Abstract
Antimicrobial resistance is a major health issue induced by the overuse of antibiotics and disinfection reagents, e.g. chlorine. Resistant bacteria thus occur in water supply systems, and they transfer genes to other microbial populations, including pathogens. Treatment and inactivation of resistant bacteria are difficult in complex systems because the behaviour of resistant bacteria in such systems is poorly known, as most previous investigations are commonly performed in pure media. Therefore, we tested here the effect of 0.5 mg/mL chlorine and pipe materials made of polyvinyl chloride (PVC), copper and cement, on microbial populations in biofilms, during 5 days. Bacterial survival was monitored by viable counts, and resistant genes were analysed by quantitative polymerase chain reaction (qPCR). Results show that, in 56% of the cases, resistant bacteria became immediately enriched into biofilms due to chlorine exposure. Higher proportion of resistant bacteria were found in biofilms on PVC and copper pipes. Our findings imply that resistant microbial strains are very rapidly selected and that the pipe material has an influence on microbial selection.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Antimicrobial resistance has emerged globally as an environmental problem, but it can cross into being a food/water issue (Rusu et al. 2015; Daghrir and Drogui 2013). Water treatment plants and distribution systems use filtration and disinfection to control micro-organisms and maintain the potability of water (Morente et al. 2013). While generally effective, bacteria (including those resistant) invade water systems via line disruptions or compromised cisterns, which can introduce additional bacterial hazards. One cannot fully eradicate antimicrobial-resistant bacteria at normal treatment dosages; rather, much higher treatment dosages often become required. Unfortunately, higher concentrations produce odour and taste problems and are difficult to achieve and maintain in water. While disinfectants help attenuate bacteria, nonetheless, they can selectively cause robust bacteria to proliferate (e.g. Aeromonas; McBain et al. 2004) and enrich antimicrobial-resistant bacteria.
The emergence, enrichment and transfer of resistance traits among bacteria depend on the microbial conditions in water distribution system. For example, biofilm-forming bacteria facilitate the attachment of micro-organisms along the distribution system and create community structures with greater complexity in terms of bacterial species than those in finished water (Chu et al. 2003), enhancing the capability to retain and protect pathogens. Furthermore, biofilm structures, with increased cell densities, facilitate the horizontal gene transfer of resistance genes. Moreover, bacterial physiology and metabolic rates vary along biofilm depth and reflect the gradients of nutrients versus chemical toxicants; the consequential reduced growth rates impact biocide efficacy (Boe-Hansen et al. 2002). Protective layers of biofilm bacteria provide better tolerance to metal ions and other toxic substances and improve survivability. Ultimately, biofilms in distribution systems become a generator and repository of resistance traits.
Conditions within water systems impact micro-organisms and further contribute to the formation, composition and proliferation of biofilms, e.g. source water (Zeng et al. 2013), excretion of extracellular polymeric substances (Fish et al. 2015), physico-chemical conditions of water, hydraulic operations and flow rates (Douterelo et al. 2016) and composition of pipe materials (Nguyen et al. 2012).
Pipes comprise of various metals: galvanized iron, lead, copper, cast iron, steel (Nguyen et al. 2012); plastic materials: chlorinated polyvinyl chloride, polypropylene, polyethylene, polyvinyl chloride and other materials: cementitious composites, such as concrete (Niquette et al. 2000); and elasto-materials. Pipe materials and biofilms differentially affect each other; e.g. the presence of slime-forming bacteria facilitates metal tolerance, and biofilm bacteria produce organic acids, which contribute to the dissolution of metals and cement. Consequently, exposure to leached materials from pipes can enhance antimicrobial resistance development, as frequently observed in contaminated environments in nature (e.g. Rodgers et al. 2018; Abdu et al. 2017). Ultimately, disinfection strategies and surface exposures may effectively attenuate most bacteria in distribution networks (e.g. Dong et al. 2018; Li et al. 2017), but could the stressors selectively contribute to resistance problem?
Here, we aimed to determine whether disinfectants and pipe material differentially affect bacteria with antimicrobial resistance. We employed experimental microcosms to examine how factors selectively influenced bacterial populations in a municipal water supply system, which included the formation of biofilms. These microcosms represented miniaturized analogues of actual distribution systems, and the bacteria were harvested from tap water; we hypothesized that different pipe material and disinfectant residuals impact microbial communities to select resistant bacteria and biofilm formation.
Materials and methods
Microcosm set-up
Experimental design involved laboratory microcosms with autoclaved phosphate buffer saline in Millipore water (10 mM, pH 7) having polyvinyl chloride, copper and cement pipes (internal volume 102 cm3, surface area 240 cm2) versus presence/absence of free chlorine (0.5 mg/mL), and as such six independent experiments were conducted. Peristaltic pumps (MP model M312) provided continuous recirculation (2 mL/min). Before each experiment, the microcosms, pipes and tubing were disinfected twice by 500 mg/L of sodium hypochlorite for 2 h.
Bacterial strains
Bacillus, Paenibacillus and Micrococcus were isolated from drinking water, except Bacillus subtilis R2 (the National Collection of Type Cultures, NCTC 10400, UK). The presence of antibiotic-resistant genes (sul1 and sul2), mobile genetic elements (intI1) and chlorine susceptibility were determined as described previously (Khan et al. 2016a, b). The paired microcosm assays (chlorinated and chlorine free) were run with two populations of each genus having different levels of resistance: sul1 (Bacillus and Paenibacillus), sul2 and intI1 (Micrococcus).
Experimental operation
Overnight culture in 50 mL Luria–Bertani broth was centrifuged 3 × (3000 rpm, 10 min), washed with phosphate buffer saline, added (final concentration 1 × 105 cell/mL) to microcosms and recirculated for 5 days at 20 °C (± 2), with 1 mL harvested from each pipe on days 0, 3 and 5 for viable counts (Nutrient agar). Biofilm samples were taken on day 5 from 4 cm2 area using sterile swabs, and 50 mL water was filtered through 0.22-µm membrane filter (Millipore, UK) on days 0 and 5. Filters and swabs were stored at − 20 °C. An additional 30–50 mL was collected on days 3 and 5 to measure temperature, pH (Model S40 SevenMulti™, Mettler Toledo™), dissolved oxygen (DO200 meter, VWR), total organic carbon (Teledyne Tekmar Apollo 9000 Combustion TOC Analyzer) and free chlorine (N,N-diethyl-p-phenylenediamine colorimetric method, Hach).
DNA extraction and quantitative polymerase chain reaction
DNA was extracted using the ISOLATE II Genomic DNA kit (Bioline, UK) as per manufacturer’s instructions. Quantitative polymerase chain reaction (qPCR) was used to enumerate Bacillus (DeClerck et al. 2004), Paenibacillus (Pettersson et al. 1999) and Micrococcus species (Walcott and Gitaitis 2000). qPCR involves the amplification (copying) of specific DNA sequences in a manner with timing of fluorescent detection to count the number of genes in a sample. Strains were discerned with primers associated with their antibiotic resistance genes (sul1, sul2; Pei et al. 2006) or mobile genetic element (intI1; Luo et al. 2010); 16S-rRNA genes were quantified as a surrogate measure of ‘total bacteria’. Serially diluted DNA standards (108–102 genes/mL) and a control without DNA template were run with each qPCR assay.
Data analyses
From qPCR data, we calculated per cent resistance [resistance genes]/[total genus] and bacterial abundances on pipe surfaces and water. Fold increases in total bacteria and %resistance were calculated by comparing abundances between days 0 and 5. Abundances were log-transformed for presentation and statistical (e.g. normality) purposes. Data were analysed by Excel (Microsoft Office 2010) and Minitab version 17.
Results and discussion
Abundances of bacteria in biofilm
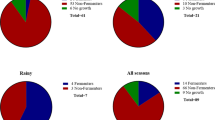
Microbial populations responded almost immediately by attaching to surfaces, with a greater percentage of those having resistant traits—suggesting that conditions in drinking water can select for bacterial contaminants with antimicrobial resistance. Surface swabs on day 5 for biofilm formation indicated that in two third of the cases bacteria adhered themselves onto surfaces (Fig. 1). Bacterial viability declines under stressful conditions, which resulted in differences in plate (viability) and genetic counts; however, the likely cause for declining gene counts in the water is the surface attachment of cells, which would escape detection in the water. One exception was Bacillus, where only 35% of total bacteria and 65% of the resistant strain were found in biofilms in the polyvinyl chloride microcosms. Further, only the resistant strains of Micrococcus and Paenibacillus were found on copper surfaces. Percentages of total and resistant populations were equivalent for other treatments.
Here, up to 106.5 cfu/cm2 was found on surfaces in 5 days, more than reported previously (i.e. 105 cfu/cm2 Frias et al. 2001). Bacteria showed different biofilm potentials: Paenibacillus > Bacillus > Micrococcus (Fig. 1) depending on pipe materials. Reported trend is as follows: polyvinyl chloride < copper < steel < iron < cement < plastic (e.g. polyethylene) < elastomeric surfaces (Yu et al. 2010), with some variation in adhesion potentials (Niquette et al. 2000). Conversely in this study, polyvinyl chloride and copper pipes allowed more bacterial attachment with chlorine. Biofilm formation was faster on plastic pipes than on copper pipes, due to copper toxicity (Yu et al. 2010); however, in this experiment, phosphate levels may have confounded effects of copper toxicity.
Waterborne resistant and total populations
Each pair of bacteria was added (totalling 2 × 105 cfu/mL, log 5.3) into the microcosms; heterotrophic plate counts determined viabilities (Table 1). In unchlorinated systems, viable cell counts declined by various orders of magnitude, except Micrococcus, which showed greater viability between days 3 and 5 in polyvinyl chloride and copper pipes. Chlorination compromised viabilities by 1–2 orders greater reduction than untreated systems (Table 1). No viable Micrococcus were recovered from any chlorinated microcosm on day 3, although Paenibacillus and Micrococcus showed some recovery by day 5 in copper and polyvinyl chloride pipes, respectively.
Both the susceptible and resistant strains were simultaneously challenged in the microcosms and monitored by quantitative PCR to differentiate strains. Their presence in water declined across all treatments (Table 2), except for Paenibacillus and Micrococcus spp. in the polyvinyl chloride and cement treatments. In 44.4% of the cases, the resistant strains also declined, but at a lesser rate—resulting in mixed populations skewed towards a greater proportion of resistant bacteria.
Viability counts (Table 1) and gene detection (non-viable) are not analogous. Genetic (resistance and 16S rRNA) traits indicate the presence of bacteria in the system, whereas plate counts indicate whether bacteria are capable of growing. Although not growing, their presence in the water (even with chlorine) could increase the prevalence of resistant genes if conditions change.
Low concentrations of disinfectants exert selective pressure on resistant populations, but another driving force, i.e. pipe materials, showed differential selection patterns in Micrococcus species (Table 1). Further, pipe materials impacted bacterial concentrations; e.g. polyvinyl chloride pipes had greatest abundances of bacteria, and copper, the least, which may be attributable to its antimicrobial effects. Depending on the bacteria, resistant populations also proliferated in circulating water, e.g. Micrococcus and Paenibacillus (Table 2).
Water chemistry
Water conditions remained similar: 3.3 mg/L dissolved oxygen (± 0.7, 95% confidence intervals); 87.4 ± 58.4 mg/L total organic carbon; and initial chlorine 0.50 mg/L, but declined to 0.15 (± 0.06) and 0.06 (± 0.03) on days 3 and 5, respectively. Initial pHs were 7.6 (± 0.1); however, elevated pHs were observed in copper (9.4 ± 1.8) and cement microcosms (9.0 ± 1.3). Neither pH nor dissolved oxygen was impacted by inoculations or chlorination.
The dissolved oxygen and total organic carbon were within World Health Organization guidelines for drinking water. Dissolved oxygen influences corrosion in water pipes and bacterial activity, while the organic carbon increases chlorine demand and microbial growth. Here, any residual organic carbon likely reduced the free chlorine concentration as observed.
Relationship between physico-chemical properties, abundances of genes and bacteria
Comparison of population abundances and viabilities to the physico-chemical conditions (Table 3) indicated that dissolved oxygen increased bacterial abundances and might have an inverse impact on relative abundance of resistant strains (though significant in Paenibacillus assays only). Occasional correlations occurred between organic carbon and bacterial abundances. Further, elevated pH reduced total bacteria, which were observed by inverse correlations in absolute abundances, and positive correlations in relative numbers—suggesting that presence of resistant strains were less impacted. There were no significant correlations between viable counts and resistant gene abundances, nor chlorine and resistance genes; while interesting, this is not surprising as gene presence does not represent viability, as discussed previously.
Conclusion
This study demonstrated the factors affecting the fate of antimicrobial resistance in water distribution systems; pipe material, residual disinfectant and water age shift community dynamics. Resistant bacteria developed biofilm on polyvinyl chloride and copper pipes (66.7%) more than on cement pipes (16.7%). Biofilm-forming bacteria behave differently than planktonic cells, and due to physiological adaptations, they exhibit greater resistance to disinfectants as 55.6% cases showed more resistant population in biofilm in the presence of chlorine. In terms of total population, more bacteria were in biofilms. Further, increased population densities and selective pressures by sub-inhibitory concentrations of disinfectants and pipes create conditions, under which micro-organisms acquire new traits and become more resistant.
References
Abdu N, Abdullahi AA, Abdulkadir A (2017) Heavy metals and soil microbes. Environ Chem Lett 15(1):65–84. https://doi.org/10.1007/s10311-016-0587-x
Boe-Hansen R, Albrechtsen H-J, Arvin E, Jorgensen C (2002) Bulk water phase and biofilm growth in drinking water at low nutrient conditions. Water Res 36:4477–4486
Chu C, Lu C, Lee C, Tsai C (2003) Effects of chlorine level on the growth of biofilm in water pipes. J Environ Sci Health Part A 38(7):1377–1388. https://doi.org/10.1081/ese-120021132
Daghrir R, Drogui P (2013) Tetracycline antibiotics in the environment: a review. Environ Chem Lett 11(3):209–227. https://doi.org/10.1007/s1031-013-0404-8
DeClerck E, Van Mol K, Jannes G, Rossau R, De Vos P (2004) Design of a 5′ exonuclease-based real-time PCR assay for simultaneous detection of Bacillus licheniformis, members of the ‘B. cereus group’ and B. fumarioli in gelatine. Lett Appl Microbiol 39(1):109–115. https://doi.org/10.1111/j.1472-765x.2004.01550.x
Dong F, Li C, Lin Q, Duan H (2018) Effect of pipe materials on disinfection by-products and bacterial communities during sulfamethazine chlorination in a pilot-scale water distribution system. Environ Chem Lett. https://doi.org/10.1007/s10311-018-00823-3
Douterelo I, Jackson M, Solomon C, Boxall J (2016) Microbial analysis of in situ biofilm formation in drinking water distribution systems: implications for monitoring and control of drinking water quality. Appl Microbiol Biotechnol 100(7):3301–3311. https://doi.org/10.1007/s00253-015-7155-3
Fish KE, Collins R, Green NH, Sharpe RL, Douterelo I, Osborn AM, Boxall JB (2015) Characterisation of the physical composition and microbial community structure of biofilms within a model full-scale drinking water distribution system. PLoS ONE 10(2):e0115824. https://doi.org/10.1371/journal.pone.0115824
Frias J, Ribas F, Lucena F (2001) Effects of different nutrients on bacterial growth in a pilot distribution system. Antonie Van Leeuwenhoek 80:129–138. https://doi.org/10.1023/A:1012229503589
Khan S, Beattie TK, Knapp CW (2016a) Relationship between antibiotic- and disinfectant-resistance profiles in bacteria harvested from tap water. Chemosphere 152:132–141. https://doi.org/10.1016/j.chemosphere.2016.02.086
Khan S, Knapp CW, Beattie TK (2016b) Antibiotic resistant bacteria found in municipal drinking water. Environ Process 3:541–552. https://doi.org/10.1007/s40710-016-0149-z
Li C, Feilong D, Feng L, Zhao J, Zhang T, Cizmas L, Sharma VK (2017) Bacterial community structure and microorganism inactivation following water treatment with ferrate(VI) or chlorine. Environ Chem Lett 15(3):525–530
Luo Y, Mao D, Rysz M, Zhou Q, Zhang H, Xu L, Alvarez PJJ (2010) Trends in antibiotic resistance genes occurrence in the Haihe River, China. Environ Sci Technol 44(19):7220–7225. https://doi.org/10.1021/es100233w
McBain AJ, Ledder RG, Moore LE, Catrenich CE, Gilbert P (2004) Effects of quaternary-ammonium-based formulations on bacterial community dynamics and antimicrobial susceptibility. Appl Environ Microbiol 70(6):3449–3456. https://doi.org/10.1128/aem.70.6.3449-3456.2004
Morente OE, Fernandez-Fuentes MA, Burgos MJG, Abriouel H, Pulido RP, Galvez A (2013) Biocide tolerance in bacteria. Int J Food Microbiol 162(1):13–25. https://doi.org/10.1016/j.ijfoodmicro.2012.12.028
Nguyen C, Elfland C, Edwards M (2012) Impact of advanced water conservation features and new copper pipe on rapid chloramine decay and microbial regrowth. Water Res 46(3):611–621. https://doi.org/10.1016/j.watres.2011.11.006
Niquette P, Servails P, Savoir R (2000) Impacts of pipe materials on densities of fixed bacterial biomass in a drinking water distribution system. Water Res 34(6):1952–1956. https://doi.org/10.1016/s0043-1354(99)00307-3
Pei R, Kim S-C, Carlson KH, Pruden A (2006) Effect of river landscape on the sediment concentrations of antibiotics and corresponding antibiotic resistance genes (ARG). Water Res 40(12):2427–2435. https://doi.org/10.1016/j.watres.2006.04.017
Pettersson B, Rippere KE, Yousten AA, Priest FG (1999) Transfer of Bacillus lentimorbus and Bacillus popilliae to the genus Paenibacillus with emended descriptions of Paenibacillus lentirnorbus comb. nov. and Paenibacillus popilliae comb. nov. Int J Syst Bacteriol 49:531–540
Rodgers K, McLellan I, Peshkur T, Williams R, Tonner R, Hursthouse AS, Knapp CW, Henriquez FL (2018) Can the legacy of industrial pollution influence antimicrobial resistance in estuarine sediments? Environ Chem Lett. https://doi.org/10.1007/s10311-018-0791-y
Rusu A, Hancu G, Uivaroşi V (2015) Fluoroquinolone pollution of food, water and soil, and bacterial resistance. Environ Chem Lett 13:21. https://doi.org/10.1007/s10311-014-0481-3
Walcott RR, Gitaitis RD (2000) Detection of Acidovorax avenae subsp. citrulli in watermelon seed using immunomagnetic separation and the polymerase chain reaction. Plant Dis 84(4):470–474. https://doi.org/10.1094/PDIS.2000.84.4.470
Yu J, Kim D, Lee T (2010) Microbial diversity in biofilms on water distribution pipes of different materials. Water Sci Technol 61(1):163–171. https://doi.org/10.2166/wst.2010.813
Zeng DN, Fan ZY, Chi L, Wang X, Qu WD, Quan ZX (2013) Analysis of the bacterial communities associated with different drinking water treatment processes. World J Microbiol Biotechnol 29(9):1573–1584. https://doi.org/10.1007/s11274-013-1321-5
Acknowledgements
The research is supported by Schlumberger Foundation’s Faculty for the Future in the form of funding for SK’s Ph.D.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Khan, S., Beattie, T.K. & Knapp, C.W. Rapid selection of antimicrobial-resistant bacteria in complex water systems by chlorine and pipe materials. Environ Chem Lett 17, 1367–1373 (2019). https://doi.org/10.1007/s10311-019-00867-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10311-019-00867-z




 Total bacteria.
Total bacteria.
 Resistant bacteria
Resistant bacteria