated worldwide by heavy metals (He et al. 2015). And more important than that beyond broad contamination, heavy metals cannot be decomposed by biological systems and many terrain organisms are constantly exposed to these heavy metals (Sarwar et al. 2010; Vijgen et al. 2011). Decontamination/removing or after being reduced form releases of contaminants spread over gains great importance for health of nature as well as humanity (Schnoor et al. 1995; Susarla et al. 2002).
Heavy metal entry into the food chain is of great concern due to threatening human health; therefore, there is a great worldwide effort to find out effective ways for removing of heavy metals from soil as well as from other contaminated layers of biosphere (Ghori et al. 2016). According to the recent literature, the most common and popular strategy for removing of heavy metals from contaminated areas is phytoremediation (Sun et al. 2018b; Nehnevajova et al. 2019; Zhao et al. 2019a; Farid et al. 2020; Saleh et al. 2020; Zehra et al. 2020). Various types of remediation strategies bearing electrical, chemical, physical, biological and their integrated approaches are available for cleaning up contaminated sites. These cleanup strategies include: surface capping and encapsulation under context of physical approach; electro-kinetics and vitrification under the context of electrical approach; soil flushing and immobilization under the context of chemical approach; and finally phytoremediation under the context of biological approach, respectively (Liu et al. 2018). A common concern about employing chemical and physical methods for cleaning up the contaminated areas is related to suitability due to economic consideration and estimation for being harmful due to chemical reagent utilization causing further contamination in the soil (Heckenroth et al. 2016; Sarwar et al. 2017; Fasani et al. 2018; Sun et al. 2018a). Therefore, phytoremediation as an in situ safe and cost-effective way of cleaning up these organic/inorganic contaminants primarily from soil using plants and some plant-associated microorganisms is the most convenient choice nowadays (Arslan et al. 2017; Feng et al. 2017; Gerhardt et al. 2017). Some of trace elements and heavy metals, including Ag, As, Au, Bi, Cd, Ce, Co, Cr, Cu, Fe, Hg, Mn, Ni, Pb, Sb, Sn, Te, Tl, U, V and Zn, could be removed from contaminated area via application of phytoremediation processes involving employing of certain plant species (Ozyigit and Dogan 2015).
Here we review the absorption and removal of heavy metals from the environment using various phytoremediation methods including the mechanisms, especially focusing on the explanation of the phytoremediation efficiency boosted by genetically modified plants. We discuss the use of transgenic plants, and we compare conventional and the cutting-edge methods.
Mechanisms of phytoremediation
Phytoremediation is mainly divided into seven processes in which physiological mechanisms are phytoextraction, rhizofiltration, phytostabilization, phytodegradation, rhizodegradation, phytovolatilization and hydraulic control, as shown in Fig. 1a (Ozyigit and Dogan 2015; Muthusaravanan et al. 2018).
A Main phytoremediation strategies, modified from Ozyigit and Dogan 2015, B transporter proteins used during the uptake of heavy metal into plant, NRAMP natural resistance-associated macrophage protein, OPT oligopeptide transporters, MATE multidrug and toxic compound extrusion (MATE) family of transporters, ZIP ZTR/IRT-related proteins, HMA4 heavy metal ATPase 4 (copyrighted illustration from Prof. Ozyigit and Dr. Can), C detoxification of heavy metals taken into the plant, HM heavy metal, PC phytochelatins, MT metallothionein, GSH glutathione, PAL phenylalanine ammonia-lyase, oxML oxy monolignol, LMWC low molecular weight compounds (amino acid/organic acid) (copyrighted illustration from Prof. Ozyigit and Dr. Can)
Phytoextraction allows ecological restoration of soils contaminated by toxic metals in a way of transportation of them from roots to above-ground parts of plants, resulting accumulation of them in plants; thus, phytoextraction provides reducing of toxic metal concentrations in soils via harvesting plant biomass and subsequent incineration for removing toxic metals permanently (Cherian and Oliveira 2005; Zhang et al. 2010; Ali et al. 2013; Ozyigit and Dogan 2015).
In general, phytoextraction is classified into two categories: chelate-assisted or induced phytoextraction and long-term or continuous phytoextraction. First one is potentially more convenient in use and currently being utilized commercially (Jan et al. 2015). Phytoextraction approach can be utilized to remove heavy metals, including Ag, Cd, Co, Cr, Cu, Hg, Mo, Ni, Pb and Zn from contaminated areas (DalCorso et al. 2019).
All soils contain heavy metals in different concentrations depending on the conditions and pollution status of soil. Plants showing ability to survive or accumulate heavy metals in large amounts are known as hyperaccumulators and commonly utilized in phytoextraction technology. Slow growth, a shallow root system and low biomass are the famous features of hyperaccumulator plants (Surriya et al. 2015).
Some well-known plants used in phytoextraction technology are listed as: Brassica juncea (Salido et al. 2003), Lactuca sativa and Lolium perenne (Hernández et al. 2019), Mesembryanthemum criniflorum (Manzoor et al. 2019), Nicotiana tabacum (Yang et al. 2019b), Pelargonium hortorum (Gul et al. 2019; Manzoor et al. 2019), Sedum alfredii (Ning et al. 2019), Solanum nigrum (He et al. 2019), Thlaspi caerulescens (Cosio et al. 2004), Xanthium strumarium (Khalid et al. 2019) and Zea mays (Huang et al. 2019),
Based on rhizofiltration, contaminated aquatic environments (damp soils and ground and/or surface waters) due to mainly heavy metal exposure can be treated in purpose of removing pollutants via using the way of adsorption or precipitation onto roots or other submerged organs of metal tolerant aquatic plants (Kvesitadze et al. 2006; Jadia and Fulekar 2009). Experiments showed that efficient uptakes of radionuclides and heavy metals are being realized using aquatic hyperaccumulator plants through induction of hairy roots (Nedelkoska and Doran 2000; Eapen et al. 2003). There are reports about appearing successful results being employing rhizofiltration for extraction of Cd, Cr, Cu, Ni, Pb and Zn (Henry 2000; Surriya et al. 2015). Approaches in rhizofiltration can be performed as either floating rafts on ponds or tank systems. The main disadvantage of rhizofiltration is that the plants being used in rhizofiltration are grown in a greenhouse first and are then transferred to the remediation site (Surriya et al. 2015).
Some plants used in rhizofiltration are: Azolla caroliniana (Favas et al. 2012), Eichhornia crassipes (Saleh 2012; Rai 2019), Lemna minor, L. gibba (Pratas et al. 2012) and L. valdiviana (Souza et al. 2019), Phleum pretense, Salix matsudana (Wang et al. 2019), Sagittaria montevidensis (Demarco et al. 2019) and Typha angustifolia (Chandra and Yadav 2010).
The application of phytostabilization (phytoimmobilization) in alleviating heavy metal contamination is based on performing immobilization of contaminants in soils by sorption, precipitation and complexation using plants having ability. Creating an environment supporting contaminant immobilization by these processes, leaching and groundwater pollution can be prevented and also soil erosion and migration of sediments can be minimized (Kvesitadze et al. 2006; Ali et al. 2013; Ozyigit and Dogan 2015). Although each having a different mechanism, toxic metal removal can be achieved by applying phytoremediative techniques other than phytostabilization, which involves using plants having ability to stabilize rather than remove metal contaminants from a site and this provides relatively safe environment and reduce the risk for human health. (Arthur et al. 2005; Jan et al. 2015). Additionally, plants can serve as natural barrier in some extent for the aeolian dispersion of metals into the environment due to the vegetative cover (Sabir et al. 2015). Phytostabilization is deployed successfully for remediation of soils contaminated with As, Cd, Cr Cu and Pb (Surriya et al. 2015).
Athyrium wardii (Zhan et al. 2019), Cynodon dactylon (Leung et al. 2007), Eucalyptus urophylla, E. saligna (Magalhães et al. 2011), Eupatorium cannabinum (Gonzalez et al. 2019), Kosteletzkya pentacarpos (Zhou et al. 2019), Salix sp. (Yang et al. 2019a), Solanum nigrum (Ferraz et al. 2012; Li et al. 2019), Sorghum sp. (Jadia and Fulekar 2008) and Vigna unguiculata (Kshirsagar and Aery 2007) have been employed in phytostabilization applications.
Efficient elimination of organic pollutants via phytodegradation can be accomplished through access of organic pollutants into plant tissues or the rhizosphere and following decomposition by internal or secreted plant enzymes or products (Pilon-Smits 2005; Peer et al. 2006; Ozyigit and Dogan 2015). Organic compounds degraded by use of this approach are included herbicides, insecticides, chlorinated solvents and inorganic contaminants (Pivetz 2001). Dehalogenase, nitrilase, phosphalase, nitroreductase and oxidoreductase are the plant enzymes commonly involved in degradation of those organic contaminants (Kumar et al. 2019).
Blumea malcolmii (Kagalkar et al. 2011), Chlorella pyrenoidosa (Headley et al. 2008), Datura innoxia (Lucero et al. 1999), Erythrina crista-galli (De Farias et al. 2009), Ipomoea carnea (Jha et al. 2016), Leucaena leucocephala (Doty et al. 2003), Lycopersicon peruvianum (Schnoor et al. 1995), Phragmites australis (He et al. 2017), Pontederia crassipes (Gong et al. 2019) and Spirodela polyrhiza (Singh et al. 2019) are some plants utilized in phytodegradation processes.
Microorganisms-assisted degradation of organic pollutants (e.g., xenobiotics) in the vicinity of rhizosphere is known as rhizodegradation (Sangeeta and Maiti 2010; Ali et al. 2013; Ozyigit and Dogan 2015). Degradation or transformation for a compound involves in breakdown or alteration of it into its smaller constituents or a metabolite; therefore, rhizodegradation is important in solving the pollution problem caused by organic toxics through the remediation process (Arthur et al. 2005).
Rhizodegradation comprises an association between plants and microorganisms so that remediation can be achieved, which requires the presence of a series of low molecular weight organic acids, carbon and nitrogen compounds. Plants produce these compounds in order to nourish microorganisms in the rhizosphere and this makes contaminants in the vicinity of rhizosphere more bioavailable and exudates released by plants also enhance degradation of contaminants through stimulation of biochemical pathways within microorganisms (Leigh et al. 2002; White et al. 2003; Jan et al. 2015). To achieve to have higher yield in degradation process, a two-step enrichment approach proposed by Kuiper et al. (2004) was realized through bacteria harvested from the roots of plants grown in contaminated sites have used to stimulate biodegradation potential by re-colonizing plant roots.
Bromus inermis, Dactylis glomerata, Desmanthus illinoensis, Festuca arundinacea, Lolium perenne, Panicum virgatum, Tripsacum dactyloides (Lin et al. 2011), Cynodon dactylon (Nguemte et al. 2018), Kandelia candel (Lu et al. 2011), Melia azedarach (Kotoky and Pandey 2019), Rubus fruticosus (Alagić et al. 2016), Salix nigra (Yifru and Nzengung 2008), Sesbania cannabina (Maqbool et al. 2012), Triticum aestivum, Cucurbita ssp. (Ely and Smets 2017) and Zea mays (Zamani et al. 2018) are some plants employed in rhizodegradation process.
By phytovolatilization, uptake of many environmental contaminants, including organics (tetrachloroethane, trichloromethane and tetrachloromethane) and/or certain metals (As, Hg and Se), by plants can be carried out and they are released into the atmosphere through transpiration either in their original form or after metabolic modification (Susarla et al. 2002; Ali et al. 2013; Ozyigit and Dogan 2015). This technique is applicable for transforming the metal contaminants into less toxic elemental forms. For example, the species of mercury are converted into other less toxic forms. Elimination of heavy metals and organic solvents is successfully accomplished via using phytovolatilization technique, in which the use of transpiration on parts of plants is important for efficiency depending on climatic conditions (Surriya et al. 2015).
There is a major consideration for phytovolatilization. Though it is often thought to be beneficial, a risk assessment should be done for toxic volatile contaminants released into the atmosphere during the process. However, some reports say that despite exposure potential existing for volatile toxic contaminants due to substantial dispersion and dilution in the atmosphere they pose no environmental risk (Lin et al. 2000).
Azolla sp., Oryza sativa, Polypogon monspeliensis, Salicornia sp. (Hansen et al. 1998; Pilon-Smits et al. 1999; Lin et al. 2000; Hooda 2007; Ruppert et al. 2013), Juncus effuses (Wiessner et al. 2013), Phragmites australis (San Miguel et al. 2013), Brassica juncea, Astragalus sp. (Bañuelos et al. 1997; Raskin et al. 1997), Juncus xiphioides, Myriophyllum brasiliense, Scirpus robustus and Typha latifolia (Pilon-Smits et al. 1999; Arthur et al. 2005) are suitable plants for removing certain metals from the soil by this approach.
Due to nature of phytovolatilization, the process goes slowly. Therefore, optimization steps are put in practice as using novel plant species having high rates of transpiration and enzymes overexpressing (Van Huysen et al. 2003) and also technology is used to produce transgenic plants having efficient volatilization features (LeDuc et al. 2004; Malik et al. 2015).
Hydraulic control utilizes abilities of plants to absorb large amounts of contaminated water, and through transpiration, contaminants are kept in the plant bodies preventing the spread of contaminants (Kvesitadze et al. 2006; Ozyigit and Dogan 2015). For the application of this approach, poplar, birch, willow and eucalyptus are the most suitable plant species (Gatliff 1994; Pivetz 2001; Kvesitadze et al. 2006). Hydraulic control can affect movement of a plume of contaminated groundwater, minimize or suppress infiltration and leaching, and stimulate upward water movement from the water table (Hoffman et al. 1982; Olson and Fletcher 2000).
Uptake, transport and accumulation of heavy metal ions in plants
Released toxic agents are of having tendency for entering into the plants via various ways, commonly by soil (Cutillas-Barreiro et al. 2016), groundwater (Abid et al. 2016) and air/foliar (Bondada et al. 2004). Besides, soil-based entrance basically is the most pervasive and well-known way, irrigation practices and air-based entrance into the plants shall not be ignored when compared with soil-based entrance (Akguc et al. 2010; Zhao et al. 2019b). When one of essential or nonessential metals reaches the certain level in soil, potential threats arise not only for plants but also for other living organisms. One dramatic instance showing negative effects on human health is of taking excessive amounts of one of essential or nonessential metals by consumption of edible parts of plants via the food chain (Gaur et al. 2014; Dixit et al. 2015). Heavy metal accumulation is a complex event consisting of several steps that allows transport of heavy metals into the root cells via membrane carrier proteins, loading of heavy metals into xylem and detoxification/compartmentalization of heavy metals in appropriate aerial parts of the plant (Lombi et al. 2002).
This complex phenomenon can be better followed by explaining issues related to bioavailability and transportation of heavy metals to the root active sorbing zone. Metal accumulation having beneficial or toxic effects in plants starts with metals existing in bioavailable ionic forms in the soil (Jabeen et al. 2009; Maestri et al. 2010). Excreting of protons (H+) from organic acids (phytochelatins), amino acids and enzymes (known as acidification) present in the root rhizosphere environment increases bioavailability or mobility of metal ions whether being beneficial or toxic (Yang et al. 2005b). Metal ions (heavy or other beneficial ones) move throughout root active sorbing zone via the way of mass flow of soil liquid phase driven by transpiration force and ion diffusion pace of root surface cell (Barber 1962). In connection therewith: (1) soil-based entrance of metal ions into the root cells is catalyzed by a group of membrane transporter proteins (Fig. 1b) (Baxter et al. 2003). The excess amounts of nonessential elements in soils have potential for entering into the plants in terms of their similarities to the essential microelements (Zhou et al. 2012) and the membrane transporter proteins having weak target specificities (Schaaf et al. 2006) for nonessential and essential elements. Examples are: Ca and Cd intakes are catalyzed by the same calcium transporter (Perfus-Barbeoch et al. 2002) and iron-regulated transporter proteins (Korshunova et al. 1999); natural resistance-associated macrophage protein, copper transporter and zinc/iron transporter-related proteins generate major driving force for the entrance of metalloids into the cells in association with some of aquaporin transporters (nodulin 26-like intrinsic proteins and aquaglyceroporin) (Yin et al. 2016); natural resistance-associated macrophage protein and zinc-iron permease membrane transporter protein families have tendency for transportations of divalent cations between the inner and outer parts of the plant cells (DalCorso et al. 2013); low-affinity cation transporter conduct transportation of divalent heavy metals simultaneously with Ca in wheat (Clemens et al. 1998) and heavy metal chelate complexes are catalyzed by multidrug-resistance protein for transportation into the vacuoles (Tommasini et al. 1998); some other transporter proteins for carrying out heavy metal transportation into plant cells have also been shown by recent studies, e.g., cyclic nucleotide-gated (Moon et al. 2019); and a casket of phosphate transporter also allows As transportation into the plant cells (Kumar and Trivedi 2016). (2) When heavy metals are transported inside, they move from one place to another in the cell by exploiting of non-specific for essential or nonessential metal P1B-type transporters, also known as heavy metal-transporting ATPases. Organelles having intracellular membranes, such as Golgi apparatus, vacuole and endoplasmic reticulum also bear P1B-type of transporters for the selective compartmentalization (Williams et al. 2000). Besides, there are protein families, including natural resistance-associated macrophage protein (Belouchi et al. 1997), cation diffusion facilitator (Maser et al. 2001), zinc–iron permease (Van der Zaal et al. 1999), multidrug and toxin efflux (Kramer et al. 2007) and oligopeptide transporters (DalCorso et al. 2013) responsible for translocating heavy metals from root to shoot cells in plants (Singh et al. 2016). (3) Detoxification of metals/metalloids can be mediated by: metal binding agents, including metallothioneins, phytochelatins, involving chelation by organic acids and amino acids (Kumar et al. 2016); glutathione function and subsequently production of phosphate derivatives (Hernández et al. 2015); activation of reactive oxygen species enzyme production (Hossain et al. 2012); accumulation via compartmentalization (Sharma et al. 2016); and binding by the cell walls (Fig. 1c) (Mari and Lebrun 2006; Krzesłowska 2011). Under stress circumstances, the plant cell wall structure plays a role, acting as a storage reservoir for heavy metals (Le Gall et al. 2015) and re-organize itself for lignification (Loix et al. 2018).
Physiological aspects of heavy metal toxicity in plants
Physiologically, over excessive amounts of essential and nonessential metal intakes by the plant cells cause detrimental effects arising on various pathways that are extremely critical for the continuity of life. The most emergent symptoms due to excessive metal (Cd, Cu, Ni and Pb) accumulation by plants are losses of biomass and limited growth activities (Meyer et al. 2015; Krzeslowska et al. 2016; Asgari Lajayer et al. 2017; Roy et al. 2017; Dubey et al. 2018). Disruptions on photosynthetic machinery due to the negative influences affecting carbon management are the main reason not to seize normal biomass production and growth rate (Myśliwa-Kurdziel et al. 2004; Ozyigit et al. 2016; Sorrentino et al. 2018). Besides malfunctioning of photosynthetic mechanism, germination, plant growth, water-electrolyte balance, stomatal conductance, transpiration rate, leaf relative water contents, nutritional status of plant, carbon-based primary metabolite mechanism, genotoxicity and integrities of key enzymes/biomolecules related events are gradually collapsed due to the presence of excessive amounts of metals (DalCorso et al. 2013; Latef 2013; Choppala et al. 2014; Sarwar et al. 2017; Yadav et al. 2019). There are 4 levels in sense of execute evaluation for occurrences of disruptive effects of metalloids in plants. Some metalloids, arsenate (AsO43−), have great competition with phosphate (PO43−), selenate (SeO42−) and chromate (CrO42−) as well as with sulfate (SO42−) anions because of relatedness between their chemical properties. (1) Because of the chemical similarities between heavy metals and basic macronutrients, firstly plant nutrition-related metabolism is hampered as a result of this competition (Vetterlein et al. 2007; Schiavon et al. 2012). (2) Another type of competition exists between enzymes consisting of sulfhydryl and carboxyl groups. In particular, cysteine-rich sites in enzymes are important for the secondary, tertiary and quaternary folding and structure stabilization via folding creates centers playing important roles in terms of catalyzing state of existence-based reactions in enzymes. The binding of heavy metals, including As, Cd, Hg and Pb, to these stabilization centers leads to appearances of restrictions on enzyme activities; hence, the reactions are catalyzed in low volumes (Van Assche and Clijsters 1990; Quig 1998; Sharma and Dubey 2005). (3) The third event interrupting regular plant growth and development in plants is the productions of direct or indirect reactive oxygen species. Superoxide radicals (O2−), hydrogen peroxide (H2O2) and hydroxyl radicals (OH−) are common forms of reactive oxygen species produced in response to oxidative stress arising from many normal/impaired cellular reactions following expression of stress genes and pose danger for biomolecules, including lipids, proteins and nucleic acids (Ghori et al. 2019). Active metals in redox reactions, including Cu, Fe and Zn can produce reactive oxygen species directly as a result of Haber–Weiss/Fenton reactions. Also, reactive oxygen species can be generated indirectly by stimulation of nicotinamide adenine dinucleotide phosphate oxidases having suppressive ability on enzymes related to detoxification of reactive oxygen species in the presence of some of nonessential metals, including Cd, Hg and Pb in cells (DalCorso 2012; Shahid et al. 2014). (4) And finally, essential macro- and microelements (Fe, Mg, Mn, P and Zn) that play important roles in biological processes can be replaced by some nonessential elements (As, Cd, Cr and Pb) in terms of occurrences of interactions between elements and biomolecules based on affinity. Those kinds of metallic exchanges cause: altered enzymatic activities and gene expressions in some cases (Sanita di Toppi and Gabbrielli 1999); interruptions of signal transduction pathways (calcium dependent cellular signal transduction) (DalCorso et al. 2008); and blockages on chlorophyll production (Sharma and Sharma 1996; Yusuf et al. 2011). Under circumstances of exceeding acceptable limits of detoxification capacity of cells having, the reactive oxygen species cause cells to fall into the oxidative stress (Yuan et al. 2013) and first sing for measurable oxidative stress damage comes from DNA, lipid and protein monitoring (Berni et al. 2018). The reactive oxygen species are of key players of some physiological processes within the cellular acceptable limits (Yuan et al. 2013).
Phytochelatins
In biological systems, converting heavy metals into less dangerous and more mobile forms accompanying with binding to cysteine-rich polypeptides is the most frequent way of detoxification process following compartmentalization arising right after loading of the complexes into the vacuoles (Fig. 2a) (Liu et al. 2015c). Metal chelation agents consist of functional groups bearing thiol, amino, or hydroxyl groups in their structures having strong affinity for heavy metals (Rea et al. 2004). Phytochelatin is defined as s-rich, thiolate peptides and formulated as [γ-glutamyl-cysteinyl]n-X where n stands for numbers between 2 and 11 depending on organism and X stands for amino acids, including glycine, serine, β-alanine, glutamate or glutamine (Cobbett and Goldsbrough 2002). It is categorized according to having how many -Glu-Cys units such as PC2, PC3, PC4, PC5 (Rauser 1995). Phytochelatins are synthesized enzymatically in the presence of heavy metals from glutathione via phytochelatin synthases activity (Zenk 1996; Vatamaniuk et al. 2004; Filiz et al. 2019b). They are only found in plants and certain microorganisms (Hanikenne 2003; Chaurasia et al. 2008), and their synthesis also is initiated/transformed in the entity of anionic (Ag, Au, Cd, Cu, Hg, Pb and Zn) and cationic (As) metal(loid)s (Shukla et al. 2016). Among these metal(loid)s, particularly As and Cd are major inducers of phytochelatin gene activity (Verkleij et al. 2003). Glutathione is a kind of precursor and is considered playing role together with chelating agents in detoxification of free radicals existing in the cells because of heavy metal activities (Anjum et al. 2014).
A Detoxifying heavy metals by phytochelatins (copyrighted illustration from Prof. Ozyigit and Dr. Can), (Cys cysteine, GSH glutathione, Glu glutamate, GSH1 gamma-glutamylcysteine synthetase, GSH2 glutathione synthetase, JA jasmonic acid, LMWPC-HM low molecular weight phytochelatin-heavy metal, HMWPC-HM high molecular weight phytochelatin-heavy metal, PC phytochelatin, ɣ-EC gamma-glutamylcysteine), B detoxifying heavy metals by metallothioneins (apoMTF1 metal-responsive transcription factor 1, Zn-MTF1 zinc-metal-responsive transcription factor 1, CAT catalase, GPX glutathione peroxidase, MT metallothionein, SOD superoxide dismutase) (copyrighted illustration from Prof. Ozyigit and Dr. Can)
Cysteine residues of phytochelatin consisting of thiols or sulfonyl groups have great potential for occurrences of interactions with heavy metals, and phytochelatin–metal stability depends on intracellular deposition of metals (Le Faucheur et al. 2005; Filiz et al. 2019a). After formation of the phytochelatin–metal complex, shipping of it into the plant cellular vacuole is conducted immediately via activity of an ATP-binding cassette transporter family or an organic solute transporter family (Pal and Rai 2010; Solanki and Dhankhar 2011; Ghori et al. 2019). Specific transporter families (cation diffusion facilitator and natural resistance-associated macrophage protein) also are involved in sequestration process of heavy metals from cytosol to vacuole (Singh et al. 2016). This is not the only way plants cope with detoxification of heavy metals simultaneous escalation in phytochelatins activity enhances antioxidant production in Brassica chinensis under Cd stress (Chen et al. 2008). Phytochelatins stimulate antioxidant enzyme formations (superoxide dismutase, ascorbate peroxidase, glutathione peroxidase, glutathione and glutathione reductase) and might also compensate reactive oxygen species generation resulting from heavy metal stress in plants (Mishra et al. 2006; Filiz et al. 2019a) (Fig. 2-A).
Brassicaceae species are widely preferred in using transgenic phytoremediation strategies for the following reasons: (1) multiple cultivation practices can be executed within a season due to their rapid growth and development properties; (2) they have wide adaptation capabilities for particular environmental conditions; (3) they also have tolerance against heavy metal accumulation; therefore, they are suitable for different transformation approaches for efficient heavy metal remediation from soils (Agnihotri and Seth 2019).
Some following influential cases are exemplified along with the species belonging to Brassicaceae family and some other species (Table 1). A phytochelatin synthase gene from Brassica napus was cloned and transformed into Arabidopsis thaliana AtPCs1 mutant cad1–3 via Agrobacterium-mediated transformation floral dip method after generation of three genetically engineered lines, a morphological and physiological evaluation was done for these transgene lines and A. thaliana AtPCs1 mutant under excessive level of Cd treatment. In this work, the transgenic lines were found to be highly efficient in remediation (Bai et al. 2019).
Despite being in many plants, not all phytochelatin synthase genes have been identified and characterized at functional and molecular levels. Fan et al. (2018) identified two phytochelatin synthase genes from Morus alba and transformed them into Arabidopsis and tobacco plants via employing Agrobacterium-mediated floral dip method for validating their heavy metal accumulation capabilities. When transgenic Arabidopsis and tobacco plants reached the seedling stage, their gaining biomass and Zn/Cd accumulation capability rates were analyzed using total root length and fresh weight readings and atomic absorption spectrophotometry data. According to their results, identified phytochelatin synthase genes were classified as promising genetic resources for phytoremediation of heavy metals.
Vicia sativa phytochelatin synthase genes 1 homolog and its functionality under Cd stress have been investigated after transforming it into the A. thaliana. Amplified PCs1 gene from total RNA was inserted into the pCAMBIA1304 vector under control of cauliflower mosaic virus 35 promoter. After insertion, Agrobacterium-mediated floral dip method was utilized for transformation of PCs1 gene. Besides atomic absorption spectrophotometer that was employed for determining Cd concentrations, cysteine, glutathione and phytochelatin measurements were also taken via determining thiols using ultra-performance liquid chromatography system for analyses of functionality and expression levels. The exact intracellular Cd localization was shown using Cd-sensing fluorescent dyes and ultraview vox confocal microscope. At the end of this work, an inference was made as VcPCs1, upgraded of our understanding of phytochelatin-mediated heavy metal tolerance in higher plants (Zhang et al. 2018).
During the generation of new genetically modified plants, the new genes discovered from different types of organisms (animals, plants and bacteria) are exploited in possible manipulation of accumulating or degrading heavy metals (Seth 2012). An interesting transgenic research by Kühnlenz et al. (2015) was carried out using phytochelatin synthase genes originally from Caenorhabditis elegans (Vatamaniuk et al. 2001) and Schizosaccharomyces pombe (Clemens et al. 1999) for detoxification of heavy metals in Arabidopsis utilized as study material. In that research, AtPCs1 mutant line was transformed with CePCs genes to perform investigation on Cd accumulation/CePCs transcript/phytochelatin contents.
Besides the members of brassicaceae family, poplar (Populus tomentosa) is another widely distributed model plant species actively used in phytoremediation of heavy metals. After transferring PtPCs gene to tobacco using Agrobacterium-mediated leaf disk method, wild and transgenic types were used to conduct investigation for Cd stress in terms of determination of morphological and physiological indices (leaf relative electrolyte leakage, malondialdehyde content, total superoxide dismutase activity, chlorophyll content and root activity) and Cd tolerance rate. At the end, the authors pointed out that PtPCs may involve in Cd tolerance and accumulation but not in Cd transport (Chen et al. 2015b).
In transgenics, glutathione-dependent phytochelatin production is not achieved only by transferring phytochelatin synthase genes but also gained by induction of transferred genes that are dependent on glutathione. After cloning of a kind of inducer gene, XCD1 also known as MAN3 from A. thaliana Heynh columbia-0 was transferred to Arabidopsis mutant genotypes and tested for Cd accumulation and tolerance. Gene transcripts and protein expressions, Cd accumulation, glutathione and phytochelatin contents were analyzed in transgenic and mutant Arabidopsis lines. XCD1 has a regulatory function on glutathione-dependent phytochelatin production by cross-talking with related genes (Chen et al. 2015a).
Metallothioneins
Once one of the heavy metals enters through the plant cell, firstly productions of kinds of glutathione-based thiol derivatives, such as malic acid, citric acid and malonic acid, called phytochelatins are stimulated to prevent possible damages via binding to heavy metals resulting in compensation of their effects and increasing of metal tolerance in plants (Filiz et al. 2019a; Pandey et al. 2019). Metallothioneins are classified as low molecular weight, cysteine-rich proteins/polypeptides, and they have ability to bind essential or nonessential metals; thereby, they maintain cellular homeostasis (Fasani et al. 2018). Metallothionein genes are found in a wide variety of organisms, including fungi, invertebrates, mammals, plants and some prokaryotes (Du et al. 2012). Metallothioneins having mercaptide bonds arising between metallothioneins and metal(loid)s are produced as low-weight biomolecules (4–14 kDa) from transcriptionally gene encoded mRNAs, which are relatively different from enzymatically synthesized phytochelatins (Cobbett and Goldsbrough 2002). The stimulation of transcript productions from metallothionein genes is realized via releasing of plant hormones due to existing abiotic stress factors, including cytotoxic agents and heavy metals such as Ag, Au, Bi, Cd, Co, Cu, Hg, Ni and Zn (Yang et al. 2005a) (Fig. 2b). Metallothioneins, a group of chelators, carry out various tasks, which involved in homeostasis of essential metals, compartmentalization of nonessential metals and detoxification of reactive oxygen species (Gasic and Korban 2006; Hossain et al. 2012). Also, metallothioneins actively participate in restoring of existing functional and structural cellular damages such as those related to membrane, cell division, growth and developmental events (Mishra and Dubey 2006) and DNA (Grennan 2011). Metallothioneins are divided into three main classes (Zhou et al. 2006), and four distinct subgroups are defined for plant metallothioneins according to conformations of cysteine residues (Huang and Wang 2009). The arrangements of three types of cysteine motifs are shown as cysteine–cysteine, cysteine–X–cysteine and cysteine–X–X–cysteine used for classification of metallothioneins where X indicates one of 20 amino acids (Cobbett and Goldsbrough 2002; Hossain et al. 2012). Plants, fungi and invertebrate metallothioneins are generally evaluated as under the context of class II (Robinson et al. 1993). Types of plant metallothioneins genes show tissue-specific expression profiles as in roots, leaves, shoots, fruits and seeds (Shukla et al. 2016).
Metallothioneins are effective not only for elimination of metal stress but also for suppression of drought stress in plants. Metallothionein gene from chickpea was transferred to Arabidopsis using Agrobacterium-mediated floral dip method and expression of its product (30-fold higher), and some physiological properties, enzymatic and non-enzymatic antioxidant productions were found to be better than wild-type Arabidopsis (Dubey et al. 2019).
A study performed for determining of heavy metal tolerance rates of transgenic tobacco, of which Lm-SAP gene (a member of the stress-associated protein gene family) from Lobularia maritima transformed and their findings from physiological (total shoot biomass and root length), biochemical (oxidative stress markers and antioxidant enzyme activities) and transcript (real time polymerase chain reaction) analyzes showed that under Cd, Cu, Mn and Zn stresses, the expressions of metallothionein genes (Met1, Met2, Met3, Met4, and Met5) as well as the expression of other stress-related genes in transgenic tobacco were stimulated (Saad et al. 2018).
After cloning a metallothionein gene (SsMT2) from Suaeda salsa which was grown commonly in saline/alkaline soils, the characterization of its product was done in transgenic Saccharomyces cerevisiae and Arabidopsis thaliana showing distribution nearly in all organs and tissues, except flowers. And also, induced levels for its product were observed after applications of different stress conditions generated using CdCl2, NaCl, NaHCO3 and H2O2. SsMT2-transgenic S. cerevisiae and A. thaliana showed significant resistance to metal, salt and oxidant stresses. Results were as follows: high accumulation for Cd and low accumulation for Na, and maintaining low level of H2O2 in comparison with wild non-transgenic types. The results encourage the utilization of these transgenics in phytoremedial cases (Jin et al. 2017).
An investigation was carried out by Zhang et al. (2017) dealing with Cd and Cu stresses in point of applications of CuSO4 and CdCl2 at cellular level with respect to comparisons between the generated transgenic cell line from rice cultivar Nipponbare by in use of Agrobacterium-mediated transformation and wild type in terms of measuring H2O2 levels and cell death rates. Descended cell death rate and H2O2 level in the transgenic cell line were observed by means of expression of ricMT gene. Low H2O2 level via expression of ricMT gene plays a crucial role for having tolerance for Cd and Cu stresses.
Transformation of the metallothionein gene (OsMT2c) from Oryza sativa to Arabidopsis carried out by Liu et al. (2015b) provided tolerance for Cu stress. Expression product of OsMT2c (a kind of type II metallothionein gene) is usually seen in the leaves and roots of plants. Transcription of the gene is stimulated by the presence of H2O2 and Cu. OsMT2c gene was introduced to Arabidopsis by Liu et al. (2015b). After its transformation, its efficiency was tested for Cu stress and the results showed that tolerance for Cu stress and efficient neutralization for ROS were observed in the transgenic Arabidopsis in comparison with wild type.
IlMT2b, a metallothionein gene from Iris lactea var. chinensis, was cloned by Gu et al. (2015) and introduced to A. thaliana ecotype columbia-0 using floral dip method. Real-time polymerase chain reaction was used for validation of transformation for IlMT2b gene. Cu contents and H2O2 levels were analyzed in transgenic and wild types after application of Cu stress. Besides scale-up Cu accumulation and scale-down H2O2 level observations, significant root elongation (in ranges of 1.5–3-fold increase in comparison with wild type) was noticed under Cu stress; therefore, this feature might be useful for remediation of copper-rich soils.
Plant species for optimum phytoremediation
Although plant species having high tolerance ability to metals and hyper-accumulation capacity can likely be found naturally in soils bearing rich metal contents, plant species living in such soils almost all are not suitable for use in phytoremediation because of usually being in small size and biomass, whereas plant species showing large growth capabilities generally exhibit weak metal deposition ability and low tolerance to heavy metals. For high capacity accumulation of heavy metals, the following features borne by a plant are: exhibiting fast growth ability and building eminent biomass; having ability and high toleration capacity for accumulation of heavy metal(s); and being easily handle for harvest (Karenlampi et al. 2000). Therefore, creating transgenic plants carrying specific features for phytoremediation is perhaps inevitable. In general sense, new or improved phenotypic characters in plants can be brought into existence through genetic engineering in terms of transferring genes from other organisms (Clemens et al. 2002; Shah and Nongkynrih 2007). Heterotrophic organisms, bacteria and mammals, have metabolic pathways consisting of unique enzymes for complete digestion of organic substances; hence, these enzymes can be employed in plants for improvements of their degradation capabilities (Eapen et al. 2007; Van Aken 2008).
First attempts were historically related to providing enhance tolerance to plants for heavy metals in connection with phytoremedial practices, including transgenic tobacco and Arabidopsis thaliana created by introducing a yeast metallothionein gene and a mercuric ion reductase gene for higher tolerance to Cd and Hg, respectively (Misra and Gedamu 1989; Rugh et al. 1996). Though they produced satisfactory results at laboratory scale, because of their competence at field applications they were not found to be suitable. Alternatively, poplars (Populus sp.) are considered as good choice for phytoremediation due to fast growing ability and producing eminent biomass (Misra and Gedamu 1989; Rugh et al. 1996). Although, in years, Agrobacterium tumefaciens with tumor-inducing (Ti) plasmid has been recognized as a favorite vector system, effectively used for introducing genes to plants, challenges for A. tumefaciens-mediated transformation of forest trees arise. That is why only a few studies have been reported about the genetic innovations of poplars (Han et al. 2000). The first transgenic poplars developed for phytoremedial practices were realized for treating chloroacetanilide herbicides by the overexpression of a gamma-glutamylcysteine synthetase involving in glutathione synthesis (Gullner et al. 2001). Since there may be a specific mechanism for uptake of a heavy metal into plants, it is important for considering appropriate strategies for each heavy metal when developing transgenic plants (Eapen and D’Souza 2005).
The plant species having capabilities of accumulating large amounts of heavy metals show extraordinary storage abilities in their epidermal cells. Accordingly, feasibility for improving tolerance and accumulation capacity to heavy metals in plants can be met through productions of metallothioneins, phytochelatins and metal chelators in terms of inserting and/or overexpressing of the genes playing roles in tolerance and accumulation of heavy metals as mentioned above (Eapen and D’Souza 2005; Hassan and Aarts 2011). Also, improving heavy metal tolerance and accumulation capacities in plants could be accomplished by modification of metal transporter genes. The translocations of heavy metals from root to shoot are an important step in accumulator plants, and the way of stimulating translocation from root to shoot can be managed by intense metal driven in shoots, improved xylem loading and repressed metal sequestration in root vacuoles (Hassan and Aarts 2011; Soleimani et al. 2011). In the following section, some information about direct and indirect gene transfer techniques widely used as well as the cutting-edge gene transfer approaches is given.
Effective gene transfer methods for phytoremediation
Conventional gene transfer methods
Agrobacterium-mediated transformation
Agrobacterium-mediated genetic transformation is the most common technique used for the development of transgenic plants (Ozyigit et al. 2013). The species from genus Agrobacterium cause different types of neoplastic diseases, including crown gall (A. tumefaciens and A. vitis), hairy root (A. rhizogenes) and cane gall (A. rubi) by inserting T-DNA into a large group of plant species, including dicotyledons and monocotyledons (Ozyigit 2012). Agrobacterium strains containing native T-DNA allow us to produce an efficient mechanism for creating transgenic plants in terms of introducing and stabilizing of foreign gene(s) in plants. For this reason, Agrobacterium-mediated transformation as a genetic engineering tool has been utilized for constructions of new plasmids and strains throughout of exploring new ways for developing different transgenic plants (Ahmad et al. 2017).
Tumor-inducing plasmid found in the soil bacterium A. tumefaciens used as a vector for transferring functional foreign genes to plants is a powerful tool developed so far. The wild-type plasmid carrying oncogenic genes encode the enzymes playing roles in biosynthesis of auxins and cytokines. Opines secreted in plant crown gall cells as a result of condensation of amino acid and sugars through in related biosynthetic pathway catalyzed by specific enzymes encoded by genes of tumor-inducing plasmid are exploited by pathogenic bacteria of the genus Agrobacterium as carbon and nitrogen sources (Vladimirov et al. 2015). The 25-bp repeat sequences known as the left and right borders (LB and RB, respectively) are located on T-DNA region defining the boundaries. Virulence (vir) genes, virA, virB, virC, virD, virE, virF, virG, and virH positioned outside the T-DNA region within plasmid and chvA, chvB, and chvF positioned on bacterial chromosome in the forms of operons are crucial for T-DNA transfer (Zupan et al. 2000; Barampuram and Zhang 2011). The regulation mechanism of T-DNA transfer and integration is carried out via the proteins encoded by the vir genes found as 8 operons located on vir region of the tumor-inducing plasmid (Subramoni et al. 2014; Alok et al. 2017).
Agrobacterium-mediated transformation used for genetic modification of plants relies on five essential steps: (1) inciting of vir genes expression; (2) generation of T-DNA complex; (3) introducing of T-DNA from Agrobacterium to the plant cell nucleus; (4) integration of T-DNA into the plant genome; and (5) expression of T-DNA genes (Pitzschke and Hirt 2010; Gelvin 2012; Alok et al. 2017). virB/D4 type IV secretion system is involved in transportation of T-DNA complex and several vir proteins into the plant cell, and the following last step is integration of T-DNA into the plant chromosome (Christie 2004; Alok et al. 2017).
Ti plasmid being responsible for formation of crown gall disease in plants is adapted by scientists in purposes of introduction of foreign gene(s) to plants and of constructing stable plants providing expression of the gene(s) in heritable fashion in non-tumorous individuals (Ozyigit 2012). Adaptation includes modification of tumor-inducing plasmid in way of the elimination of its tumor-promoting properties in order to be able to regenerate plants from cells transformed with T-DNA in culture. Expression of introduced foreign gene(s) can be turn on under the control of their own normal regulatory signals (Kado 2014).
As a neoplastic disease, hairy root arising due to root-inducing plasmid found in A. rhizogenes is the second commonly employed bacterium through using Agrobacterium-mediated gene transfer. As tumor-inducing plasmid found in A. tumefaciens, root-inducing (Ri) plasmid found in A. rhizogenes consists T-DNA having oncogenic and opine catabolism genes (Ozyigit 2012). Relying on hairy root induction, culturing of some plant species through employing A. rhizogenes-mediated transformation is very practical in productions of some secondary metabolites and/or recombinant proteins, generally utilized in phytoremediation and healthcare industry (Fig. 3a) (Makhzoum et al. 2013; Pala et al. 2016; Alok et al. 2017).
A Agrobacterium-mediated transformation (left) (modified from Ozyigit 2012; Yuksek 2018), crown galls by A. tumefaciens (right up) (copyrighted picture from Prof. Ozyigit), hairy root formations by A. rhizogenes (right down) (with the permission of Prof. Dr. Sule Ari and Assoc. Prof. Dr. Semra Hasancebi), B particle bombardment (Biolistic)-Helios® Gene Gun System (with the permission of Bio-Rad Laboratories, Inc.) and particle bombardment treatment (illustrated by Prof. Ozyigit), C Electroporation-Gene Pulser Xcell™ Electroporation Systems (with the permission of Bio-Rad Laboratories, Inc.) (right) and 1-Membrane before electric pulse, 2-Electropore formations during the pulse and transferring of DNA, RNA, enzyme, antibody, and some chemicals from cell membrane, 3-transferred chemicals and resealing-recovering after the pulse (left) (modified from Ozyigit 2020), D 1-zinc-finger nucleotide (ZFN), 2-TALEN and 3-CRISPRCas/9 mediated transformation (from Prof. Ozyigit)
Particle bombardment
As an alternative to Agrobacterium-mediated gene transformation method, the particle bombardment is an accepted major gene transfer method, commonly used for the transformation of plant species. Due to recalcitrancy for some plant species in use of Agrobacterium-mediated transformation, biolistic is utilized to transform these species at the nuclear level although having some disadvantages including multiple foreign gene insertions frequently seen and difficulties for defining of the DNA segment inserted in the host. First drawback is being causing silencing events, and the latter one leads to obliging for detailed screening of the transformants to rescue those having the full-length DNA of interest (Kikkert et al. 2004).
Biolistics is a technique based on a mechanism involving propelling DNA-coated metal beads (tungsten or gold particles, 1–2 μm sized) at high speed into the target plant material by firing an explosive in terms of releasing the genetic load within the cells and integrating the genetic load into the genome (Slater et al. 2008). The important step in this technique is ensuring proper coating of the metal beads with the DNA of interest being carried out under following circumstances: in the beginning, DNA of interest is precipitated with calcium chloride (CaCl2) in order to provide a positive charged surface in terms of the adhering of the DNA to the metal bead; and finally, a cationic polyamine, spermidine, is added in purpose of protecting DNA from degradation of cellular nucleases and allowing DNA adsorption onto the metal beads (Brune et al. 1991; Thomas et al. 1996; Márquez-Escobar et al. 2018) (Fig. 3b).
For DNA delivery to plant cells using biolistic process, the plant tissue, used in transformation, is positioned in an evacuated chamber below a retaining plate for stopping particles. The metal beads coated with vector DNA on a macroprojectile are placed above the retaining plate. And, a firing disk is found above them. Acceleration of metal beads coated with DNA of interest is gained by a shock wave driven via pressure of helium (He). After application of driving force to propel the macroprojectile with metal beads coated with DNA of interest, the chamber goes down and stops at the retaining plate, while the metal beads coated with DNA of interest pass through hitting the plant tissue (Bradshaw 2016). Primary explants as tissues, cell suspensions, or callus cultures from plants are used for bombardment. Metal beads loaded with the DNA of interest are driven into the plant cell and then integration of the DNA of interest into host genome occurs (Kikkert et al. 2004; Thakur and Shankar 2017). Biolistics is generally employed for introducing foreign genetic material into the nuclear genome, but also through transformation, it can be used for introducing foreign genetic material into chloroplast for providing high levels of gene expressions and synthesis of active proteins (Bradshaw 2016).
Electroporation
Transformation of prokaryotic and eukaryotic cells by applying electroporation is a highly efficient system using electrical field for creating transient microscopic pores in plasma membranes of cells allowing integration of DNA of interest into the host genome following the passage of it into cells (Khan 2010; Kotnik et al. 2015; Thakur and Shankar 2017; Ozyigit 2020). After electroporation application using optimized electric pulse, created microscopic pores on the membranes of cells can be sealed and the cells can be recovered. However, deviating from application of suitable electrical current, one important dynamic of electroporation could cause the cell deaths due to extreme overheat leading to deterioration of structure and permeabilization of the membranes (Kotnik et al. 2019).
By stimulation of an electrical stress, in vitro electrophoretic DNA transfer into cells is a fast process done through employing electroporation. At the beginning, the procedure was set up for protoplast change but was also approved to be applicable for plant cells too for DNA transfer (Ozyigit 2020). A general setting for electroporation is for voltage at 25 mV and for amperage at 0.5 mA for 15 min, but the other parameters including DNA concentration at cell surface and strengths of cells to membrane permeation can have impacts on electroporation performance (Kotnik et al. 2015, 2019). Transforming protoplast of both monocotyledonous and dicotyledonous plants using the electroporation approach, remarkable improvements have been gained (Ahmad et al. 2017). Transgenic rice plants produced by electroporation-mediated DNA insertion were first reported to be used into embryogenic protoplasts (Zhang et al. 1988) (Fig. 3c).
Other direct and indirect techniques
For carrying out genetic transformation of plants, there are direct and indirect approaches other than mentioned above. They are: dextran-mediated transformation, liposome-mediated, pollen-mediated, polyethylene glycol-mediated, and silicon carbide fiber-mediated genetic transformations; genetic transformations by microinjection and macroinjection; genetic manipulation by laser microbeam; sonication-assisted; and desiccation and virus-induced gene transfers (Subramanyam et al. 2011; Ahmad et al. 2017; Mendel and Hänsch 2017; Gosal and Wani 2018).
Target-specific genome editing
Introducing targeted mutagenesis, precise gene editing and site-specific gene insertion can be done by genome-editing techniques using chimeric mega nucleases, including zinc-finger nucleases (ZFN), transcription activator-like effector nucleases (TALEN), and RNA-guided DNA endonucleases found in clustered regularly interspaced short palindromic repeat (CRISPR)/Cas (CRISPR-associated) systems. After inducing of double-strand DNA breakages at specific sites by participations of these tools, plant DNA repair mechanism is mobilized for repairing DNA double-strand breaks created. Either involving homologous recombination or non-homologous end joining, the broken DNAs are joined again (Chen and Gao 2014). Generating transgene-free genetically modified plants by using these tools, not needed for seeking permissions for regulatory issues is the main advantage for this type of works (Zhang et al. 2016; Alok et al. 2017).
Zinc-finger nucleases
The cysteine2-histidine2-type zinc-finger domains as DNA-binding motifs distributed among eukaryotes are found to be involved in sequence-specific DNA binding and mediate targeted genetic modification in many plants; therefore, over last two decades, due to application potentials, this type of nucleases has received attention for genetic modifications (Lee and Ezura 2016). Each of zinc finger consisting approximately 30 amino acids exists in ββα configuration and is stabilized by a zinc ion. The zinc finger has feature for recognizing and binding to a specific 3-bp DNA sequence through interaction occurring between α-helix structure of zinc finger and the major groove of the DNA double helix (Pabo et al. 2001). A DNA-binding domain having a tandem array of zinc fingers and the non-specific DNA cleavage domain of the FokI restriction endonuclease are fused for building of zinc-finger nucleases (Durai et al. 2005; Alok et al. 2017) (Fig. 3d-1).
5-GNN-3, 5-ANN-3 and 5-TNN-3 are the triplets, mostly recognized and bond by each nuclease domain of zinc finger (Dreier et al. 2001; Segal 2002). Mainly ZFN (zinc-finger nucleotide) pairs are able to work precisely in targeting the binding sites when the binding sites are 6 bp apart from each other (Bibikova et al. 2001). In case of having a ZFN recognition sequence, it could be used only 5-GNN-3 triplets, and it also could be made up of the following frame 5-NNCNNCNNC(N5-6)GNNGNNGNN-3. Possible DNA target sites are seemingly available in every 0.5–1.0 kb for the published zinc-finger domain, resulting in being able to target most plant genes (Segal 2002; Lee and Ezura 2016). The ascending outcome is an enzyme having ability for recognizing a unique DNA sequence and ability to induce targeted DNA double stranded breaks (Alok et al. 2017).
Transcription activator-like effector nucleases
Engineered transcription activator-like effector nucleases having a transcription activator-like effector DNA-binding domain and a DNA cleavage domain deployed for creating double-strand DNA breaks in a non-specific manner are hybrid nucleases (Joung and Sander 2013). After deploying first strategies using transcription activator-like effector nucleases for achieving genomic modifications, successful demonstrations have been increased rapidly in this field (Christian et al. 2012). Recognitions of specific DNA sites by zinc-finger nucleases and transcription activator-like effector nucleases are carried through in similar fashion; however, each motif in transcription activator-like effector nucleases recognizes a single nucleotide, while each motif in zinc-finger nucleases recognizes 3–4 nucleotides (Deng et al. 2014). Plant pathogen effector proteins, transcription activator-like effectors, can turn on transcription activator-like effector-specific host genes involved in disease formation following binding of the promoters of these genes after entering the nucleus of host cell (Marois et al. 2002; Kay et al. 2007; Lee and Ezura 2016) (Fig. 3d-2).
Transcription activator-like effectors have translocation signal at the N-terminus and nuclear localization signal and transcription activator domain at the C-terminus and interact with DNA sequences specific to binding domains referred to as the repeat domains found in the central part of transcription activator-like effectors, in which each includes a number of tandem 33–34 amino acid repeats except repeats in positions 12 and 13 that are highly variable and are, therefore, referred to as repeat-variable di-residues exhibiting nucleotide preference (Yang et al. 2000; Marois et al. 2002; Gurlebeck et al. 2006; Kay et al. 2007; Boch et al. 2009). It allows the allowance of targeting any sequence through the assembly of repeat-variable di-residues on specific DNA sequences (Lee and Ezura 2016).
Clustered regularly interspaced short palindromic repeats-associated nuclease 9
Lately, the RNA-guided CRISPR/Cas9-mediated genome editing as a tool for the manipulations of genomes of organisms has brought promises in the field. This applied system is required at least two components for the site-specific cleavage of DNA, identified as a Cas9 endonuclease protein and a chimeric RNA (chiRNA) comprising CRISPR RNA (crRNA) and trans-activating crRNA (tracrRNA) (Lee and Ezura 2016). The CRISPR/Cas system as part of an adaptive defense system is identified in bacteria and archaea functions so as to cope with invading nucleic acids by targeting them and leading them for degradation (Wiedenheft et al. 2012; Bradshaw 2016).
CRISPR/Cas systems are classified mainly into three groups, but among them type II appears to be the simplest one (Bhaya et al. 2011; Alok et al. 2017). The type II CRISPR system consisting of crRNA and tracrRNA generated by insertion of short DNA fragments into a CRISPR locus forms ribonucleoprotein complexes along with cas proteins (Cong et al. 2013). The crRNA-tracrRNA fusion part of the complex called the guide RNA (gRNA) and the Cas9 endonuclease from Streptococcus pyogenes are integrated to form a structure capable of recognizing the specific sequences on the target site in the presence of a downstream protospacer adjacent motif with the sequence 5′-NGG-3′ (Jinek et al. 2012). By the presence of the protospacer adjacent motif serving as targeting component, the recognition and subsequent cleavage of invading DNA can be accomplished through distinguishing self from non-self-DNA (Raina et al. 2018). A single guide RNA (sgRNA) can be designed, and instead of the tracrRNA:crRNA heteroduplex, it can be replaced for reprogramming the Cas9/sgRNA system to a specific complementary target site (Mali et al. 2013; Sardesai and Subramanyam 2018). By targeting the first 20 nucleotides in the sgRNA, specific sgRNA variants can be purposefully generated for various genome-editing applications (Sander and Joung 2014). Also, due to frequent protospacer adjacent motif occurrences in genomes, nearly any gene bearing a protospacer adjacent motif could be targeted by a Cas9/sgRNA complex (Sardesai and Subramanyam 2018). After introduction of Cas9 nuclease with appropriate gRNAs into a cell, targeted modifications can be accomplished as cutting DNA at specific locations within genome under guidance of appropriate gRNAs and removal of that existing DNA piece from genome and/or integration of a new DNA piece into genome via homologous recombination (Cetintas et al. 2017; Pandotra et al. 2018). Activity of the system depends upon cell type and the approach used in introduction. Introduction of gRNA and Cas9 nuclease into cells can be executed via relying on direct insertion or transforming as transgenes on a separate chromosome to the targeted locus by cloning. The effects of unintended outcomes appear to be rare and by well-designed specific gRNA sequences in terms of on target accuracy should minimize the risk of off-target genome modifications (Bradshaw 2016) (Fig. 3-d-3).
The power of CRISPR/Cas9 system comes from its simplicity, efficiency and versatility. After target specifies are designated for organisms in terms of Cas9 optimization, the suitable constructs can be generated using cloning techniques (Jiang et al. 2013; Feng et al. 2014; Shan et al. 2014). Zinc-finger nucleases, transcription activator-like effector nucleases and CRISPR/Cas9 as powerful genome-editing tools similarly target specific sites in genome, and the last system has an attractive advantage over the first two systems in generating targeted genetic elements; hence; CRISPR/Cas9 has rapidly become a widely used choice in laboratories (Lee and Ezura 2016).
Heavy metal phytoremediation by genetically engineered plants
Remediation researches are getting great attention recently and offer worthy strategies for cleaning up soils from contaminants. In particular, using transgenic plants for this purpose is a promising approach in this section. Exploiting of transgenic approach is not only limited to transformation of functional genes but also certain defined promoters that elevate existed gene functions related to accumulation/translocation/detoxification mechanisms of heavy metals can be introduced to the target plants. Here, some examples related to detoxification of heavy metal contaminants involving transgenics are given in table.
In one such case, transgenic canola varieties were generated from wild-type spring canola (Brassica napus L. cv. Westar) using a rice gene, OsMyb4, consisting Arabidopsis thaliana COR15a stress-inducible promoter (Raldugina et al. 2018). Singh et al. (2002) reported that under different stress conditions, myeloblastosis protein family members provide confers for continuity of plant development and protecting plants. Transgenic spring canola plants grown under high levels of Cu (as 150 μM CuSO4) and Zn (as 5000 μM ZnSO4) concentrations were shown to have extended surviving ability (more than 15 days) compared with wild types. It means that OsMyb4 gene plays a crucial role in survival mechanism related to free radical elimination in plants exposed excessive levels of Cu and Zn. Consequently, Raldugina et al. (2018) suggested that OsMyb4 may play a role as a positive regulator of phenylpropanoid pathway and proline synthesis and may also have potential in phytoremedial applications.
As stated by Boudet (2007), lignin builds up the first line of defense against biotic and abiotic stresses as a result of interaction between non-cellulosic polysaccharides that give strength to plant cell wall structures. Zhang et al. (2014) revealed that lignin biosynthesis was enhanced via overexpression of CCoAOMT and also Rui et al. (2016) concluded that application of Cd stress in Vicia sativa caused stimulated lignin production. These above-mentioned results were used for generation of a transgenic A. thaliana by cloning and transferring of VsCCoAOMT gene analog from V. sativa for the phytoremediation applications in terms of enhancing lignin biosynthesis (Xia et al. 2018). At the end of this work, the products of VsCCoAOMT gene were found to be playing crucial roles related to providing enhanced tolerance in transgenic A. thaliana for Cd stress. Increased biomass production and enhanced loading and transportation processes for Cd, and especially accumulation of Cd in cell wall structures were observed as results herein that study giving promise in phytoremedial applications (Xia et al. 2018).
Heavy metal ATPases were exploited in a research for improving Cd tolerance in transgenic tobacco created by Wang et al. (2018) as model organism. Heavy metal ATPase is a kind of plant-based P1B-type ATPase protein, defined as Cd exchanger between root and shoot cells (Takahashi et al. 2012). Heavy metal ATPases were discovered in various plants, including Oryza sativa L. (Takahashi et al. 2012), Hordeum vulgare L. (Williams and Mills 2005), Thlaspi caerulescens L. (Papoyan and Kochian 2004) and Populus tomentosa Carr. (Wang et al. 2018). Using information from Populus trichocarpa genome, degenerate primer pairs were designed and used for screening/cloning of heavy metal ATPase cDNA. After validation of heavy metal ATPase gene, PtoHMA5, it was cloned, sequenced and then transformed into N. tobaccum. After that, the transgenic N. tabacum was tested by application of Cd stress and a 25.05% increase in Cd accumulation was detected in leaves of the plant. Wang et al. (2018) concluded that PtoHMA5 and PtPCs were found to be as useful gene sources for phytoremediation.
Certain types of genes isolated from endophytic bacteria are occasionally used in transformations of target plants done for scaling up phytoremediation capabilities. A research was carried out using nettle (Urtica dioica) seeds as research material and seedlings generated from the seeds after sowing stage were grown in As, Cd, Pb and Zn contaminated soils (Viktorova et al. (2017) and thereafter, Bacillus shackletonii and Streptomyces badius, endophytic bacteria, were isolated from these plants using appropriate protocols. Such endophytic bacteria are valuable sources for genes related to detoxification pathways of heavy metals for their various types of energy source practices (Uhlik et al. 2012). And, the transgenic plants containing the genes derived from endophytic bacteria are very useful tools for removing of heavy metals from soils. CUP and bphC genes under control of cauliflower mosaic virus 35 promoter were introduced to the nettle plants to increase heavy metal accumulation performance and after transformation, heavy metal accumulation capabilities of these transgenics grown in contaminated soils were evaluated. By this work, first transgenic nettle plants were developed by Viktorova et al. (2017) for phytoremedial purposes and tested in polychlorinated biphenyls contaminated soils. According to their results, the declination rate of polychlorinated biphenyls in soils was up to 33% and the rate of heavy metal cleanup in soils reached up to 8%.
The cells of eukaryotic organisms have various detoxification mechanisms for metal stresses; one of them is glutathione synthesis (Filiz et al. 2019a). Glutathione involves in homeostasis of redox reactions in plant cells by two ways: participate in core redox reactions or establish a cross-connections between bioenergetic-signaling pathways in plants (Foyer and Noctor 2011). Glutathione is synthesized generally via γ-glutamylcysteine synthetase and glutathione synthetase activities (Li et al. 2004). Recently, a new glutathione synthetase enzyme, γ-glutamylcysteine synthetase-glutathione synthetase gene (StGCS-GS), was discovered in Streptococcus thermophiles (Liedschulte et al. 2010). StGCS-GS exhibits high tolerance capability for heavy metals and ability for considerable amounts of accumulations of Cd, Cu and Zn in E. coli (Liu et al. 2013). Because of the given data, the coding sequence of StGCS-GS gene was inserted into the appropriate vector by Liu et al. (2015a) and then cloned into sugar beet (Beta vulgaris) for determining a possible increase in heavy metal accumulation capability and also defining specifications of expression patterns of inserted genes. Transgenic sugar beets demonstrated dominant ability to retain Cd, Cu and Zn ions and also exhibited enhanced glutathione and phytochelatin activities under the applications of heavy metal stresses compared with wild-type sugar beets. Liu et al. (2015a) stated that the created transgenic as a promising tool for phytoremediation could work fairly well.
Besides phytochelatin and metallothionein, nicotianamine also is another and alternative metal binding complex, produced from S-adenosylmethionine via the activity of nicotianamine synthetase (Higuchi et al. 1994). Yellow stripe-like family members are metal transporter proteins responsible for uptake and transportation of various metals in the plants (Curie et al. 2009). They exploit phytosiderophores- and nicotianamine–metal complexes as their substrates for either up-taking or translocating of these complexes from one place to another in plants. A yellow stripe-like gene from Miscanthus sacchariflorus was transferred into Arabidopsis by Chen et al. (2018). Analyses were conducted for transgenic Arabidopsis for the following parameters: phenotypic characteristics, biochemical features and metal accumulation/transportation ability. Chen et al. (2018) concluded that MsYSL gene specifically involved in translocation of Cd from root to shoot and in detoxification of Cd in the above parts of the plant. Some plants have ability to accumulate heavy metals in their roots. Translocating of accumulated heavy metals from roots to above parts can serve to enhance the capacity of phytoremediation in plants.
In a recent study done by Sun et al. (2018b), poplar was utilized in aiming of remediation of Hg from soils via using a class of ATP-binding cassette transporter gene. To achieve this goal, the transformation of ATP-binding cassette transporter gene (PtABCC1) from Populus trichocarpa to wild-type Arabidopsis was realized. The rates of Hg accumulation were found to be as 26–72% and 7–160% for transgenic Arabidopsis and poplar compared with wild types, respectively. Metal transporters have always been an important part of phytoremediation studies, and PtABCC1 gene playing a key role in enhancing green cleaning technology is given as an example for this.
In addition to metal transporter proteins, there are also some other functional genes related to remediation capabilities. Arsenic reductase 2 gene from A. thaliana was cloned by Nahar et al. (2017) for phytoremediation purposes in their study. Transformation of previously defined 1356 bp AtACR2 open reading frame was done via A. tumefaciens using Nicotiana tabacum var. Samsun leaf disks. Transgenics were grown for a 45-day period in arsenic containing medium. After harvesting of transgenics and non-transgenic, As contents were determined using inductively coupled plasma-mass spectroscopy. As levels in the roots of transgenic tobacco were found to be significantly higher in comparison with wild types. On the other hand, As levels in the shoots of transgenic tobacco were determined as quite low compared with As levels found in roots suggesting transformation of ACR2 gene could be profitable choice for phytoremedial purposes.
Scientists working in phytoremedial research are eager to use CRISPR-mediated gene expressions to increase the volumes for the synthesis of various molecules, including metal ligands (metallothioneins and phytochelatins), metal transport proteins (cation diffusion facilitator, heavy metal ATPases, multidrug efflux transporters, yellow stripe-like and zinc-regulated transporter families), plant growth hormones (auxins, cytokinins, and gibberellic acid) and root exudates (particularly low molecular weight organic acid and siderophores) (Basharat et al. 2018).
Environmental implications
In field applications, the most important prerequisite in phytoremediation for the effectiveness relies on the usability of experimental works. Without using in field applications, no matter how good and effective a method developed with employing a genetically modified organism it remains experimental and it is the greatest limitation for it. For removing heavy metals from the soil effectively, the priority should be given to the species used in remediation. The well-documented experimental studies utilizing transgenic plants were described in the previous section. The studies given below not experimental but rather include practical examples.
Anh et al. (2017) reported that plant species including some grass types and Pteris vittata, Pityrogramma calomelanos, and Vetiveria zizanioides were used to remove heavy metals from soil in Vietnam and some local species were found to be applicable for field conditions. Another phytoremedial application for wastewater treatment was successfully done using agricultural species including Medicago sativa, Zea mays, Helianthus annus and Sorghum bicolor (Atia et al. 2019). Cameselle et al. (2019) revealed promising results from the large-scale phytoremedial field applications using Brassica rapa subsp rapa and Lolium perenne. Also, phytoremedial applications were performed using Elodea canadensis for Co removal from wastewater (Mosoarca et al. 2018) and Plantago lanceolate for removal of Cd and As from the environment. Furthermore, Salvia sclarea (Chand et al. (2015), Sedum plumbizincicola (Deng et al. (2016), and S. alfredii and P. vittata Wan et al. (2016) were used for large-scale remediation of numerous heavy metals. These studies are the examples of eco-compatible environmental applications using wild-type plant species and these species bear great potential for removal of heavy metal accumulations via modification of their genetic structures with the genes having phytoremedial features.
Despite many plant species including Alyssum sp., Thlaspi sp., Brassica juncea, Viola calaminaria used in remediation of heavy metal removing strategies, new hyperaccumulator plants should be identified and used for creating genetically modified ones having better heavy metal accumulation capabilities. Based on the relevant studies, one of the most effective plants in this regard is the plants produced for energy (Shah and Pathak 2019). Plants used in bioenergy production should be genetically modified and grown in soils with high heavy metal content to make phytoremediation feasible. Also, to increase biomass of existing transgenic, hyperaccumulator or bioenergy plants, they should also be cultivated with microorganisms including Agrobacterium sp., Alcaligenes sp., Arthrobacter sp., Azospirillum sp., Bacillus sp., Burkholderia sp., Pseudomonas sp. Rhizobium sp. and Serretia sp., in these soils in considering mycorrhizal relationships. Aromatic plants, native wild plants or invasive plants could be considered as targets for creating transgenic plants in purpose of using in remediation of heavy metals, and also research on these targets should be carried out for identifying new genes related to heavy metal accumulation and detoxification processes. There are few studies published regarding this subject. There are plants that are well adapted to such contaminated environments and are used in transgenic and phytoremedial research because of their ability to produce photosynthesis and biomass in large scales.
The basis for creating transgenic involves the transfer of a gene, being not in its genome, from a different species to an organism or modifying genetic material of that organism without any genetic material transfer for some purposes. For latter, DNA shuffling approach is used for generating new chimeric genes without any gene transfer from a foreign organism, and after that, step examination for phytoremedial features of these chimeric genes is performed. In order to make development in this area, having extensive knowledge about the working dynamics of genes is a necessity. Also synthetic promoter designing could be an alternative strategy in an effort to enhance activity of a gene, which is related to phytoremediation. Designing a promoter region would be easier than that of shuffling approach.
Another method for escalating the removal of heavy metal accumulation in the environment is to modify plants genetically to gain ability to absorb heavy metals from both the soil and air. Phylloremediation is the name for the strategy for air phytoremediation (Wei et al. 2017). The crops genetically modified for the absorption of heavy metals from both air and soil are used in agricultural areas by the ways of intercropping or mixed cropping practising in terms of using plant growth regulators to increase the effectiveness of phytoremediation (Vamerali et al. 2010). Using genetically engineered organisms at appropriate distances in crops in agricultural farming areas will reduce heavy metal entry into the food chain and will help to remove heavy metals from soil. Here, the important point is that transgenic species to be used have not to make competition with those agricultural plants and should be easily removed from the farming land after their missions are completed. These features should be considered in the intercropping approach. From the agricultural lands left to fallowing, heavy metals can be removed via cultivating genetically modified organisms (especially showing growing ability with given low amounts of nutrients) without disturbing the soil. Finally, genetically modified organisms make contributions to research on phytoremediation with applications of many different approaches and provide promoting force for the effectiveness of research in this field.
Despite having all the advantages explained here, there are some serious concerns about the environmental applications of transgenic plants. The one is that the genetically modified plants could become dominant species in the application area due to their adaptation and competition capabilities, provided by having the gene or genes introduced compared to the native plant species (Ellstrand and Schierenbeck 2006; Auer 2008). Compared to the corresponding native species, they have potential for spreading out largely and due to this fact that they are considered as invasive species. Another serious concern is about the gene flow from the genetically modified plants to the corresponding wild types (Gunarathne et al. 2019). Thus, in some extent, deterioration on ecosystem as well as biodiversity present is also seen by these types of applications. For these reasons, creating transgenic plants is highly sensitive subject and after very careful analyzing and considering the region and biodiversity can be realized.
As a result, heavy metals pose threats for human health, primarily by consumption of drinking water and foods contaminated. While nanomaterial-based filtration systems are used effectively for cleaning drinking water contaminated or wastewater, phytoremediation remains as an environmentally friendly and effective method for removing heavy metals from the areas used for the productions of foods. In this review, a summary including the experimental and practical applications is presented related to how phytoremediation approaches developed involve phytochelatins and metallothioneins for the removing of heavy metals.
Abbreviations
- AtPCs1:
-
Arabidopsis thaliana phytochelatin synthase 1
- AtACBP1:
-
Arabidopsis thaliana acyl-CoA-binding domain-containing protein 1
- bphC:
-
Biphenyl-2,3-diol 1,2-dioxygenase
- cad1–3:
-
Arabidopsis thaliana knockout mutant for phytochelatin synthase 1
- CarMT:
-
Cicer arietinum metallothionein
- CCoAOMT:
-
Caffeoyl-CoA O-methyltransferase
- CePCs:
-
Caenorhabditis elegans phytochelatin synthase 1
- chv:
-
Chromosomal virulence
- CKX2:
-
Cytokinin dehydrogenase 2
- COR15a:
-
Cold regulated 15 a promoter
- IlMT2b:
-
Iris lactea var. chinensis metallothionein 2b
- Lm-SAP:
-
Lobularia maritima stress-associated protein
- MAN3:
-
Endo-1,4-β-Mannanase 3
- merC:
-
Mercury transporter
- Met:
-
Metallothionein
- MsYSL:
-
Miscanthus sacchariflorus yellow stripe-like
- OsARM1:
-
Oryza sativa arsenite-responsive myeloblastosis 1
- OsMyb4:
-
Oryza sativa myeloblastosis 4
- OsMT2c:
-
Oryza sativa metallothionein 2c
- PCs1:
-
Phytochelatin synthase 1
- PtoHMA5:
-
Populus tomentosa heavy metal ATPase 5
- PtPCs:
-
Populus tomentosa phytochelatin synthase
- PjHMT:
-
Prosopis juliflora heavy metal ATPase peptide
- PtoEXPA12:
-
Populus tomentosa alpha expansin 12
- PtPCs:
-
Populus tomentosa phytochelatin synthase
- ricMT:
-
Rice metallothionein
- SaCAD:
-
Sedum alfredii cinnamyl alcohol dehydrogenase
- SsMT2:
-
Suaeda salsa metallothionein 2
- VcPCs1:
-
Vicia sativa phytochelatin synthase 1
- vir:
-
Virulence
- VsCCoAOMT:
-
Vicia sativa caffeoyl-CoA O-methyltransferase
- XCD1:
-
XVE system-induced cadmium-tolerance 1
References
Abid M et al (2016) Arsenic(V) biosorption by charred orange peel in aqueous environments. Int J Phytoremediat 18:442–449. https://doi.org/10.1080/15226514.2015.1109604
Agnihotri A, Seth CS (2019) Chapter 11—Transgenic Brassicaceae: a promising approach for phytoremediation of heavy metals. In: Prasad MNV (ed) Transgenic plant technology for remediation of toxic metals and metalloids. Academic Press, New York, pp 239–255. https://doi.org/10.1016/B978-0-12-814389-6.00011-0
Ahmad MM, Ali A, Siddiqui S, Kamaluddin, Abdin MZ (2017) Methods in Transgenic Technology. In: Abdin MZ, Kiran U, Kamaluddin, Ali A (eds) Plant Biotechnology: Principles and Applications. Springer, Singapore, pp 93–115. https://doi.org/10.1007/978-981-10-2961-5_4
Akguc N, Ozyigit II, Yarci C (2008) Pyracantha coccinea roem. (Rosaceae) as a biomonitor for Cd, Pb and Zn in Mugla Province (TURKEY). Pak J Bot 40:1767–1776
Akguc N, Ozyigit II, Yasar U, Leblebici Z, Yarci C (2010) Use of Pyracantha coccinea Roem. as a possible biomonitor for the selected heavy metals. Int J Environ Sci Technol 7:427–434. https://doi.org/10.1007/bf03326152
Alagić SČ, Jovanović VPS, Mitić VD, Cvetković JS, Petrović GM, Stojanović GS (2016) Bioaccumulation of HMW PAHs in the roots of wild blackberry from the Bor region (Serbia): phytoremediation and biomonitoring aspects. Sci Total Environ 562:561–570. https://doi.org/10.1016/j.scitotenv.2016.04.063
Ali H, Khan E, Sajad MA (2013) Phytoremediation of heavy metals-concepts and applications. Chemosphere 91:869–881
Alok A, Sharma S, Kumar J, Verma S, Sood H (2017) Engineering in plant genome using agrobacterium: progress and future. In: Kalia VC, Saini AK (eds) Metabolic engineering for bioactive compounds: strategies and processes. Springer, Singapore, pp 91–111. https://doi.org/10.1007/978-981-10-5511-9_5
Anh BTK, Ha NTH, Danh LT, Van Minh V, Kim DD (2017) Phytoremediation applications for metal-contaminated soils using terrestrial plants in Vietnam. In: Ansari AA, Gill SS, Gill R, Lanza GR, Newman L (eds) Phytoremediation: management of environmental contaminants, vol 5. Springer, Cham, pp 157–181. https://doi.org/10.1007/978-3-319-52381-1_6
Anjum NA, Aref IM, Duarte AC, Pereira E, Ahmad I, Iqbal M (2014) Glutathione and proline can coordinately make plants withstand the joint attack of metal(loid) and salinity stresses. Front Plant Sci 5:662. https://doi.org/10.3389/fpls.2014.00662
Arslan M, Imran A, Khan QM, Afzal M (2017) Plant-bacteria partnerships for the remediation of persistent organic pollutants. Environ Sci Pollut Res 24:4322–4336
Arthur EL, Rice PJ, Rice PJ, Anderson TA, Baladi SM, Henderson KLD, Coats JR (2005) Phytoremediation—an overview. Crit Rev Plant Sci 24:109–122. https://doi.org/10.1080/07352680590952496
Asgari Lajayer B, Ghorbanpour M, Nikabadi S (2017) Heavy metals in contaminated environment: destiny of secondary metabolite biosynthesis, oxidative status and phytoextraction in medicinal plants. Ecotoxicol Environ Saf 145:377–390. https://doi.org/10.1016/j.ecoenv.2017.07.035
Atia FAM, Al-Ghouti MA, Al-Naimi F, Abu-Dieyeh M, Ahmed T, Al-Meer SH (2019) Removal of toxic pollutants from produced water by phytoremediation: applications and mechanistic study. J Water Process Eng 32:100990. https://doi.org/10.1016/j.jwpe.2019.100990
Auer C (2008) Ecological risk assessment and regulation for genetically-modified ornamental plants. Crit Rev Plant Sci 27:255–271
Bai J, Wang X, Wang R, Wang J, Le S, Zhao Y (2019) Overexpression of three duplicated BnPCS genes enhanced Cd accumulation and translocation in Arabidopsis thaliana mutant cad1–3. Bull Environ Contam Toxicol 102:146–152. https://doi.org/10.1007/s00128-018-2487-1
Bañuelos GS, Ajwa HA, Terry N, Zayed A (1997) Phytoremediation of selenium laden soils: a new technology. J Soil Water Conserv 52:426–430
Barampuram S, Zhang ZJ (2011) Recent advances in plant transformation. Methods Mol Biol (Clifton, NJ) 701:1–35. https://doi.org/10.1007/978-1-61737-957-4_1
Barber SA (1962) A diffusion and mass-flow concept of soil nutrient availability. Soil Sci 93:39–49
Basharat Z, Novo LAB, Yasmin A (2018) Genome editing weds CRISPR: what is in it for phytoremediation? Plants 7:51
Baxter I et al (2003) Genomic comparison of P-type ATPase ion pumps in Arabidopsis and rice. Plant Physiol 132:618–628. https://doi.org/10.1104/pp.103.021923
Belouchi A, Kwan T, Gros P (1997) Cloning and characterization of the OsNramp family from Oryza sativa, a new family of membrane proteins possibly implicated in the transport of metal ions. Plant Mol Biol 33:1085–1092. https://doi.org/10.1023/a:1005723304911
Berni R et al (2018) Reactive oxygen species and heavy metal stress in plants: impact on the cell wall and secondary metabolism. Environ Exp Bot. https://doi.org/10.1016/j.envexpbot.2018.10.017
Bhaya D, Davison M, Barrangou R (2011) CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu Rev Genet 45:273–297. https://doi.org/10.1146/annurev-genet-110410-132430
Bibikova M, Carroll D, Segal DJ, Trautman JK, Smith J, Kim YG, Chandrasegaran S (2001) Stimulation of homologous recombination through targeted cleavage by chimeric nucleases. Mol Cell Biol 21:289–297. https://doi.org/10.1128/mcb.21.1.289-297.2001
Boch J et al (2009) Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326:1509–1512. https://doi.org/10.1126/science.1178811
Bondada BR, Tu S, Ma LQ (2004) Absorption of foliar-applied arsenic by the arsenic hyperaccumulating fern (Pteris vittata L.). Sci Total Environ 332:61–70. https://doi.org/10.1016/j.scitotenv.2004.05.001
Boudet AM (2007) Evolution and current status of research in phenolic compounds. Phytochemistry 68:2722–2735
Bradshaw JE (2016) Genetically modified crops. In: Bradshaw JE (ed) Plant breeding: past, present and future. Springer, Berlin. https://doi.org/10.1007/978-3-319-23285-0
Brune B, Hartzell P, Nicotera P, Orrenius S (1991) Spermine prevents endonuclease activation and apoptosis in thymocytes. Exp Cell Res 195:323–329
Cameselle C, Gouveia S, Urréjola S (2019) Benefits of phytoremediation amended with DC electric field. Application to soils contaminated with heavy metals. Chemosphere 229:481–488. https://doi.org/10.1016/j.chemosphere.2019.04.222
Cetintas VB, Kotmakcı M, Kaymaz BT (2017) From the immune response to the genome design; CRISPR-Cas9 system: review. Turkiye Klinikleri J Med Sci 37:27–42. https://doi.org/10.5336/medsci.2016-54153
Chand S, Yaseen M, Rajkumari Patra DD (2015) Application of heavy metal rich tannery sludge on sustainable growth, yield and metal accumulation by clarysage (Salvia sclarea L.). Int J Phytoremediat 17:1171–1176. https://doi.org/10.1080/15226514.2015.1045128
Chandra R, Yadav S (2010) Potential of Typha angustifolia for phytoremediation of heavy metals from aqueous solution of phenol and melanoidin. Ecol Eng 36:1277–1284. https://doi.org/10.1016/j.ecoleng.2010.06.003
Chaurasia N, Mishra Y, Rai LC (2008) Cloning expression and analysis of phytochelatin synthase (pcs) gene from Anabaena sp. PCC 7120 offering multiple stress tolerance in Escherichia coli. Biochem Biophys Res Commun 376:225–230. https://doi.org/10.1016/j.bbrc.2008.08.129
Chen L, Guo Y, Yang L, Wang Q (2008) Synergistic defensive mechanism of phytochelatins and antioxidative enzymes in Brassica chinensis L. against Cd stress. Chin Sci Bull 53:1503. https://doi.org/10.1007/s11434-008-0062-6
Chen J et al (2015a) MAN3 gene regulates cadmium tolerance through the glutathione-dependent pathway in Arabidopsis thaliana. New Phytol 205:570–582. https://doi.org/10.1111/nph.13101
Chen YK, Liu YX, Ding YN, Wang XT, Xu JC (2015b) Overexpression of PtPCS enhances cadmium tolerance and cadmium accumulation in tobacco. Plant Cell Tissue Organ 121:389–396
Chen SS, Han XJ, Fang J, Lu ZC, Qiu WM, Liu MY, Sang J, Jiang J, Zhuo RY (2017) Sedum alfredii sanramp6 metal transporter contributes to cadmium accumulation in transgenic arabidopsis thaliana. Sci Rep-Uk 7
Chen H, Zhang C, Guo H, Hu Y, He Y, Jiang D (2018) Overexpression of a Miscanthus sacchariflorus yellow stripe-like transporter MsYSL1 enhances resistance of Arabidopsis to cadmium by mediating metal ion reallocation. Plant Growth Regul 85:101–111. https://doi.org/10.1007/s10725-018-0376-6
Cherian S, Oliveira MM (2005) Transgenic plants in phytoremediation: recent advances and new possibilities. Environ Sci Technol 39:9377–9390
Choppala G et al (2014) Cellular mechanisms in higher plants governing tolerance to cadmium toxicity. Crit Rev Plant Sci 33:374–391. https://doi.org/10.1080/07352689.2014.903747
Christian ML et al (2012) Targeting G with TAL effectors: a comparison of activities of TALENs constructed with NN and NK repeat variable di-residues. Plos One 7:e45383. https://doi.org/10.1371/journal.pone.0045383
Christie PJ (2004) Type IV secretion: the agrobacterium VirB/D4 and related conjugation systems. Biochim Biophys Acta 1694:219–234. https://doi.org/10.1016/j.bbamcr.2004.02.013
Clemens S, Antosiewicz DM, Ward JM, Schachtman DP, Schroeder JI (1998) The plant cDNA mediates the uptake of calcium and cadmium in yeast. Proc Natl Acad Sci 95:12043–12048. https://doi.org/10.1073/pnas.95.20.12043
Clemens S, Kim EJ, Neumann D, Schroeder JI (1999) Tolerance to toxic metals by a gene family of phytochelatin synthases from plants and yeast. EMBO J 18:3325–3333. https://doi.org/10.1093/emboj/18.12.3325
Clemens S, Palmgren MG, Krämer U (2002) A long way ahead: understanding and engineering plant metal accumulation. Trends Plant Sci 7:309–315. https://doi.org/10.1016/S1360-1385(02)02295-1
Cobbett C, Goldsbrough P (2002) Phytochelatins and metallothioneins: roles in heavy metal detoxification and homeostasis. Annu Rev Plant Biol 53:159–182. https://doi.org/10.1146/annurev.arplant.53.100301.135154
Cong L et al (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339:819–823. https://doi.org/10.1126/science.1231143
Cosio C, Martinoia E, Keller C (2004) Hyperaccumulation of cadmium and zinc in Thlaspi caerulescens and Arabidopsis halleri at the leaf cellular level. Plant Physiol 134:716–725. https://doi.org/10.1104/pp.103.031948
Curie C et al (2009) Metal movement within the plant: contribution of nicotianamine and yellow stripe 1-like transporters. Ann Bot 103:1–11. https://doi.org/10.1093/aob/mcn207
Cutillas-Barreiro L et al (2016) Lithological and land-use based assessment of heavy metal pollution in soils surrounding a cement plant in SW Europe. Sci Total Environ 562:179–190. https://doi.org/10.1016/j.scitotenv.2016.03.198
DalCorso G (2012) Heavy metal toxicity in plants. In: Furini A (ed) Plants and heavy metals. Springer, Heidelberg, pp 1–26
DalCorso G, Farinati S, Maistri S, Furini A (2008) How plants cope with cadmium: staking all on metabolism and gene expression. J Integr Plant Biol 50:1268–1280
DalCorso G, Manara A, Furini A (2013) An overview of heavy metal challenge in plants: from roots to shoots. Metallomics 5:1117–1132
DalCorso G, Fasani E, Manara A, Visioli G, Furini A (2019) Heavy metal pollutions: state of the art and innovation in phytoremediation. Int J Mol Sci 20:3412
Das N, Bhattacharya S, Maiti MK (2016) Enhanced cadmium accumulation and tolerance in transgenic tobacco overexpressing rice metal tolerance protein gene osmtp1 is promising for phytoremediation. Plant Physiol Bioch 105:297–309
De Farias V et al (2009) Phytodegradation potential of Erythrina crista-galli L., Fabaceae, in petroleum-contaminated soil. Appl Biochem Biotechnol 157:10–22. https://doi.org/10.1007/s12010-009-8531-1
Demarco CF, Afonso TF, Pieniz S, Quadro MS, Camargo FAD, Andreazza R (2019) Phytoremediation of heavy metals and nutrients by the Sagittaria montevidensis into an anthropogenic contaminated site at Southern of Brazil. Int J Phytoremediat 21:1145–1152. https://doi.org/10.1080/15226514.2019.1612843
Deng D, Yan C, Wu J, Pan X, Yan N (2014) Revisiting the TALE repeat. Protein Cell 5:297–306. https://doi.org/10.1007/s13238-014-0035-2
Deng L et al (2016) Long-term field phytoextraction of zinc/cadmium contaminated soil by Sedum plumbizincicola under different agronomic strategies. Int J Phytoremediat 18:134–140. https://doi.org/10.1080/15226514.2015.1058328
Dharupaneedi SP, Nataraj SK, Nadagouda M, Reddy KR, Shukla SS, Aminabhavi TM (2019) Membrane-based separation of potential emerging pollutants. Sep Purif Technol 210:850–866. https://doi.org/10.1016/j.seppur.2018.09.003
Dixit R et al (2015) Bioremediation of heavy metals from soil and aquatic environment: an overview of principles and criteria of fundamental processes. Sustainability 7:2189
Doty SL, Shang TQ, Wilson AM, Moore AL, Newman LA, Strand SE, Gordon MP (2003) Metabolism of the soil and groundwater contaminants, ethylene dibromide and trichloroethylene, by the tropical leguminous tree, Leuceana leucocephala. Water Res 37:441–449
Dreier B, Beerli RR, Segal DJ, Flippin JD, Barbas CF 3rd (2001) Development of zinc finger domains for recognition of the 5′-ANN-3′ family of DNA sequences and their use in the construction of artificial transcription factors. J Biol Chem 276:29466–29478. https://doi.org/10.1074/jbc.m102604200
Du J, Yang JL, Li CH (2012) Advances in metallotionein studies in forest trees. Plant Omics 5:46–51
Du ZY, Chen MX, Chen QF, Gu JD, Chye ML (2015) Expression of arabidopsis acyl-coa-binding proteins atacbp1 and atacbp4 confers pb(ii) accumulation in brassica juncea roots. Plant Cell Environ 38:101–117
Dubey S, Shri M, Gupta A, Rani V, Chakrabarty D (2018) Toxicity and detoxification of heavy metals during plant growth and metabolism. Environ Chem Lett 16:1169–1192. https://doi.org/10.1007/s10311-018-0741-8
Dubey AK et al (2019) Over-expression of CarMT gene modulates the physiological performance and antioxidant defense system to provide tolerance against drought stress in Arabidopsis thaliana L. Ecotoxicol Environ Saf 171:54–65. https://doi.org/10.1016/j.ecoenv.2018.12.050
Durai S, Mani M, Kandavelou K, Wu J, Porteus MH, Chandrasegaran S (2005) Zinc finger nucleases: custom-designed molecular scissors for genome engineering of plant and mammalian cells. Nucleic Acids Res 33:5978–5990. https://doi.org/10.1093/nar/gki912
Eapen S, D’Souza SF (2005) Prospects of genetic engineering of plants for phytoremediation of toxic metals. Biotechnol Adv 23:97–114. https://doi.org/10.1016/j.biotechadv.2004.10.001
Eapen S, Suseelan KN, Tivarekar S, Kotwal SA, Mitra R (2003) Potential for rhizofiltration of uranium using hairy root cultures of Brassica juncea and Chenopodium amaranticolor. Environ Res 91:127–133
Eapen S, Singh S, D’Souza SF (2007) Advances in development of transgenic plants for remediation of xenobiotic pollutants. Biotechnol Adv 25:442–451. https://doi.org/10.1016/j.biotechadv.2007.05.001
Ellstrand NC, Schierenbeck KA (2006) Hybridization as a stimulus for the evolution of invasiveness in plants? Euphytica 148:35–46
Ely CS, Smets BF (2017) Bacteria from wheat and cucurbit plant roots metabolize PAHs and aromatic root exudates: implications for rhizodegradation. Int J Phytoremediat 19:877–883. https://doi.org/10.1080/15226514.2017.1303805
Fan W et al (2018) Two mulberry phytochelatin synthase genes confer zinc/cadmium tolerance and accumulation in transgenic Arabidopsis and tobacco. Gene 645:95–104
Farid M et al (2020) Efficacy of Zea mays L. for the management of marble effluent contaminated soil under citric acid amendment; morpho-physiological and biochemical response. Chemosphere 240:124930. https://doi.org/10.1016/j.chemosphere.2019.124930
Fasani E, Manara A, Martini F, Furini A, DalCorso G (2018) The potential of genetic engineering of plants for the remediation of soils contaminated with heavy metals. Plant Cell Environ 41:1201–1232
Favas PJ, Pratas J, Prasad MN (2012) Accumulation of arsenic by aquatic plants in large-scale field conditions: opportunities for phytoremediation and bioindication. Sci Total Environ 433:390–397. https://doi.org/10.1016/j.scitotenv.2012.06.091
Feng Z et al (2014) Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc Natl Acad Sci USA 111:4632–4637. https://doi.org/10.1073/pnas.1400822111
Feng NX et al (2017) Efficient phytoremediation of organic contaminants in soils using plant-endophyte partnerships. Sci Total Environ 583:352–368
Ferraz P, Fidalgo F, Almeida A, Teixeira J (2012) Phytostabilization of nickel by the zinc and cadmium hyperaccumulator Solanum nigrum L. Are metallothioneins involved? Plant Physiol Biochem PPB 57:254–260. https://doi.org/10.1016/j.plaphy.2012.05.025
Filiz E, Ozyigit II, Saracoglu IA, Uras ME, Sen U, Yalcin B (2019a) Abiotic stress-induced regulation of antioxidant genes in different Arabidopsis ecotypes: microarray data evaluation. Biotechnol Biotechnol Equip 33:128–143. https://doi.org/10.1080/13102818.2018.1556120
Filiz E, Saracoglu IA, Ozyigit II, Yalcin B (2019b) Comparative analyses of phytochelatin synthase (PCS) genes in higher plants. Biotechnol Biotechnol Equip 33:178–194. https://doi.org/10.1080/13102818.2018.1559096
Foyer CH, Noctor G (2011) Ascorbate and glutathione: the heart of the redox hub. Plant Physiol 155:2–18
Gasic K, Korban SS (2006) Heavy metal stress. In: Madhava Rao KV, Raghavendra AS, Janardhan Reddy K (eds) Physiology and molecular biology of stress tolerance in plants. Springer, Dordrecht, pp 219–254. https://doi.org/10.1007/1-4020-4225-6_8
Gatliff EG (1994) Vegetative remediation process offers advantages over traditional pump-and-treat technologies. Remediat J 4:343–352. https://doi.org/10.1002/rem.3440040307
Gaur N, Flora G, Yadav M, Tiwari A (2014) A review with recent advancements on bioremediation-based abolition of heavy metals. Environ Sci Process Impacts 16:180–193. https://doi.org/10.1039/c3em00491k
Gelvin SB (2012) Traversing the cell: agrobacterium T-DNA’s journey to the host genome front. Plant Sci 3:52. https://doi.org/10.3389/fpls.2012.00052
Gerhardt KE, Huang XD, Glick BR, Greenberg BM (2009) Phytoremediation and rhizoremediation of organic soil contaminants: potential and challenges. Plant Sci 176:20–30
Gerhardt KE, Gerwing PD, Greenberg BM (2017) Opinion: taking phytoremediation from proven technology to accepted practice. Plant Sci 256:170–185
Ghori Z, Iftikhar H, Bhatti MF, Nasar um M, Sharma I, Kazi AG, Ahmad P (2016) Chapter 15-Phytoextraction: the use of plants to remove heavy metals from soil. In: Ahmad P (ed) Plant metal interaction. Elsevier, Amsterdam, pp 385–409. https://doi.org/10.1016/B978-0-12-803158-2.00015-1
Ghori N-H, Ghori T, Hayat MQ, Imadi SR, Gul A, Altay V, Ozturk M (2019) Heavy metal stress and responses in plants. Int J Environ Sci Technol. https://doi.org/10.1007/s13762-019-02215-8
Gomes MAD, Hauser-Davis RA, de Souza AN, Vitoria AP (2016) Metal phytoremediation: general strategies, genetically modified plants and applications in metal nanoparticle contamination. Ecotoxicol Environ Saf 134:133–147
Gong Y, Chen JP, Pu RP (2019) The enhanced removal and phytodegradation of sodium dodecyl sulfate (SDS) in wastewater using controllable water hyacinth. Int J Phytoremediat 21:1080–1089. https://doi.org/10.1080/15226514.2019.1606779
Gonzalez H, Fernandez-Fuego D, Bertrand A, Gonzalez A (2019) Effect of pH and citric acid on the growth, arsenic accumulation, and phytochelatin synthesis in Eupatorium cannabinum L., a promising plant for phytostabilization. Environ Sci Pollut Res 26:26242–26253. https://doi.org/10.1007/s11356-019-05657-2
Gosal SS, Wani SH (2018) Plant genetic transformation and transgenic crops: methods and applications. In: Gosal SS, Wani SH (eds) Biotechnologies of crop improvement, volume 2: transgenic approaches. Springer, Cham, pp 1–23. https://doi.org/10.1007/978-3-319-90650-8_1
Grennan AK (2011) Metallothioneins, a diverse protein family. Plant Physiol 155:1750–1751. https://doi.org/10.1104/pp.111.900407
Gu C-S, Liu L-Q, Deng Y-M, Zhu X-D, Huang S-Z, Lu X-Q (2015) The heterologous expression of the Iris lactea var chinensis type 2 metallothionein IlMT2b gene enhances copper tolerance in Arabidopsis thaliana. Bull Environ Contam Toxicol 94:247–253. https://doi.org/10.1007/s00128-014-1444-x
Gul I, Manzoor M, Silvestre J, Rizwan M, Hina K, Kallerhoff J, Arshad M (2019) EDTA-assisted phytoextraction of lead and cadmium by Pelargonium cultivars grown on spiked soil. Int J Phytoremediat 21:101–110. https://doi.org/10.1080/15226514.2018.1474441
Gullner G, Komives T, Rennenberg H (2001) Enhanced tolerance of transgenic poplar plants overexpressing gamma-glutamylcysteine synthetase towards chloroacetanilide herbicides. J Exp Bot 52:971–979. https://doi.org/10.1093/jexbot/52.358.971
Gunarathne V, Mayakaduwa S, Ashiq A, Weerakoon SR, Biswas JK, Vithanage M (2019) Transgenic plants: benefits, applications, and potential risks in phytoremediation. Transgenic plant technology for remediation of toxic metals and metalloids. Elsevier, Amsterdam, pp 89–102
Gurlebeck D, Thieme F, Bonas U (2006) Type III effector proteins from the plant pathogen Xanthomonas and their role in the interaction with the host plant. J Plant Physiol 163:233–255. https://doi.org/10.1016/j.jplph.2005.11.011
Han K-H, Meilan R, Ma C, Strauss SH (2000) An Agrobacterium tumefaciens transformation protocol effective on a variety of cottonwood hybrids (genus Populus). Plant Cell Rep 19:315–320. https://doi.org/10.1007/s002990050019
Hanikenne M (2003) Chlamydomonas reinhardtii as a eukaryotic photosynthetic model for studies of heavy metal homeostasis and tolerance. New Phytol 159:331–340. https://doi.org/10.1046/j.1469-8137.2003.00788.x
Hansen D, Duda PJ, Zayed A, Terry N (1998) Selenium removal by constructed wetlands: role of biological volatilization. Environ Sci Technol 32:591–597. https://doi.org/10.1021/es970502l
Hassan Z, Aarts MGM (2011) Opportunities and feasibilities for biotechnological improvement of Zn, Cd or Ni tolerance and accumulation in plants. Environ Exp Bot 72:53–63. https://doi.org/10.1016/j.envexpbot.2010.04.003
He Z, Shentu J, Yang X, Baligar VC, Zhang T, Stoffella PJ (2015) Heavy metal contamination of soils: sources, indicators, and assessment. J Environ Indic 9:17–18
He YJ et al (2017) Metabolism of Ibuprofen by Phragmites australis: uptake and phytodegradation. Environ Sci Technol 51:4576–4584. https://doi.org/10.1021/acs.est.7b00458
He X et al (2019) Polyaspartate and liquid amino acid fertilizer are appropriate alternatives for promoting the phytoextraction of cadmium and lead in Solanum nigrum L. Chemosphere 237:124483. https://doi.org/10.1016/j.chemosphere.2019.124483
Headley JV, Peru KM, Du JL, Gurprasad N, McMartin DW (2008) Evaluation of the apparent phytodegradation of pentachlorophenol by Chlorella pyrenoidosa. J Environ Sci Health Part A Toxic/Hazard Subst Environ Eng 43:361–364. https://doi.org/10.1080/10934520701795491
Heckenroth A, Rabier J, Dutoit T, Torre F, Prudent P, Laffont-Schwob I (2016) Selection of native plants with phytoremediation potential for highly contaminated Mediterranean soil restoration: tools for a non-destructive and integrative approach. J Environ Manag 183:850–863
Henry JR (2000) An overview of the phytoremediation of lead and mercury. Office of Solid Waste and Emergency Response, Technology Innovation Office, Washington, DC
Hernández LE, Sobrino-Plata J, Montero-Palmero MB, Carrasco-Gil S, Flores-Cáceres ML, Ortega-Villasante C, Escobar C (2015) Contribution of glutathione to the control of cellular redox homeostasis under toxic metal and metalloid stress. J Exp Bot 66:2901–2911. https://doi.org/10.1093/jxb/erv063
Hernández A, Loera N, Contreras M, Fischer L, Sánchez D (2019) Comparison between Lactuca sativa L and Lolium perenne: phytoextraction capacity of Ni, Fe, and Co from galvanoplastic industry. Energy technology 2019. Springer, Cham, pp 137–147
Higuchi K, Kanazawa K, Nishizawa N-K, Chino M, Mori S (1994) Purification and characterization of nicotianamine synthase from Fe-deficient barley roots. Plant Soil 165:173–179. https://doi.org/10.1007/bf00008059
Hoffman FO, Garten CT, Huckabee JW, Lucas DM (1982) Interception and retention of technetium by vegetation and soil. J Environ Qual 11:134–141. https://doi.org/10.2134/jeq1982.00472425001100010030x
Hooda V (2007) Phytoremediation of toxic metals from soil and waste water. J Environ Biol 28:367–376
Hossain MA, Piyatida P, da Silva JAT, Fujita M (2012) Molecular mechanism of heavy metal toxicity and tolerance in plants: central role of glutathione in detoxification of reactive oxygen species and methylglyoxal and in heavy metal chelation. J Bot 2012:37. https://doi.org/10.1155/2012/872875
Huang G-Y, Wang Y-S (2009) Expression analysis of type 2 metallothionein gene in mangrove species (Bruguiera gymnorrhiza) under heavy metal stress. Chemosphere 77:1026–1029. https://doi.org/10.1016/j.chemosphere.2009.07.073
Huang X et al (2019) Insights into heavy metals leakage in chelator-induced phytoextraction of Pb- and Tl-contaminated soil. Int J Environ Res Public Health 16:1328
Jabeen R, Ahmad A, Iqbal M (2009) Phytoremediation of heavy metals: physiological and molecular mechanisms. Bot Rev 75:339–364. https://doi.org/10.1007/s12229-009-9036-x
Jadia CD, Fulekar MH (2008) Phytoremediation: the application of vermicompost to remove zinc, cadmium, copper, nickel and lead by sunflower plant. Environ Eng Manag J 7:547–558
Jadia CD, Fulekar MH (2009) Phytoremediation of heavy metals: recent techniques. Afr J Biotechnol 8:921–928
Jan AT, Ali A, Rizwanul Haq QM (2015) Chapter 3-Phytoremediation: a promising strategy on the crossroads of remediation. In: Hakeem KR, Sabir M, Öztürk M, Mermut AR (eds) Soil remediation and plants. Academic Press, San Diego, pp 63–84. https://doi.org/10.1016/B978-0-12-799937-1.00003-6
Jha P, Jobby R, Desai NS (2016) Remediation of textile azo dye acid red 114 by hairy roots of Ipomoea carnea Jacq. and assessment of degraded dye toxicity with human keratinocyte cell line. J Hazard Mater 311:158–167. https://doi.org/10.1016/j.jhazmat.2016.02.058
Jiali H, Hong L, Chaofeng M, Yanli Z, Andrea P, Heinz R, Xingqi C, Zhi-Bin L (2015) Overexpression of bacterial γ-glutamylcysteine synthetase mediates changes in cadmium influx, allocation and detoxification in poplar. New Phytol 205:240–254. https://doi.org/10.1111/nph.13013
Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA (2013) RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol 31:233–239. https://doi.org/10.1038/nbt.2508
Jin S, Xu C, Li G, Sun D, Li Y, Wang X, Liu S (2017) Functional characterization of a type 2 metallothionein gene, SsMT2, from alkaline-tolerant Suaeda salsa. Sci Rep UK 7:17914. https://doi.org/10.1038/s41598-017-18263-4
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337:816–821. https://doi.org/10.1126/science.1225829
Joung JK, Sander JD (2013) TALENs: a widely applicable technology for targeted genome editing. Nat Rev Mol Cell Biol 14:49–55. https://doi.org/10.1038/nrm3486
Kado CI (2014) Historical account on gaining insights on the mechanism of crown gall tumorigenesis induced by Agrobacterium tumefaciens. Front Microbiol 5:340–340. https://doi.org/10.3389/fmicb.2014.00340
Kagalkar AN, Jadhav MU, Bapat VA, Govindwar SP (2011) Phytodegradation of the triphenylmethane dye Malachite Green mediated by cell suspension cultures of Blumea malcolmii Hook. Biores Technol 102:10312–10318. https://doi.org/10.1016/j.biortech.2011.08.101
Kang JW (2014) Removing environmental organic pollutants with bioremediation and phytoremediation. Biotechnol Lett 36:1129–1139
Karenlampi S, Schat H, Vangronsveld J, Verkleij JA, van der Lelie D, Mergeay M, Tervahauta AI (2000) Genetic engineering in the improvement of plants for phytoremediation of metal polluted soils. Environ Pollut (Barking, Essex: 1987) 107:225–231
Kay S, Hahn S, Marois E, Hause G, Bonas U (2007) A bacterial effector acts as a plant transcription factor and induces a cell size regulator. Science 318:648–651. https://doi.org/10.1126/science.1144956
Keeran NS, Ganesan G, Parida AK (2017) A novel heavy metal ATPase peptide from prosopis juliflora is involved in metal uptake in yeast and tobacco. Transgenic Res 26:247–261
Kelishadi R, Moeini R, Poursafa P, Farajian S, Yousefy H, Okhovat-Souraki AA (2014) Independent association between air pollutants and vitamin D deficiency in young children in Isfahan, Iran. Paediatr Int Child Health 34:50–55. https://doi.org/10.1179/2046905513y.0000000080
Khalid N, Noman A, Aqeel M, Masood A, Tufail A (2019) Phytoremediation potential of Xanthium strumarium for heavy metals contaminated soils at roadsides. Int J Environ Sci Technol 16:2091–2100. https://doi.org/10.1007/s13762-018-1825-5
Khan K (2010) Gene transfer technologies and their applications: roles in human diseases. Asian J Exp Biol Sci 1:208–221
Kikkert JR, Vidal JR, Reisch BI (2004) Stable transformation of plant cells by particle bombardment/biolistics. In: Peña L (ed) Transgenic plants: methods and protocols. Humana Press, Totowa, pp 61–78. https://doi.org/10.1385/1-59259-827-7:061
Korshunova YO, Eide D, Clark WG, Guerinot ML, Pakrasi HB (1999) The IRT1 protein from Arabidopsis thaliana is a metal transporter with a broad substrate range. Plant Mol Biol 40:37–44
Kotnik T, Frey W, Sack M, Haberl Meglic S, Peterka M, Miklavcic D (2015) Electroporation-based applications in biotechnology. Trends Biotechnol 33:480–488. https://doi.org/10.1016/j.tibtech.2015.06.002
Kotnik T, Rems L, Tarek M, Miklavcic D (2019) Membrane electroporation and electropermeabilization: mechanisms and models. In: Dill KA (ed) Annual review of biophysics, vol 48. Annual Reviews, Palo Alto, pp 63–91. https://doi.org/10.1146/annurev-biophys-052118-115451
Kotoky R, Pandey P (2019) Rhizosphere mediated biodegradation of benzo(A)pyrene by surfactin producing soil bacilli applied through Melia azadirachta rhizosphere. Int J Phytoremediat. https://doi.org/10.1080/15226514.2019.1663486
Koutavarapu R et al (2020) ZnO nanosheets-decorated Bi2WO6 nanolayers as efficient photocatalysts for the removal of toxic environmental pollutants and photoelectrochemical solar water oxidation. J Environ Manag 265:110504. https://doi.org/10.1016/j.jenvman.2020.110504
Kramer U, Talke IN, Hanikenne M (2007) Transition metal transport. FEBS Lett 581:2263–2272. https://doi.org/10.1016/j.febslet.2007.04.010
Krzesłowska M (2011) The cell wall in plant cell response to trace metals: polysaccharide remodeling and its role in defense strategy. Acta Physiol Plant 33:35–51. https://doi.org/10.1007/s11738-010-0581-z
Krzeslowska M et al (2016) Pectinous cell wall thickenings formation—a common defense strategy of plants to cope with Pb. Environ Pollut (Barking, Essex: 1987) 214:354–361. https://doi.org/10.1016/j.envpol.2016.04.019
Kshirsagar S, Aery NC (2007) Phytostabilization of mine waste: growth and physiological responses of Vigna unguiculata (L.) Walp. J Environ Biol 28:651–654
Kühnlenz T, Westphal L, Schmidt H, Scheel D, Clemens S (2015) Expression of Caenorhabditis elegans PCS in the AtPCS1-deficient Arabidopsis thaliana cad1-3 mutant separates the metal tolerance and non-host resistance functions of phytochelatin synthases. Plant Cell Environ 38:2239–2247. https://doi.org/10.1111/pce.12534
Kuiper I, Lagendijk EL, Bloemberg GV, Lugtenberg BJJ (2004) Rhizoremediation: a beneficial plant-microbe interaction. Mol Plant Microbe Interact 17:6–15. https://doi.org/10.1094/mpmi.2004.17.1.6
Kumar S, Trivedi PK (2016) Chapter 25-Heavy metal stress signaling in plants. In: Ahmad P (ed) Plant metal interaction. Elsevier, Amsterdam, pp 585–603. https://doi.org/10.1016/B978-0-12-803158-2.00025-4
Kumar R et al (2016) Chapter 13-Detoxification and tolerance of heavy metals in plants. In: Ahmad P (ed) Plant metal interaction. Elsevier, Amsterdam, pp 335–359. https://doi.org/10.1016/B978-0-12-803158-2.00013-8
Kumar N, Jeena N, Kumar N, Gangola S, Singh H (2019) Chapter 15-Phytoremediation facilitating enzymes: an enzymatic approach for enhancing remediation process. In: Bhatt P (ed) Smart bioremediation technologies. Academic Press, New York, pp 289–306. https://doi.org/10.1016/B978-0-12-818307-6.00015-9
Kvesitadze G, Khatisashvili G, Sadunishvili T, Ramsden JJ (2006) Uptake, translocation and effects of contaminants in plants. In: Biochemical mechanisms of detoxification in higher plants: basis of phytoremediation. Springer, Berlin, pp 55–102. https://doi.org/10.1007/3-540-28997-6_2
Latef AAA (2013) Growth and some physiological activities of pepper (Capsicum annuum L.) in response to cadmium stress and mycorrhizal symbiosis. J Agric Sci Technol Iran 15:1437–1448
Le Faucheur S, Behra R, Sigg L (2005) Phytochelatin induction, cadmium accumulation, and algal sensitivity to free cadmium ion in Scenedesmus vacuolatus. Environ Toxicol Chem 24:1731–1737
Le Gall H, Philippe F, Domon J-M, Gillet F, Pelloux J, Rayon C (2015) Cell wall metabolism in response to abiotic stress. Plants 4:112
LeDuc DL et al (2004) Overexpression of selenocysteine methyltransferase in Arabidopsis and Indian mustard increases selenium tolerance and accumulation. Plant Physiol 135:377–383. https://doi.org/10.1104/pp.103.026989
Lee J-E, Ezura H (2016) Genome-editing technologies and their use in tomato. In: Ezura H, Ariizumi T, Garcia-Mas J, Rose J (eds) Functional genomics and biotechnology in Solanaceae and Cucurbitaceae crops. Springer, Berlin, pp 239–250. https://doi.org/10.1007/978-3-662-48535-4_14
Leigh MB, Fletcher JS, Fu X, Schmitz FJ (2002) Root turnover: an important source of microbial substrates in rhizosphere remediation of recalcitrant contaminants. Environ Sci Technol 36:1579–1583
Leung HM, Ye ZH, Wong MH (2007) Survival strategies of plants associated with arbuscular mycorrhizal fungi on toxic mine tailings. Chemosphere 66:905–915. https://doi.org/10.1016/j.chemosphere.2006.06.037
Li Y, Wei GY, Chen J (2004) Glutathione: a review on biotechnological production. Appl Microbiol Biot 66:233–242
Li XX, Zhang X, Wang XL, Cui ZJ (2019) Phytoremediation of multi-metal contaminated mine tailings with Solanum nigrum L. and biochar/attapulgite amendments. Ecotoxicol Environ Saf 180:517–525. https://doi.org/10.1016/j.ecoenv.2019.05.033
Liedschulte V, Wachter A, An ZG, Rausch T (2010) Exploiting plants for glutathione (GSH) production: uncoupling GSH synthesis from cellular controls results in unprecedented GSH accumulation. Plant Biotechnol J 8:807–820
Lin Z-Q, Schemenauer RS, Cervinka V, Zayed A, Lee A, Terry N (2000) Selenium volatilization from a soil—plant system for the remediation of contaminated water and soil in the San Joaquin Valley. J Environ Qual 29:1048–1056. https://doi.org/10.2134/jeq2000.00472425002900040003x
Lin CH, Lerch RN, Kremer RJ, Garrett HE (2011) Stimulated rhizodegradation of atrazine by selected plant species. J Environ Qual 40:1113–1121. https://doi.org/10.2134/jeq2010.0440
Liu D, Mao Z, An Z, Ma L, Lu Z (2013) Significant improved Escherichia coli tolerant to Cd2+, Zn2+ and Cu2+ expressing Streptococcus thermophilus StGCS-GS, compared to Arabidopsis thaliana AtGCS and AtGS. Annu Microbiol 64:961–967
Liu D, An Z, Mao Z, Ma L, Lu Z (2015a) Enhanced Heavy Metal Tolerance and Accumulation by Transgenic Sugar Beets Expressing Streptococcus thermophilus StGCS-GS in the Presence of Cd, Zn and Cu Alone or in Combination. Plos One 10:e0128824. https://doi.org/10.1371/journal.pone.0128824
Liu J, Shi X, Qian M, Zheng L, Lian C, Xia Y, Shen Z (2015b) Copper-induced hydrogen peroxide upregulation of a metallothionein gene, OsMT2c, from Oryza sativa L. confers copper tolerance in Arabidopsis thaliana. J Hazard Mater 294:99–108. https://doi.org/10.1016/j.jhazmat.2015.03.060
Liu P, Cai W-J, Yu L, Yuan B-F, Feng Y-Q (2015c) Determination of phytochelatins in rice by stable isotope labeling coupled with liquid chromatography-mass spectrometry. J Agric Food Chem 63:5935–5942. https://doi.org/10.1021/acs.jafc.5b01797
Liu LW, Li W, Song WP, Guo MX (2018) Remediation techniques for heavy metal-contaminated soils: principles and applicability. Sci Total Environ 633:206–219
Loix C, Huybrechts M, Vangronsveld J, Gielen M, Keunen E, Cuypers A (2018) Reciprocal interactions between cadmium-induced cell wall responses and oxidative stress in plants (vol 8, 1897, 2017). Front Plant Sci. https://doi.org/10.3389/fpls.2018.00391
Lombi E, Tearall KL, Howarth JR, Zhao F-J, Hawkesford MJ, McGrath SP (2002) Influence of iron status on cadmium and zinc uptake by different ecotypes of the hyperaccumulator Thlaspi caerulescens. Plant Physiol 128:1359–1367. https://doi.org/10.1104/pp.010731
Lu H, Zhang Y, Liu B, Liu J, Ye J, Yan C (2011) Rhizodegradation gradients of phenanthrene and pyrene in sediment of mangrove (Kandelia candel (L.) Druce). J Hazard Mater 196:263–269. https://doi.org/10.1016/j.jhazmat.2011.09.031
Lucero ME, Mueller W, Hubstenberger J, Phillips GC, O’Connell MA (1999) Tolerance to nitrogenous explosives and metabolism of TNT by cell suspensions of Datura innoxia. In Vitro Cell Dev Biol Plant 35:480–486. https://doi.org/10.1007/s11627-999-0072-3
Maestri E, Marmiroli M, Visioli G, Marmiroli N (2010) Metal tolerance and hyperaccumulation: costs and trade-offs between traits and environment. Environ Exp Bot 68:1–13. https://doi.org/10.1016/j.envexpbot.2009.10.011
Magalhães MOL, Amaral Sobrinho NMBd, Santos FSd, Mazur N (2011) Potencial de duas espécies de eucalipto na fitoestabilização de solo contaminado com zinco. Rev Ciênc Agron 42:805–812
Makhzoum AB, Sharma P, Bernards MA, Trémouillaux-Guiller J (2013) Hairy roots: an ideal platform for transgenic plant production and other promising applications. In: Gang DR (ed) Phytochemicals, plant growth, and the environment. Springer, New York, pp 95–142. https://doi.org/10.1007/978-1-4614-4066-6_6
Mali P et al (2013) RNA-guided human genome engineering via Cas9. Science (New York, NY) 339:823–826. https://doi.org/10.1126/science.1232033
Malik B, Pirzadah TB, Tahir I, Dar TuH, Rehman RU (2015) Chapter 6-Recent trends and approaches in phytoremediation. In: Hakeem KR, Sabir M, Öztürk M, Mermut AR (eds) Soil remediation and plants. Academic Press, San Diego, pp 131–146. https://doi.org/10.1016/B978-0-12-799937-1.00006-1
Manzoor M et al (2019) Metal tolerant bacteria enhanced phytoextraction of lead by two accumulator ornamental species. Chemosphere 227:561–569. https://doi.org/10.1016/j.chemosphere.2019.04.093
Maqbool F et al (2012) Rhizodegradation of petroleum hydrocarbons by Sesbania cannabina in bioaugmented soil with free and immobilized consortium. J Hazard Mater 237–238:262–269. https://doi.org/10.1016/j.jhazmat.2012.08.038
Mari S, Lebrun M (2006) Metal immobilization: where and how? In: Tamas MJ, Martinoia E (eds) Molecular biology of metal homeostasis and detoxification: from microbes to man. Springer, Berlin, pp 273–298. https://doi.org/10.1007/4735_103
Marois E, Van den Ackerveken G, Bonas U (2002) The xanthomonas type III effector protein AvrBs3 modulates plant gene expression and induces cell hypertrophy in the susceptible host. Mol Plant Microbe Interact MPMI 15:637–646. https://doi.org/10.1094/mpmi.2002.15.7.637
Márquez-Escobar VA, González-Ortega O, Rosales-Mendoza S (2018) Plant transformation strategies. In: MacDonald J (ed) Prospects of plant-based vaccines in veterinary medicine. Springer, Cham, pp 23–42. https://doi.org/10.1007/978-3-319-90137-4_2
Maser P et al (2001) Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiol 126:1646–1667
Mendel R, Hänsch R (2017) Gene transfer to higher plants. In: Singh RP, Singh US (eds) Molecular methods in plant pathology. CRC Press, Boca Raton, pp 189–202. https://doi.org/10.1201/9780203746523-14
Meyer C-L et al (2015) Intraspecific variability of cadmium tolerance and accumulation, and cadmium-induced cell wall modifications in the metal hyperaccumulator Arabidopsis halleri. J Exp Bot 66:3215–3227. https://doi.org/10.1093/jxb/erv144
Mishra S, Dubey RS (2006) Heavy metal uptake and detoxification mechanisms in plants. Int J Agric Res 1:122–141. https://doi.org/10.3923/ijar.2006.122.141
Mishra S, Srivastava S, Tripathi RD, Govindarajan R, Kuriakose SV, Prasad MNV (2006) Phytochelatin synthesis and response of antioxidants during cadmium stress in Bacopa monnieri L. Plant Physiol Biochem 44:25–37. https://doi.org/10.1016/j.plaphy.2006.01.007
Misra S, Gedamu L (1989) Heavy metal tolerant transgenic Brassica napus L and Nicotiana tabacum L plants. Theor Appl Genet 78:161–168. https://doi.org/10.1007/bf00288793
Moon JY, Belloeil C, Ianna ML, Shin R (2019) Arabidopsis CNGC family members contribute to heavy metal ion uptake in plants. Int J Mol Sci 20:413
Mosoarca G, Vancea C, Popa S, Boran S (2018) Adsorption, bioaccumulation and kinetics parameters of the phytoremediation of cobalt from wastewater using Elodea canadensis. Bull Environ Contam Toxicol 100:733–739. https://doi.org/10.1007/s00128-018-2327-3
Mudgal V, Madaan N, Mudgal A (2010) Heavy metals in plants: phytoremediation: plants used to remediate heavy metal pollution. Agric Biol J N Am 1:40–46
Muthusaravanan S et al (2018) Phytoremediation of heavy metals: mechanisms, methods and enhancements. Environ Chem Lett 16:1339–1359. https://doi.org/10.1007/s10311-018-0762-3
Myśliwa-Kurdziel B, Prasad MNV, Strzałtka K (2004) Photosynthesis in heavy metal stressed plants. In: Prasad MNV (ed) Heavy metal stress in plants: from biomolecules to ecosystems. Springer, Berlin, pp 146–181. https://doi.org/10.1007/978-3-662-07743-6_6
Nahar N, Rahman A, Nawani NN, Ghosh S, Mandal A (2017) Phytoremediation of arsenic from the contaminated soil using transgenic tobacco plants expressing ACR2 gene of Arabidopsis thaliana. J Plant Physiol 218:121–126
Nanasato Y et al (2016) Biodegradation of gamma-hexachlorocyclohexane by transgenic hairy root cultures of Cucurbita moschata that accumulate recombinant bacterial LinA. Plant Cell Rep 35:1963–1974. https://doi.org/10.1007/s00299-016-2011-1
Nedelkoska TV, Doran PM (2000) Hyperaccumulation of cadmium by hairy roots of Thlaspi caerulescens. Biotechnol Bioeng 67:607–615
Nehnevajova E, Ramireddy E, Stolz A, Gerdemann-Knörck M, Novák O, Strnad M, Schmülling T (2019) Root enhancement in cytokinin-deficient oilseed rape causes leaf mineral enrichment, increases the chlorophyll concentration under nutrient limitation and enhances the phytoremediation capacity. BMC Plant Biol 19:83. https://doi.org/10.1186/s12870-019-1657-6
Nguemte PM, Wafo GVD, Djocgoue PF, Noumsi IMK, Ngnien AW (2018) Potentialities of six plant species on phytoremediation attempts of fuel oil-contaminated soils. Water Air Soil Pollut 229:18. https://doi.org/10.1007/s11270-018-3738-9
Ning Y, Liu N, Song Y, Luo J, Li T (2019) Enhancement of phytoextraction of Pb by compounded activation agent derived from fruit residue. Int J Phytoremediat. https://doi.org/10.1080/15226514.2019.1633266
Olson PE, Fletcher JS (2000) Ecological recovery of vegetation at a former industrial sludge basin and its implications to phytoremediation. Environ Sci Pollut Res 7:195–204. https://doi.org/10.1007/bf02987348
Osma E, Ozyigit II, Leblebici Z, Demir G, Serin M (2012) Determination of heavy metal concentrations in tomato (Lycopersicon esculentum Miller) grown in different station types. Rom Biotechnol Lett 17:6962–6974
Ozturk A, Yarci C, Ozyigit II (2017) Assessment of heavy metal pollution in Istanbul using plant (Celtis australis L.) and soil assays. Biotechnol Biotechnol Equip 31:948–954. https://doi.org/10.1080/13102818.2017.1353922
Ozyigit II (2012) Agrobacterium tumefaciens and its use in plant biotechnology. In: Ashraf M, Öztürk M, Ahmad MSA, Aksoy A (eds) Crop production for agricultural improvement. Springer, Dordrecht, pp 317–361. https://doi.org/10.1007/978-94-007-4116-4_12
Ozyigit II (2020) Gene transfer to plants by electroporation: methods and applications. Mol Biol Rep 47:3195–3210. https://doi.org/10.1007/s11033-020-05343-4
Ozyigit II, Dogan I (2015) Chapter 9-Plant–microbe interactions in phytoremediation. In: Hakeem KR, Sabir M, Öztürk M, Mermut AR (eds) Soil remediation and plants. Academic Press, San Diego, pp 255–285. https://doi.org/10.1016/B978-0-12-799937-1.00009-7
Ozyigit II, Dogan I, Artam Tarhan E (2013) Agrobacterium rhizogenes-mediated transformation and its biotechnological applications in crops. In: Hakeem KR, Ahmad P, Ozturk M (eds) Crop improvement: new approaches and modern techniques. Springer, Boston, pp 1–48. https://doi.org/10.1007/978-1-4614-7028-1_1
Ozyigit II, Dogan I, Igdelioglu S, Filiz E, Karadeniz S, Uzunova Z (2016) Screening of damage induced by lead (Pb) in rye (Secale cereale L.)—a genetic and physiological approach. Biotechnol Biotechnol Equip 30:489–496. https://doi.org/10.1080/13102818.2016.1151378
Ozyigit II, Uyanik OL, Sahin NR, Yalcin IE, Demir G (2017) Monitoring the pollution level in Istanbul coast of the Sea of Marmara using algal species Ulva lactuca L. Pol J Environ Stud 26:773–778. https://doi.org/10.15244/pjoes/66177
Ozyigit II, Yalcin B, Turan S, Saracoglu IA, Karadeniz S, Yalcin IE, Demir G (2018) Investigation of heavy metal level and mineral nutrient status in widely used medicinal plants’ leaves in Turkey: insights into health implications. Biol Trace Elem Res 182:387–406. https://doi.org/10.1007/s12011-017-1070-7
Pabo CO, Peisach E, Grant RA (2001) Design and selection of novel Cys2His2 zinc finger proteins. Annu Rev Biochem 70:313–340. https://doi.org/10.1146/annurev.biochem.70.1.313
Pal R, Rai JP (2010) Phytochelatins: peptides involved in heavy metal detoxification. Appl Biochem Biotechnol 160:945–963. https://doi.org/10.1007/s12010-009-8565-4
Pala Z, Shukla V, Alok A, Kudale S, Desai N (2016) Enhanced production of an anti-malarial compound artesunate by hairy root cultures and phytochemical analysis of Artemisia pallens Wall. 3 Biotech 6:182. https://doi.org/10.1007/s13205-016-0496-5
Pandey VC, Pandey DN, Singh N (2015) Sustainable phytoremediation based on naturally colonizing and economically valuable plants. J Clean Prod 86:37–39. https://doi.org/10.1016/j.jclepro.2014.08.030
Pandey AK, Gautam A, Dubey RS (2019) Transport and detoxification of metalloids in plants in relation to plant-metalloid tolerance Plant. Gene 17:100171. https://doi.org/10.1016/j.plgene.2019.100171
Pandotra P et al (2018) Plant-bacterial partnership: a major pollutants remediation approach. In: Oves M, Zain Khan M, Ismail IMI (eds) Modern age environmental problems and their remediation. Springer, Cham, pp 169–200. https://doi.org/10.1007/978-3-319-64501-8_10
Papoyan A, Kochian LV (2004) Identification of Thlaspi caerulescens genes that may be involved in heavy metal hyperaccumulation and tolerance. Characterization of a novel heavy metal transporting ATPase. Plant Physiol 136:3814–3823
Peer WA, Baxter IR, Richards EL, Freeman JL, Murphy AS (2006) Phytoremediation and hyperaccumulator plants. In: Tamas MJ, Martinoia E (eds) Molecular biology of metal homeostasis and detoxification: from microbes to man. Springer, Berlin, pp 299–340. https://doi.org/10.1007/4735_100
Perfus-Barbeoch L, Leonhardt N, Vavasseur A, Forestier C (2002) Heavy metal toxicity: cadmium permeates through calcium channels and disturbs the plant water status. Plant J Cell Mol Biol 32:539–548
Pilon-Smits E (2005) Phytoremediation. Annu Rev Plant Biol 56:15–39. https://doi.org/10.1146/annurev.arplant.56.032604.144214
Pilon-Smits EAH, de Souza MP, Hong G, Amini A, Bravo RC, Payabyab ST, Terry N (1999) Selenium volatilization and accumulation by twenty aquatic plant species. J Environ Qual 28:1011–1018. https://doi.org/10.2134/jeq1999.00472425002800030035x
Pitzschke A, Hirt H (2010) New insights into an old story: agrobacterium-induced tumour formation in plants by plant transformation. EMBO J 29:1021–1032. https://doi.org/10.1038/emboj.2010.8
Pivetz B (2001) Phytoremediation of contaminated soil and ground water at hazardous waste sites. U.S. Environmental Protection Agency, Washington, DC
Pratas J, Favas PJ, Paulo C, Rodrigues N, Prasad MN (2012) Uranium accumulation by aquatic plants from uranium-contaminated water in Central Portugal. Int J Phytoremediat 14:221–234. https://doi.org/10.1080/15226514.2011.587849
Qiu W, Song X, Han X, Liu M, Qiao G, Zhuo R (2018) Overexpression of sedum alfredii cinnamyl alcohol dehydrogenase increases the tolerance and accumulation of cadmium in arabidopsis. Environ Exp Bot 155:566–577. https://doi.org/10.1016/j.envexpbot.2018.08.003
Quig D (1998) Cysteine metabolism and metal toxicity. Altern Med Rev J Clin Ther 3:262–270
Rai PK (2019) Heavy metals/metalloids remediation from wastewater using free floating macrophytes of a natural wetland. Environ Technol Innov 15:100393. https://doi.org/10.1016/j.eti.2019.100393
Raina M et al (2018) Genetic engineering and environmental risk. In: Oves M, Zain Khan M, Ismail IMI (eds) Modern age environmental problems and their remediation. Springer, Cham, pp 69–82. https://doi.org/10.1007/978-3-319-64501-8_4
Raldugina GN et al (2018) Expression of rice OsMyb4 transcription factor improves tolerance to copper or zinc in canola plants. Biol Plant 62:511–520
Ramasahayam S, Jaligama S, Atwa SM, Salley JT, Thongdy M, Blaylock BL, Meyer SA (2017) Megakaryocyte expansion and macrophage infiltration in bone marrow of rats subchronically treated with MNX, N-nitroso environmental degradation product of munitions compound RDX (hexahydro-1,3,5-trinitro-1,3,5-triazine). J Appl Toxicol 37:913–921. https://doi.org/10.1002/jat.3439
Raskin I, Smith RD, Salt DE (1997) Phytoremediation of metals: using plants to remove pollutants from the environment. Curr Opin Biotechnol 8:221–226. https://doi.org/10.1016/S0958-1669(97)80106-1
Rauser WE (1995) Phytochelatins and related peptides (structure, biosynthesis, and function). Plant Physiol 109:1141–1149. https://doi.org/10.1104/pp.109.4.1141
Rea PA, Vatamaniuk OK, Rigden DJ (2004) Weeds, worms, and more. Papain’s long-lost cousin, phytochelatin synthase. Plant Physiol 136:2463–2474. https://doi.org/10.1104/pp.104.048579
Reddy CV, Reddy IN, Akkinepally B, Harish VVN, Reddy KR, Jaesool S (2019) Mn-doped ZrO2 nanoparticles prepared by a template-free method for electrochemical energy storage and abatement of dye degradation. Ceram Int 45:15298–15306. https://doi.org/10.1016/j.ceramint.2019.05.020
Reddy CV, Reddy IN, Harish VVN, Reddy KR, Shetti NP, Shim J, Aminabhavi TM (2020) Efficient removal of toxic organic dyes and photoelectrochemical properties of iron-doped zirconia nanoparticles. Chemosphere 239:124766. https://doi.org/10.1016/j.chemosphere.2019.124766
Robinson NJ, Tommey AM, Kuske C, Jackson PJ (1993) Plant metallothioneins. Biochem J 295:1–10
Roy SK et al (2017) Proteome characterization of copper stress responses in the roots of sorghum. Biometals 30:765–785. https://doi.org/10.1007/s10534-017-0045-7
Rugh CL, Wilde HD, Stack NM, Thompson DM, Summers AO, Meagher RB (1996) Mercuric ion reduction and resistance in transgenic Arabidopsis thaliana plants expressing a modified bacterial merA gene. Proc Natl Acad Sci USA 93:3182–3187. https://doi.org/10.1073/pnas.93.8.3182
Rui HY, Chen C, Zhang XX, Shen ZG, Zhang FQ (2016) Cd-induced oxidative stress and lignification in the roots of two Vicia sativa L. varieties with different Cd tolerances. J Hazard Mater 301:304–313
Ruppert L, Lin ZQ, Dixon RP, Johnson KA (2013) Assessment of solid phase microfiber extraction fibers for the monitoring of volatile organoarsinicals emitted from a plant-soil system. J Hazard Mater 262:1230–1236. https://doi.org/10.1016/j.jhazmat.2012.06.046
Saad RB, Hsouna AB, Saibi W, Hamed KB, Brini F, Ghneim-Herrera T (2018) A stress-associated protein, LmSAP, from the halophyte Lobularia maritima provides tolerance to heavy metals in tobacco through increased ROS scavenging and metal detoxification processes. J Plant Physiol 231:234–243. https://doi.org/10.1016/j.jplph.2018.09.019
Sabir M, Waraich EA, Hakeem KR, Öztürk M, Ahmad HR, Shahid M (2015) Chapter 4-Phytoremediation: mechanisms and adaptations. In: Hakeem KR, Sabir M, Öztürk M, Mermut AR (eds) Soil remediation and plants. Academic Press, San Diego, pp 85–105. https://doi.org/10.1016/B978-0-12-799937-1.00004-8
Saleh HM (2012) Water hyacinth for phytoremediation of radioactive waste simulate contaminated with cesium and cobalt radionuclides. Nucl Eng Des 242:425–432. https://doi.org/10.1016/j.nucengdes.2011.10.023
Saleh HM, Moussa HR, Mahmoud HH, El-Saied FA, Dawoud M, Abdel Wahed RS (2020) Potential of the submerged plant Myriophyllum spicatum for treatment of aquatic environments contaminated with stable or radioactive cobalt and cesium. Prog Nucl Energy 118:103147. https://doi.org/10.1016/j.pnucene.2019.103147
Salido AL, Hasty KL, Lim JM, Butcher DJ (2003) Phytoremediation of arsenic and lead in contaminated soil using Chinese brake ferns (Pteris vittata) and Indian mustard (Brassica juncea). Int J Phytoremediat 5:89–103. https://doi.org/10.1080/713610173
San Miguel A, Ravanel P, Raveton M (2013) A comparative study on the uptake and translocation of organochlorines by Phragmites australis. J Hazard Mater 244:60–69. https://doi.org/10.1016/j.jhazmat.2012.11.025
Sander JD, Joung JK (2014) CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol 32:347–355. https://doi.org/10.1038/nbt.2842
Sangeeta M, Maiti SK (2010) Phytoremediation of metal enriched mine waste: a review. Am Eurasian J Agric Environ Sci 9:560–575
Sanita di Toppi L, Gabbrielli R (1999) Response to cadmium in higher plants. Environ Exp Bot 41:105–130
Sardesai N, Subramanyam S (2018) Agrobacterium: a genome-editing tool-delivery system. Curr Top Microbiol Immunol 418:463–488. https://doi.org/10.1007/82_2018_101
Sarwar N, Saifullah, Malhi SS, Zia MH, Naeem A, Bibi S, Farid G (2010) Role of mineral nutrition in minimizing cadmium accumulation by plants. J Sci Food Agric 90:925–937
Sarwar N et al (2017) Phytoremediation strategies for soils contaminated with heavy metals: modifications and future perspectives. Chemosphere 171:710–721. https://doi.org/10.1016/j.chemosphere.2016.12.116
Schaaf G, Honsbein A, Meda AR, Kirchner S, Wipf D, von Wiren N (2006) AtIREG2 encodes a tonoplast transport protein involved in iron-dependent nickel detoxification in Arabidopsis thaliana roots. J Biol Chem 281:25532–25540. https://doi.org/10.1074/jbc.m601062200
Schiavon M, Pittarello M, Pilon-Smits EAH, Wirtz M, Hell R, Malagoli M (2012) Selenate and molybdate alter sulfate transport and assimilation in Brassica juncea L. Czern.: implications for phytoremediation. Environ Exp Bot 75:41–51
Schnoor JL, Licht LA, McCutcheon SC, Wolfe NL, Carreira LH (1995) Phytoremediation of organic and nutrient contaminants. Environ Sci Technol 29:318a–323a. https://doi.org/10.1021/es00007a747
Segal DJ (2002) The use of zinc finger peptides to study the role of specific factor binding sites in the chromatin environment. Methods (San Diego, Calif) 26:76–83. https://doi.org/10.1016/s1046-2023(02)00009-9
Seth CS (2012) A review on mechanisms of plant tolerance and role of transgenic plants in environmental clean-up. Bot Rev 78:32–62. https://doi.org/10.1007/s12229-011-9092-x
Shah K, Nongkynrih JM (2007) Metal hyperaccumulation and bioremediation. Biol Plant 51:618–634. https://doi.org/10.1007/s10535-007-0134-5
Shah K, Pathak L (2019) Chapter 15-Transgenic energy plants for phytoremediation of toxic metals and metalloids. In: Prasad MNV (ed) Transgenic plant technology for remediation of toxic metals and metalloids. Academic Press, New York, pp 319–340. https://doi.org/10.1016/B978-0-12-814389-6.00015-8
Shahid M, Pourrut B, Dumat C, Nadeem M, Aslam M, Pinelli E (2014) Heavy-metal-induced reactive oxygen species: phytotoxicity and physicochemical changes in plants. Rev Environ Contam Toxicol 232:1–44. https://doi.org/10.1007/978-3-319-06746-9_1
Shan Q, Wang Y, Li J, Gao C (2014) Genome editing in rice and wheat using the CRISPR/Cas system. Nat Protoc 9:2395–2410. https://doi.org/10.1038/nprot.2014.157
Sharma P, Dubey RS (2005) Lead toxicity in plants. Braz J Plant Physiol 17:35–52
Sharma DC, Sharma CP (1996) Chromium uptake and toxicity effects on growth and metabolic activities in wheat, Triticum aestivum L. cv. UP 2003. Indian J Exp Biol 34:689–691
Sharma SS, Dietz K-J, Mimura T (2016) Vacuolar compartmentalization as indispensable component of heavy metal detoxification in plants. Plant Cell Environ 39:1112–1126. https://doi.org/10.1111/pce.12706
Shukla D, Trivedi PK, Nath P, Tuteja N (2016) Metallothioneins and phytochelatins: role and perspectives in heavy metal(loid)s stress tolerance in crop plants. In: Tuteja N, Gill SS (eds) Abiotic stress response in plants. Wiley, New York. https://doi.org/10.1002/9783527694570.ch12
Singh KB, Foley RC, Onate-Sanchez L (2002) Transcription factors in plant defense and stress responses. Curr Opin Plant Biol 5:430–436
Singh S, Parihar P, Singh R, Singh VP, Prasad SM (2016) Heavy metal tolerance in plants: role of transcriptomics, proteomics, metabolomics, and ionomics. Front Plant Sci. https://doi.org/10.3389/fpls.2015.01143
Singh V, Pandey B, Suthar S (2019) Phytotoxicity and degradation of antibiotic ofloxacin in duckweed (Spirodela polyrhiza) system. Ecotoxicol Environ Saf 179:88–95. https://doi.org/10.1016/j.ecoenv.2019.04.018
Slater A, Scott NW, Fowler MR (2008) Plant biotechnology: the genetic manipulation of plants. Ann Bot 94:646
Solanki R, Dhankhar R (2011) Biochemical changes and adaptive strategies of plants under heavy metal stress. Biologia 66:195–204. https://doi.org/10.2478/s11756-011-0005-6
Soleimani M, Akbar S, Hajabbasi MA (2011) Enhancing phytoremediation efficiency in response to environmental pollution stress. In: Hemanth KN, Kambiranda V, Kambiranda D (eds) Plants and environment. IntechOpen, London. https://doi.org/10.5772/22267
Sorrentino MC, Capozzi F, Amitrano C, Giordano S, Arena C, Spagnuolo V (2018) Performance of three cardoon cultivars in an industrial heavy metal-contaminated soil: effects on morphology, cytology and photosynthesis. J Hazard Mater 351:131–137
Souza TDd, Borges AC, Braga AF, Veloso RW, Teixeira de Matos A (2019) Phytoremediation of arsenic-contaminated water by Lemna valdiviana: an optimization study. Chemosphere 234:402–408. https://doi.org/10.1016/j.chemosphere.2019.06.004
Subramanyam K, Subramanyam K, Sailaja KV, Srinivasulu M, Lakshmidevi K (2011) Highly efficient Agrobacterium-mediated transformation of banana cv. Rasthali (AAB) via sonication and vacuum infiltration. Plant Cell Rep 30:425–436. https://doi.org/10.1007/s00299-010-0996-4
Subramoni S, Nathoo N, Klimov E, Yuan Z-C (2014) Agrobacterium tumefaciens responses to plant-derived signaling molecules Front. Plant Sci 5:322–322. https://doi.org/10.3389/fpls.2014.00322
Sun J, Pan L, Tsang DCW, Zhan Y, Zhu L, Li X (2018a) Organic contamination and remediation in the agricultural soils of China: a critical review. Sci Total Environ 615:724–740. https://doi.org/10.1016/j.scitotenv.2017.09.271
Sun L et al (2018b) Overexpression of PtABCC1 contributes to mercury tolerance and accumulation in Arabidopsis and poplar. Biochem Biophys Res Commun 497:997–1002. https://doi.org/10.1016/j.bbrc.2018.02.133
Surriya O, Sarah Saleem S, Waqar K, Gul Kazi A (2015) Chapter 1-Phytoremediation of soils: prospects and challenges. In: Hakeem KR, Sabir M, Öztürk M, Mermut AR (eds) Soil remediation and plants. Academic Press, San Diego, pp 1–36. https://doi.org/10.1016/B978-0-12-799937-1.00001-2
Susarla S, Medina VF, McCutcheon SC (2002) Phytoremediation: an ecological solution to organic chemical contamination. Ecol Eng 18:647–658
Takahashi R, Bashir K, Ishimaru Y, Nishizawa NK, Nakanishi H (2012) The role of heavy-metal ATPases, HMAs, in zinc and cadmium transport in rice. Plant Signal Behav 7:1605–1607. https://doi.org/10.4161/psb.22454
Tavangar T, Karimi M, Rezakazemi M, Reddy KR, Aminabhavi TM (2020) Textile waste, dyes/inorganic salts separation of cerium oxide-loaded loose nanofiltration polyethersulfone membranes. Chem Eng J 385:123787. https://doi.org/10.1016/j.cej.2019.123787
Thakur R, Shankar J (2017) Strategies for gene expression in prokaryotic and eukaryotic system. In: Kalia VC, Saini AK (eds) Metabolic engineering for bioactive compounds: strategies and processes. Springer, Singapore, pp 223–247. https://doi.org/10.1007/978-981-10-5511-9_11
Thomas TJ, Kulkarni GD, Greenfield NJ, Shirahata A, Thomas T (1996) Structural specificity effects of trivalent polyamine analogues on the stabilization and conformational plasticity of triplex DNA. Biochem J 319(Pt 2):591–599. https://doi.org/10.1042/bj3190591
Tommasini R, Vogt E, Fromenteau M, Hörtensteiner S, Matile P, Amrhein N, Martinoia E (1998) An ABC-transporter of Arabidopsis thaliana has both glutathione-conjugate and chlorophyll catabolite transport activity. Plant J 13:773–780. https://doi.org/10.1046/j.1365-313X.1998.00076.x
Tue NM et al (2014) Dioxin-related compounds in breast milk of women from Vietnamese e-waste recycling sites: levels, toxic equivalents and relevance of non-dietary exposure. Ecotoxicol Environ Saf 106:220–225
Uhlik O, Wald J, Strejcek M, Musilova L, Ridl J et al (2012) Identification of bacteria utilizing biphenyl, benzoate, and naphthalene in long-term contaminated soil. Plos One 7:e40653. https://doi.org/10.1371/journal.pone.0040653
Vamerali T, Bandiera M, Mosca G (2010) Field crops for phytoremediation of metal-contaminated land. A review. Environ Chem Lett 8:1–17. https://doi.org/10.1007/s10311-009-0268-0
Van Aken B (2008) Transgenic plants for phytoremediation: helping nature to clean up environmental pollution. Trends Biotechnol 26:225–227. https://doi.org/10.1016/j.tibtech.2008.02.001
Van Assche F, Clijsters H (1990) Effects of metals on enzyme activity in plants. Plant Cell Environ 13:195–206. https://doi.org/10.1111/j.1365-3040.1990.tb01304.x
Van der Zaal BJ, Neuteboom LW, Pinas JE, Chardonnens AN, Schat H, Verkleij JAC, Hooykaas PJJ (1999) Overexpression of a novel Arabidopsis gene related to putative zinc-transporter genes from animals can lead to enhanced zinc resistance and accumulation. Plant Physiol 119:1047–1056. https://doi.org/10.1104/pp.119.3.1047
Van Huysen T, Abdel-Ghany S, Hale KL, LeDuc D, Terry N, Pilon-Smits EA (2003) Overexpression of cystathionine-gamma-synthase enhances selenium volatilization in Brassica juncea. Planta 218:71–78. https://doi.org/10.1007/s00425-003-1070-z
Vatamaniuk OK, Bucher EA, Ward JT, Rea PA (2001) A new pathway for heavy metal detoxification in animals: phytochelatin synthase is required for Cd tolerance in Caenorhabditis elegans. J Biol Chem 276:20817–20820. https://doi.org/10.1074/jbc.C100152200
Vatamaniuk OK, Mari S, Lang A, Chalasani S, Demkiv LO, Rea PA (2004) Phytochelatin synthase, a dipeptidyltransferase that undergoes multisite acylation with γ-glutamylcysteine during catalysis: stoichiometric and site-directed mutagenic analysis of Arabidopsis thaliana PCS1-catalyzed phytochelatin synthesis. J Biol Chem 279:22449–22460. https://doi.org/10.1074/jbc.m313142200
Verkleij JAC, Sneller FEC, Schat H (2003) Metallothioneins and phytochelatins: ecophysiological aspects. In: Abrol YP, Ahmad A (eds) Sulphur in plants. Springer, Dordrecht, pp 163–176. https://doi.org/10.1007/978-94-017-0289-8_9
Vetterlein D, Szegedi K, Ackermann J, Mattusch J, Neue HU, Tanneberg H, Jahn R (2007) Competitive mobilization of phosphate and arsenate associated with goethite by root activity. J Environ Qual 36:1811–1820
Vijgen J et al (2011) Hexachlorocyclohexane (HCH) as new stockholm convention POPs-a global perspective on the management of Lindane and its waste isomers. Environ Sci Pollut Res 18:152–162
Viktorova J et al (2017) Native phytoremediation potential of Urtica dioica for removal of PCBs and heavy metals can be improved by genetic manipulations using constitutive CaMV 35S promoter (vol 11, e0167927, 2016). Plos One 12:e0187053
Vladimirov IA, Matveeva TV, Lutova LA (2015) Opine biosynthesis and catabolism genes of Agrobacterium tumefaciens and Agrobacterium rhizogenes. Russ J Genet 51:121–129. https://doi.org/10.1134/s1022795415020167
Wan X, Lei M, Chen T (2016) Cost–benefit calculation of phytoremediation technology for heavy-metal-contaminated soil. Sci Total Environ 563:796–802. https://doi.org/10.1016/j.scitotenv.2015.12.080
Wang FZ, Chen MX, Yu LJ, Xie LJ, Yuan LB, Qi H, Xiao M, Guo WX, Chen Z, Yi KK, Zhang JH, Qiu RL, Shu WS, Xiao S, Chen QF (2017) Osarm1, an r2r3 myb transcription factor, is involved in regulation of the response to arsenic stress in rice. Front Plant Sci 8:16. https://doi.org/10.3389/fpls.2017.01868
Wang XT, Zhi JK, Liu XR, Zhang H, Liu HB, Xu JC (2018) Transgenic tobacco plants expressing a P1B-ATPase gene from Populus tomentosa Carr. (PtoHMA5) demonstrate improved cadmium transport. Int J Biol Macromol 113:655–661
Wang W-W, Ke Cheng L, Hao JW, Guan X, Tian X-J (2019) Phytoextraction of initial cutting of Salix matsudana for Cd and Cu. Int J Phytoremediat 21:84–91. https://doi.org/10.1080/15226514.2016.1183574
Wei X, Lyu S, Yu Y, Wang Z, Liu H, Pan D, Chen J (2017) Phylloremediation of air pollutants: exploiting the potential of plant leaves and leaf-associated microbes. Front Plant Sci. https://doi.org/10.3389/fpls.2017.01318
White PM Jr, Wolf DC, Thoma GJ, Reynolds CM (2003) Influence of organic and inorganic soil amendments on plant growth in crude oil-contaminated soil. Int J Phytoremediat 5:381–397. https://doi.org/10.1080/15226510309359044
Wiedenheft B, Sternberg SH, Doudna JA (2012) RNA-guided genetic silencing systems in bacteria and archaea. Nature 482:331–338. https://doi.org/10.1038/nature10886
Wiessner A, Kappelmeyer U, Kaestner M, Schultze-Nobre L, Kuschk P (2013) Response of ammonium removal to growth and transpiration of Juncus effusus during the treatment of artificial sewage in laboratory-scale wetlands. Water Res 47:4265–4273. https://doi.org/10.1016/j.watres.2013.04.045
Williams LE, Mills RF (2005) P(1B)-ATPases—an ancient family of transition metal pumps with diverse functions in plants. Trends Plant Sci 10:491–502. https://doi.org/10.1016/j.tplants.2005.08.008
Williams LE, Pittman JK, Hall JL (2000) Emerging mechanisms for heavy metal transport in plants. Biochem Biophys Acta 1465:104–126
Xia Y, Liu J, Wang Y, Zhang XX, Shen ZG, Hu ZB (2018) Ectopic expression of Vicia sativa Caffeoyl-CoA O-methyltransferase (VsCCoAOMT) increases the uptake and tolerance of cadmium in Arabidopsis. Environ Exp Bot 145:47–53
Yadav A, Raj A, Purchase D, Ferreira LFR, Saratale GD, Bharagava RN (2019) Phytotoxicity, cytotoxicity and genotoxicity evaluation of organic and inorganic pollutants rich tannery wastewater from a common effluent treatment plant (CETP) in Unnao district, India using Vigna radiata and Allium cepa. Chemosphere 224:324–332. https://doi.org/10.1016/j.chemosphere.2019.02.124
Yang B, Zhu W, Johnson LB, White FF (2000) The virulence factor AvrXa7 of Xanthomonas oryzae pv. oryzae is a type III secretion pathway-dependent nuclear-localized double-stranded DNA-binding protein. Proc Natl Acad Sci USA 97:9807–9812. https://doi.org/10.1073/pnas.170286897
Yang X, Feng Y, He Z, Stoffella PJ (2005a) Molecular mechanisms of heavy metal hyperaccumulation and phytoremediation. J Trace Elem Med Biol Organ Soc Miner Trace Elem (GMS) 18:339–353. https://doi.org/10.1016/j.jtemb.2005.02.007
Yang X, Feng Y, He ZL, Stoffella PJ (2005b) Molecular mechanisms of heavy metal hyperaccumulation and phytoremediation. J Trace Elem Med Biol 18:339–353. https://doi.org/10.1016/j.jtemb.2005.02.007
Yang WD, Yang Y, Ding ZL, Yang XE, Zhao FL, Zhu ZQ (2019a) Uptake and accumulation of cadmium in flooded versus non-flooded Salix genotypes: implications for phytoremediation. Ecol Eng 136:79–88. https://doi.org/10.1016/j.ecoleng.2019.06.001
Yang Y et al (2019b) Phytoextraction of Cd from a contaminated soil by tobacco and safe use of its metal-enriched biomass. J Hazard Mater 363:385–393. https://doi.org/10.1016/j.jhazmat.2018.09.093
Yifru DD, Nzengung VA (2008) Organic carbon biostimulates rapid rhizodegradation of perchlorate. Environ Toxicol Chem 27:2419–2426. https://doi.org/10.1897/08-008.1
Yin L, Wang S, Chen D, Deng X, Cao B, Qi L (2016) Silicon-moderated K-deficiency-induced leaf chlorosis by decreasing putrescine accumulation in sorghum. Ann Bot 118:305–315. https://doi.org/10.1093/aob/mcw111
Yuan H-M, Liu W-C, Jin Y, Lu Y-T (2013) Role of ROS and auxin in plant response to metal-mediated stress. Plant Signal Behav 8:e24671–e24671. https://doi.org/10.4161/psb.24671
Yuksek A (2018) İslam hukukunna göre helal gıda ve GDO’lu ürünler (Genetiği değiştirilmiş organizmalar)
Yusuf M, Fariduddin Q, Hayat S, Ahmad A (2011) Nickel: an overview of uptake, essentiality and toxicity in plants. Bull Environ Contam Toxicol 86:1–17
Zamani J, Hajabbasi MA, Mosaddeghi MR, Soleimani M, Shirvani M, Schulin R (2018) Experimentation on degradation of petroleum in contaminated soils in the root zone of maize (Zea mays L.) inoculated with Piriformospora indica. Soil Sediment Contam 27:13–30. https://doi.org/10.1080/15320383.2018.1422693
Zehra A et al (2020) Assessment of sunflower germplasm for phytoremediation of lead-polluted soil and production of seed oil and seed meal for human and animal consumption. J Environ Sci 87:24–38. https://doi.org/10.1016/j.jes.2019.05.031
Zenk MH (1996) Heavy metal detoxification in higher plants—a review. Gene 179:21–30
Zhan J, Zhang QP, Li TX, Yu HY, Zhang XZ, Huang HG (2019) Effects of NTA on Pb phytostabilization efficiency of Athyrium wardii (Hook.) grown in a Pb-contaminated soil. J Soil Sediment 19:3576–3584. https://doi.org/10.1007/s11368-019-02308-4
Zhang HM et al (1988) Transgenic rice plants produced by electroporation-mediated plasmid uptake into protoplasts. Plant Cell Rep 7:379–384. https://doi.org/10.1007/bf00269517
Zhang BY, Zheng JS, Sharp RG (2010) Phytoremediation in engineered wetlands: mechanisms and applications. Procedia Environ Sci 2:1315–1325. https://doi.org/10.1016/j.proenv.2010.10.142
Zhang G et al (2014) The CCoAOMT1 gene from jute (Corchorus capsularis L.) is involved in lignin biosynthesis in Arabidopsis thaliana. Gene 546:398–402
Zhang Y et al (2016) Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat Commun 7:12617. https://doi.org/10.1038/ncomms12617
Zhang H, Lv S, Xu H, Hou D, Li Y, Wang F (2017) H2O2 is involved in the metallothionein-mediated rice tolerance to copper and cadmium toxicity. Int J Mol Sci 18:2083. https://doi.org/10.3390/ijms18102083
Zhang X, Rui H, Zhang F, Hu Z, Xia Y, Shen Z (2018) Overexpression of a functional Vicia sativa PCS1 homolog increases cadmium tolerance and phytochelatins synthesis in Arabidopsis. Front Plant Sci. https://doi.org/10.3389/fpls.2018.00107
Zhao H, Wang L, Zhao F-J, Wu L, Liu A, Xu W (2019a) SpHMA1 is a chloroplast cadmium exporter protecting photochemical reactions in the Cd hyperaccumulator Sedum plumbizincicola. Plant Cell Environ 42:1112–1124. https://doi.org/10.1111/pce.13456
Zhao R, Yang T, Shi C, Zhou M, Chen G, Shi F (2019b) Effects of urban–rural atmospheric environment on heavy metal accumulation and resistance characteristics of Pinus tabulaeformis in Northern China. Bull Environ Contam Toxicol 102:432–438. https://doi.org/10.1007/s00128-019-02548-7
Zhou G, Xu Y, Li J, Yang L, Liu JY (2006) Molecular analyses of the metallothionein gene family in rice (Oryza sativa L.). J Biochem Mol Biol 39:595–606
Zhou ZS, Zeng HQ, Liu ZP, Yang ZM (2012) Genome-wide identification of Medicago truncatula microRNAs and their targets reveals their differential regulation by heavy metal. Plant Cell Environ 35:86–99. https://doi.org/10.1111/j.1365-3040.2011.02418.x
Zhou MX, Ghnaya T, Dailly H, Cui GL, Vanpee B, Han RM, Lutts S (2019) The cytokinin trans-zeatine riboside increased resistance to heavy metals in the halophyte plant species Kosteletzkya pentacarpos in the absence but not in the presence of NaCl. Chemosphere 233:954–965. https://doi.org/10.1016/j.chemosphere.2019.06.023
Zupan J, Muth TR, Draper O, Zambryski P (2000) The transfer of DNA from agrobacterium tumefaciens into plants: a feast of fundamental insights. Plant J Cell Mol Biol 23:11–28
Acknowledgements
Authors are grateful to Bio-Rad Laboratories for providing the original pictures of Helios® Gene Gun and Gene Pulser Xcell™ Electroporation systems, Prof. Dr. Şule Arı from Istanbul University, Assoc. Prof. Dr. Semra Hasançebi from Trakya University and Assoc. Prof. Dr. Ali Yüksek from Ondokuz Mayis University for sharing some pictures of their previous studies.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Ozyigit, I.I., Can, H. & Dogan, I. Phytoremediation using genetically engineered plants to remove metals: a review. Environ Chem Lett 19, 669–698 (2021). https://doi.org/10.1007/s10311-020-01095-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10311-020-01095-6