Abstract
Polymyxins are used as the last-line therapy against multidrug-resistant bacteria. However, their further clinical development needs to solve problems related to the presence of heterogeneous analogs, but there is still no platform or methods that can regulate the biosynthesis of polymyxin analogs. In this study, we present an approach to swap domains in the polymyxin gene cluster to regulate the production of different analogs. Following adenylation domain swapping, the proportion of polymyxin B1 increased from 41.36 to 52.90%, while that of B1-1 decreased from 18.25 to 3.09%. The ratio of polymyxin B1 and B3 following starter condensation domain swapping changed from 41.36 and 16.99 to 55.03 and 6.39%, respectively. The two domain-swapping strains produced 62.96% of polymyxin B1, 6.70% of B3 and 3.32% of B1-1. This study also revealed the presence of overflow fluxes between acetoin, 2,3-butanediol and polymyxin. To our best knowledge, this is the first report of engineering the polymyxin synthetase gene cluster in situ to regulate the relative proportions of polymyxin analogs. This research paves a way for regulating lipopeptide analogs and will facilitate the development of novel lipopeptide derivatives.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The increasing prevalence of multidrug-resistant superbugs threatens the effective prevention and treatment of a wide scope of microbial infections [8]. Recently, lipopeptides have gained increasing attention as treatment for superbugs because they not only exhibit broad antimicrobial activity, but also have distinct modes of antimicrobial action and slower resistance development compared to conventional antibiotics [29]. Polymyxin B, which was initially abandoned due to toxicity, has resurged as a last-line therapy for multidrug-resistant Gram-negative bacterial infections [54]. The increasing need for polymyxin B is exacerbated by the lack of new antibiotics against superbugs [56].
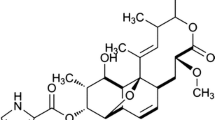
However, the crude polymyxin preparation is not a single compound, but rather a mixture of analogs [42]. In fact, polymyxin B for intravenous administration is a chemically heterogeneous mixture containing 30 related polymyxin-like lipopeptide analogs. The major components of polymyxin B are B1, B2, B3 and B1-1, which differ in the fatty acyl moiety at the N-terminus (B1: 6-methyloctanoyl; B2: 6-methylheptanonyl; B3: octanoyl), or an amino acid within the heptapeptide ring (B1, position 7: leucine; B1-1, position 7: isoleucine) [34] (Fig. 1). The minimum for the sum of polymyxins B1, B2, B3 and B1-1 proscribed by the pharmacopoeia is ≥ 80%, with the last two components (B3 and B1-1) representing no more than 15 and 6%, respectively. Despite the increase of polymyxin usage, controlling the ratio of the components in clinical and commercial preparations remains a challenge [43]. In fact, it is known that the proportion of polymyxin B analogs can vary between different brands and even batches [6, 25]. Most preclinical and clinical studies evaluated polymyxin B in aggregate, but individual polymyxin B component can behave differently compared to mixtures [21], It was shown that commercial polymyxin B mixture is slightly less active and induces more apoptosis than individual components (B1 or B2), while B3 and B1-1 are less active in vivo [44]. Combinations of polymyxins B1 and B2 are associated with the lowest probability of improved antibacterial activity [60]. It should be stressed that small differences in potency could alter the optimal polymyxin B dose [45], and the same is true for other lipopeptides [28]. Compositional variations can impact pharmacokinetics, pharmacodynamics and toxicity, and there seems to be a relationship between nephrotoxicity and the structural differences between polymyxins B1 and B2 [38, 44, 49]. Therefore, developing a platform to provide tools for generating precisely designed polymyxin derivatives via synthetic biology would be of immense value.
Polymyxins are biosynthesized by a nonribosomal peptide synthetase (NRPS) with a modular structure, which can be divided into different domains [39] (Fig. 2a). The A domain plays a decisive role in the selection of amino acid monomers [32, 48]. Researchers obtained lipopeptide derivatives with different amino acids by genetically modifying the A domain or modules containing the A domain in the biosynthetic clusters of surfactin, fusaricidin, calcium-dependent antibiotic (CDA), luminimide B, gramicidin, enterobactin, plipastatin and bacillamide [5, 16, 19, 20, 23, 24, 31, 61]. Kim SY et al. [27] first replaced the A domain in the polymyxin gene cluster to engineer B. subtilis for the production of polymyxins A, E and P, but they did not explore changes in the analog composition.
Predicted polymyxin gene cluster architecture and an outline of the experimental design. a Arrangement of pmx genes in the polymyxin B gene cluster. Modular structure of the polymyxin synthase multienzyme complexes. The specificity for adenylation domains derived by bioinformatics analysis is shown for each module. All modules contain condensation, adenylation, and thiolation subunits. Modules 3 and 6 also contain an epimerization domain, and module 10 possesses a C terminal thioesterase domain, which catalyzes the intramolecular cyclization and release of polymyxin B. b Construction of targeting vectors for homologous double-crossover. c Schematic diagram of homologous double-crossover in positive transformants
Some studies changed the fatty acids of lipopeptides through manipulation of the precursor supply [13, 55] or gene knockout [37], but there is little information on the mechanism of lipid transfer in the initial biosynthesis process. In the synthesis of daptomycin, surfactin and lipopeptide A54145, the choice of the fatty acid starter unit is controlled by the donor site of the N-terminal C domain (starter C domain) of the initiating NRPS module [30, 36, 57]. The donor site of the starter C domain has higher chain length specificity, which allows relatively fewer variations in the fatty acyl chain. However, the detailed mechanism of the lipoinitiation reaction of polymyxin is still unknown. The introduction of a polymyxin biosynthesis cluster without fatty acyl ligase into B. subtilis resulted in efficient production of polymyxin [27, 52], which means that the acylation of polymyxin may not be related to external fatty acyl ligase enzymes, because they were not localized within the cluster, nor were they identified in the biosynthetic clusters of polymyxins B, E and P [18], Moreover, the N-terminal condensation domain of the polymyxin gene cluster is similar to the starter C domain [41, 51]. The specificity of the starter C domain may potentially be exploited to produce novel lipopeptides with different chain length by introducing it into foreign NRPS enzyme complexes [10, 16].
In this study, we analyzed the polymyxin biosynthesis gene cluster, the identified polymyxin B mixture produced by P. polymyxa CJX518, and constructed recombinant P. polymyxa strains that produce higher proportions of polymyxins B1 and B2 along with lower proportions of B3 and B1-1 by swapping the A and starter C domains. To our best knowledge, this is the first report of manipulating the polymyxin synthesis gene cluster in situ to tune the polymyxin analog mixture, as well as the first study of the relationship between the starter C domain of polymyxin synthetase and fatty acid specificity.
Materials and methods
Bacterial strains, plasmids, and culture media
The strains and plasmids used in this study are listed in Table 1. E. coli and P. polymyxa CJX518 (CGMCC 7096) were cultured in Luria–Betani (LB) medium (5 g/L yeast extract, 10 g/L peptone, 10 g/L NaCl). The seed medium and fermentation medium with 20 g/L (NH4)2SO4 that were used to produce polymyxin were prepared as described previously [35]. E. coli DH5α was used for general cloning and E. coli JM110 was used for plasmids’ demethylation. P. polymyxa CJX518 was used as the parent strain for genetic engineering. When necessary, 100 μg/mL ampicillin (Amp) or 10 μg/mL erythromycin (Erm) was added.
Construction of domain-swapping chromosomal recombination plasmids
Plasmids used for individual domain substitution in the polymyxin synthetase cluster were derivatives of pMAD [2]. Substitution of the A domain and starter C domain was performed using the plasmids pMAD-△A and pMAD-△C, respectively. The primers used in this study are listed in Supplementary Table S1. To construct pMAD-△A and pMAD-△C, the A domain specific for leucine was amplified from plasmid pUC57-A using the primer pair A-F/A-R. The 5’- and 3’-flanking regions of the A7 domain specific for isoleucine were amplified from the chromosomal DNA of strain CJX518 by PCR using the primer pairs AL-F/AL-R and AR-F/AR-R, respectively. The three PCR fragments were joined by fusion PCR and cloned into the pGEM-T Easy vector to construct pT-A for cloning and sequencing. The starter C domain substitution cassette was constructed following the similar protocol, whereby the 5’- and 3’-flanking regions of starter C domain were amplified from the first domain of pmxE on the chromosome of CJX518, while the swapped starter C domain was obtained from pUC57-C. The cassette was cloned into the pGEM-T Easy vector to construct pT-C. The cassettes from pT-A or pT-C were ligated into pMAD using the SalI and BamHI cleavage sites to produce the plasmids pMAD-△A and pMAD-△C. A schematic illustration of the vector construction is shown in Fig. 2b. The A domain and starter C domain specific to leucine and the fatty acids were obtained from the polymyxin A biosynthetic gene cluster of P. polymyxa E681 by analyzing the domain specificity [40].
Construction of substitution strains
The pMAD-△A and pMAD-△C vectors were introduced into P. polymyxa CJX518 by electroporation as described previously [62]. The recombinant strains were obtained by a two-step procedure for allele swapping in P. polymyxa CJX518. In the first step, the vector was introduced into CJX518, and transformants were selected after 2 days at 30 °C on LB agar plates containing erythromycin and x-gal. In the second step, a blue colony was used to inoculate LB broth with erythromycin and grown at 42 °C overnight. The pre-culture was transferred into LB medium without antibiotics and grown for 12 h, followed by sub-culture at 42 °C, which was repeated 4–5 times. Serial dilutions of this culture were plated onto LB agar plates and grown overnight at 42 °C. Several colonies were verified for erythromycin sensitivity. To confirm the domain swapping, the strains sensitive to erythromycin were tested by colony PCR using the primer pairs A’-F/A’-R and C’-F/C’-R, which were designed to span the genome and cassette sequences. The single-domain substitution strains were designated 518-A and 518-C. Then, pMAD-△A was introduced into 518-C, and the desired strain was screened using the same method. The combined A and C domain substitution strain was named 518-AC. A schematic illustration of vector construction for homologous double-crossover in P. polymyxa CJX518 is shown in Fig. 2c.
Growth and metabolite analysis
A single colony from an LB agar plate was used to inoculate 50 mL of seed medium and grown for 24 h at a rotary shaker at 200 rpm and 30 °C. The resulting pre-culture was used to inoculate 200 mL of fresh fermentation medium to an initial OD600 of 0.2 and cultivated for 72 h at 30 °C and 200 rpm. The biomass accumulation (optical density at 600nm) was measured using a UV–visible spectrophotometer (Tianjin Kanasi Optics Analysis Instrument Co., Ltd., China). The levels of glucose, acetoin and (R,R)-2,3-butanediol in the medium were measured as described before [9], from culture supernatants obtained by centrifugation at 12000×g for 10 min and filtered through 0.22-μm, using high-performance liquid chromatography on a Waters1515 Isocratic HPLC (Waters , USA) equipped with an Aminex HPX-87H Ion Exclusion column (300 mm×7.8 mm, BIORAD, USA) and differential refractive index detector (Waters 2414). The levels of intracellular NADH and NAD+ were measured using the cycling assay [11].
Polymyxin B detection and determination
The polymyxin B in the broth was analyzed using a reverse-phase C18 column (4.6×250 mm, 4.6 μm; Alliance, USA) kept at 30 °C. The mobile phase was composed 20% acetonitrile–80% of a solution comprising 4.46 g of Na2SO4 in 900 mL of water, adjusted to pH 2.3 using dilute phosphoric acid and diluted to 1000 mL with water, at a flow rate of 1 mL/min. The elution was monitored using a spectrophotometer at 215 nm. The cell-free supernatants were further analyzed by liquid chromatography coupled with electrospray ionization mass spectrometry (Thermo Electron, San Jose, CA, USA). The ESI source was operated in positive ion mode. The ion-spray voltage was 310 kV. A C18 column (150 × 2.1 mm, 5 μm) was used, and the mobile phase comprised 10 mM trifluoroacetic water and acetonitrile (20:80). The capillary voltage, desolvation gas flow, ion source temperatures, desolvation temperature and cone voltage were 310 kV, 280 L/h, 250 °C, 115 °C, and 50 V, respectively. The mass scan range was m/z 500–1200.
Antibacterial activity assay
The fermentation broth of strain 518 and derived strains was centrifuged at 12000×g for 10 min to remove the cells and filtered through a sterile 0.22-μm filter. The antibacterial activity of the supernatant was analyzed using E. coli DH5a and P. aeruginosa BNCC186666 as indicators, which were grown in LB medium at 37 °C and homogeneously mixed into LB agar to an OD600 of 0.2. Sterile Oxford cups were placed in a Petri dish, and the mixed LB (15 mL) was poured into them. After it solidified, the Oxford cups removed, the samples were added into the well, and equal volume of sterile fermentation medium was used as a control. The plate was cultured at 37 °C.
Detection of biosurfactant produced during the fermentation
The biosurfactant produced during the fermentation was detected by measuring the surface tension. The fermentation broth was collected every 12 h for 72 h and separated from the bacterial cells by centrifugation at 5000×g for 10 min. The supernatant was used for the surface tension measurements using a QBZY series automatic surface tensiometer (Shanghai Fangrui Instrument Co., Ltd., China).
Statistical analysis
Statistical analysis was performed using Microsoft Excel 2010 software (Microsoft Corp., USA). The significance of differences in the means between the control strains and the domain-swapped strains was assessed using Student‘s t test. Differences with p values of less than 0.05 were considered statistically significant.
Results and discussion
Sequencing of the polymyxin gene cluster and identification of the polymyxin types
The combination of microbial genome screening and synthetic biology could have a major impact on combinatorial biosynthesis, allowing the application of rationally engineered NRPS to develop natural products and drugs [3, 53]. The size or base type of the polymyxin biosynthesis gene cluster (pmx) is different in different strains, and the pmx genes in different P. polymyxa strains encode enzymes that synthesize different polymyxin products (A, B, E, P) [17]. In this study, we performed genome mining to identify, annotate and evaluate the pmx gene responsible for polymyxin biosynthesis (GenBank: MN535700) (Fig. 2a). The pmx cluster sequence was found to contain five open reading frames, encompassing a total sequence length of 40.7 kb that was organized into three NRPS genes (pmxA, pmxB and pmxE) involved in polymyxin synthesis and two genes (pmxC and pmxD) encoding a transporter protein that exports the lipopeptide. The pmxA (14916 bp), pmxB (3309 bp) and pmxE (18879 bp) genes were respectively predicted to encode synthases of 4971, 1102 and 6292 amino acid residues. The specificity-conferring amino acids of the A-domain in pmx were determined using the web-based prediction programs PKS/NRPS Analysis (Fig. 2a, S1) [17, 40]. The synthetase encoded by pmxE was predicted to be composed of five modules that incorporate four 2,4-diaminobutanoic acid (Dab) monomers and one threonine (Thr). The pmxA synthetase ligates phenylalanine (Phe), isoleucine (Ile) and two Dab, while the pmxB synthetase integrates Thr. The order of these modules was collinear with the sequence of the polymyxin lipopeptide product, and the arrangement of the amino acid monomers corresponds to the structure of the polymyxin B. LC–MS analysis of the culture supernatant identified four peaks with masses consistent with polymyxins B1 ([M+2H]2+=602.4), B2 ([M+2H]2+=595.4), B3 ([M+2H]2+=595.4) and B1-1 ([M+2H]2+=602.4) (Fig. 3a, b), in agreement with the reference standard (Fig. S2). No genes in the polymyxin biosynthesis gene cluster were predicted to encode an enzyme responsible for the acylation of the peptide moiety, and no genes associated with the activation or transfer of fatty acids were found adjacent to the polymyxin cluster (Fig. S3).
Polymyxin production of wild-type CJX518 and its modified derivatives
Single- and double-domain swapping strains were obtained by homologous recombination facilitated by temperature-sensitive plasmids. The growth of wild-type P. polymyxa CJX518 and the domain-swapping strains was observed individually. As shown in Fig. 4a, in the early period of fermentation, they grew at nearly the same speed and showed strong growth in the first 24 h. However, the parent strain grew slower than the recombinant strains after 24 h. All recombinant strains reached a slightly higher biomass than wild-type CJX518 (p<0.05), which was accompanied by faster glucose depletion (p < 0.01) (Fig. 4b). After 48 h, the biomass of all strains decreased, and the glucose depletion rate was reduced. It has been published that polymyxin can kill its producer P. polymyxa [58]. Consequently, we speculated that the difference in OD600 and glucose consumption may be related to the difference in polymyxin production and residual components in the medium.
It was expected that the necessity for a functional A domain and starter C domain in polymyxin biosynthesis might be related to the regulation of analogs. To explore possible strategies for regulating polymyxin B analogs, increasing the proportion of the major components B1 and B2, as well as decreasing B3 and B1-1 as much as possible, we constructed swapping strains of domain A specific for leucine and the starter C domain related to fatty acid specificity. Earlier studies reported that polymyxin production starts in the exponential phase and continues thereafter [1, 33, 59]. The polymyxin B production of all strains is shown in Fig. 4c. There was no polymyxin B within 12 h. Then, the production rapidly increased from 12 h to 60 h, and remained almost constant in the remaining time. The polymyxin B yield of all strains reached its maximum at 60 h. Compared with the 518 strain (0.47 g/L), the yield of 518-A, 518-C and 518-AC decreased to 0.38 g/L, 0.32 g/L and 0.22g/L, respectively. Thus, the polymyxin B yield of the recombinant strains was lower than that of CJX518, whereby the yield of the 518-AC strain with two swapped domains was the lowest (p < 0.01). Previous studies showed that domain or module engineering would reduce the lipopeptide yield by destroying the structural integrity and recognized relationships between the A and C domains [5, 16, 27]. Thus, swapping the module containing the target domain as a whole should be investigated as a way of keeping domain integrity and specificity. The yield may be improved by optimizing the expression of the pmx gene cluster or the fermentation conditions.
The HPLC results of the reference standard and samples both revealed four main compounds with different retention times corresponding to B1, B2, B3 and B1-1 (Fig. S2). Swapping the A domain conferring leucine specificity led to a significant decrease in the ratio of B1-1, from 18.25 to 3.09%, as well as an increase of B1 from 41.36 to 52.90%. In addition, the proportion of B2 increased, and that of B3 decreased, even though the difference was not large. After swapping the C domain, the percentage of B2 and B3 was reduced obviously from 23.40 and 16.99 to 18.09 and 6.39%, respectively, while the ratios of B1 and B1-1 increased from 41.36 and 18.25 to 55.03 and 20.49%. After swapping both the A and C domains, the proportion of B1 increased sharply to 62.96%, while that of B2 increased from 23.40 to 27.03%. At the same time, B1-1 and B3 dropped significantly to 6.7 and 3.32% (Table S2; Fig. 4d). The A and starter C domains in the polymyxin biosynthesis gene cluster confer specificity for amino acid and fatty acid substrate, and domain swapping can lead to changes in the analog composition of polymyxin B, obviously reducing the minor and increasing the main components. Our experiments unambiguously showed that altering the specificity of the A domain by swapping could change the amino acids in the structure of the lipopeptides, and introducing a heterologous C domain can regulate the fatty acid specificity in the structure. Novel lipopeptides with different chain length and amino acids may be produced by introducing the starter C and A domains into foreign NRPS clusters.
Effects of domain swapping on overflow metabolites
P. polymyxa is also known to produce acetoin and 2,3-butanediol [11, 35], and the production of lipopeptides as secondary metabolites is related to acetoin and 2,3-butanediol [15]. As shown in Fig. 5a and b, all strains produced acetoin after 36 h, and 2,3-butanediol after 12 h. The yield of acetoin of the recombinant strains was higher than that of the wild-type strain after 48 h and reached the maximum value at 60 h. In the first 36 h, there was no significant difference in the 2,3-butanediol yield of strains 518, 518-A and 518-C, and it decreased in the remaining time except for 60 h, at which point the yield of strain 518 was lower than that of the other two strains. However, the 2,3-butanediol yield of 518-AC was much higher than that of the other strains at all time-points (Fig. 5c). The total yield of 2,3-butanediol and acetoin of the recombinant strains was higher than that of wild-type 518 from 24 h to 72 h, and reached its peak at 60 h. Although the polymyxin yield of the domain-swapping strain was lower than that of the parent strain, the total yield of acetoin and 2,3-butanediol was higher (p < 0.05 and p < 0.01, respectively). The coproduction of lipopeptides and acetoin gives more added value to the fermentation because of possible uses in crop protection, diesel biodegradation, biodiesel production and p-xylene decontamination [22]. Studies showed that acetoin and 2,3-butanediol are major overflow metabolites affecting lipopeptides production [12, 65]. Accordingly, reducing the flux toward overflow metabolites could enhance lipopeptide production [26]. Acetoin production was found to be directly related to the consumption of carbon source, which regulated the lipopeptide yield [15], since acetoin competed for acetyl-CoA [50], which can be used for the biosynthesis of straight fatty acids to generate lipopeptides. Accordingly, different strains can direct the flow of acetoin toward the production of lipopeptide isoform and vice versa [12]. It is possible that reducing the lipopeptide synthesis contributed to the accumulation of acetoin and 2,3-butanediol (Fig. 5a, b, 4c) [4]. As shown in Fig. 5c., pyruvate as an intermediate metabolite of the main metabolic flux connects the production of acetoin, 2,3-butanediol and lipopeptides [12, 64, 65]. Therefore, the yield of acetoin and 2,3-butanediol would increase when the yield of lipopeptide is decreased. The production of overflow metabolites can be reduced by deactivating the biosynthesis of acetoin and 2,3-butanediol to enhance lipopeptides production [63, 65]. Enhancing the biosynthesis of amino- and fatty acids that are direct precursors of lipopeptides would reduce the formation of acetoin and 2,3-butanediol. There was a conversion between acetoin and 2,3-butanediol at the start of acetoin production, and the 2,3-butanediol content dropped sharply from 36 h to 48 h. A low NADH/NAD+ ratio can promote acetoin production, while a high ratio favors the synthesis of 2,3-butanediol. The NADH/NAD+ ratios of the domain-swapping strains were higher than that of the control strain before 48 h. The ratio in the test group increased at 36 h, while in the control group it increased at 48 h, and at 36 h, the ratio of strain 518 was much lower than those of 518-A, -C, and -AC. After 48 h, the NADH/NAD+ ratios of all strains decreased slowly (Fig. 5d). It has been reported that the extracellular oxidation–reduction potential is closely associated with the NADH/NAD+ ratio, which in turn affects the interconversion between 2,3-butanediol and acetoin and metabolic flux partitioning [14]. Hence, swapping the A and starter C domains can regulate the ratio of polymyxin analogs, and the yield can be increased by regulating the metabolic flux toward acetoin and 2,3-butanediol.
Surface tension and antibacterial activity of polymyxin produced by wild-type CJX518 and its modified derivatives
Polymyxin and other lipopeptides are surfactants, and therefore reduce the surface tension of the culture supernatant. Consequently, we also measured the changes of surface tension during the fermentation. As shown in Fig. 6a, the surface tension of strains 518-A, -C, and -AC decreased drastically after 24 h, when polymyxin production started, and then decreased somewhat during the later stages. The surface tension of the fermentation supernatants of strains 518-A, -C, and -AC dropped faster than that of the wild-type 518. Later, the surface tension stabilized and did not continue decreasing as polymyxin production increased, which was in agreement with earlier research [46]. The surface tension of the supernatants of strains 518, 518-A, 518-C and 518-AC respectively decreased by 18.0, 18.5, 18.9 and 17.4 mN/m. Although the surface tension of the supernatant of CJX518 was higher than that of the other strains before 36 h, it fell to a similar value during later stages. Despite the decrease of polymyxin B yield after domain swapping, the drop of surface tension in the supernatants of all strains had a similar magnitude. It has been reported that there is no correlation between the yield of lipopeptides and the decrease in surface tension. When lipopeptides are produced, the surface tension rapidly drops to a minimum, and then stops decreasing with a further increase of the product yield.
Polymyxin can strongly inhibit Gram-negative bacteria. To compare the antibacterial properties of the fermentation products of P. polymyxa CJX518 with those of the domain-swapping strains 518-A, 518-AC and 518-C, E. coli and P. aeruginosa were used as an indicator strains. Culture supernatants of P. polymyxa CJX518, 518-A, 518-AC and 518-C all exhibited high antimicrobial activities against E. coli and P. aeruginosa (Fig. 6b and c), which was consistent with earlier findings that polymyxin can effectively inhibit multidrug-resistant strains of these species [7]. The inhibition zones of the CJX518 supernatants were slightly larger than that of the other strains, and the inhibition zone of 518-AC was the smallest, which may be related to the decrease of polymyxin yield (Fig. 4a). Although there was a disparity in polymyxin yield among all strains, the antibacterial activity against E. coli and P. aeruginosa was not significantly different, which may be related to the activity of polymyxin preparations with different components. The polymyxin yield of CJX518 was the highest, while that of 518-AC was the smallest, but there was only a small difference in terms of antibacterial activity. In spite of the lower yield, the inhibition zones of 518-A and 518-C were slightly smaller than that of 518, which would indicate that polymyxin B1 and B2 are more active than B3 and B1-1 [44]. To reduce the possible negative effects of polymyxin dosage, as well as decrease resistance development [47] and toxicity [49], increasing the proportion of the main components B1 and B2 should be taken into consideration.
References
Paulus H, Gray E (1964) The biosynthesus of polymyxin B by growing cultures of Bacillus Polymyxa. J Biol Chem 239:865–71
Arnaud M, Chastanet A, Debarbouille M (2004) New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl Environ Microbiol 70:6887–6891. https://doi.org/10.1128/AEM.70.11.6887-6891.2004
Baltz RH (2018) Synthetic biology, genome mining, and combinatorial biosynthesis of NRPS-derived antibiotics: a perspective. J Ind Microbiol Biotechnol 45:651–655. https://doi.org/10.1007/s10295-019-02231-0
Bao T, Zhang X, Zhao X, Rao Z, Yang T, Yang S (2015) Regulation of the NADH pool and NADH/NADPH ratio redistributes acetoin and 2,3-butanediol proportion in Bacillus subtilis. Biotechnol J 10(8):1298–1306. https://doi.org/10.1002/biot.201400577
Bian X, Plaza A, Yan F, Zhang Y, Müller R (2015) Rational and efficient site-directed mutagenesis of adenylation domain alters relative yields of luminmide derivatives in vivo. Biotechnol Bioeng 112(7):1343–1353. https://doi.org/10.1002/bit.25560
Brink AJ, Richards GA, Colombo G, Bortolotti F, Colombo P, Jehl F (2014) Multicomponent antibiotic substances produced by fermentation: Implications for regulatory authorities, critically ill patients and generics. Int J Antimicrob Agents 43(1):1–6. https://doi.org/10.1016/j.ijantimicag.2013.06.013
Brown P, Dawson MJ (2017) Development of new polymyxin derivatives for multi-drug resistant Gram-negative infections. J Antibiot 70(4):386–394. https://doi.org/10.1038/ja.2016.146
Cattoir V, Felden B (2019) Future antibacterial strategies: from basic concepts to clinical challenges. J Infect Dis 220(3):350–360. https://doi.org/10.1038/s41564-018-0205-8
Cheng JS, Liang YQ, Jiang T, Ding MZ, Cui SF, Lv XM, Yuan YJ (2013) Metabolic analysis reveals the amino acid responses of Streptomyces lydicus to pitching ratios during improving streptolydigin production. Appl Microbiol Biotechnol 97(13):5943–5954. https://doi.org/10.1007/s00253-013-4790-4
Chooi YH, Tang Y (2010) Adding the Lipo to Lipopeptides: Do more with less. Chem Biol 17(8):791–793. https://doi.org/10.1016/j.chembiol.2010.08.001
Dai JJ, Cheng JS, Liang YQ, Jiang T, Yuan YJ (2014) Regulation of extracellular oxidoreduction potential enhanced (R, R)-2,3-butanediol production by Paenibacillus polymyxa CJX518. Bioresour Technol 167(3):433–440. https://doi.org/10.1016/j.biortech.2014.06.044
Dhali D, Coutte F, Arias AA, Auger S, Bidnenko V, Chataigné G, Lalk M, Niehren J, De SJ, Versari C (2017) Genetic engineering of the branched fatty acid metabolic pathway of Bacillus subtilis for the overproduction of surfactin C14 isoform. Biotechnol J 12(7):1600574. https://doi.org/10.1002/biot.201600574
Ding L, Guo W, Chen X (2019) Exogenous addition of alkanoic acids enhanced production of antifungal lipopeptides in Bacillus amyloliquefaciens Pc3. Appl Microbiol Biotechnol 103(13):5367–5377. https://doi.org/10.1007/s00253-019-09792-1
Du C, Yan H, Zhang Y, Li Y (2006) Cao Z (2006) Use of oxidoreduction potential as an indicator to regulate 1,3-propanediol fermentation by Klebsiella pneumoniae. Appl Microbiol Biotechnol 69(5):554–563
Etchegaray A, Coutte F, Chataigne G, Bechet M, Dos Santos RH, Leclere V, Jacques P (2017) Production of Bacillus amyloliquefaciens OG and its metabolites in renewable media: valorisation for biodiesel production and p-xylene decontamination. Can J Microbiol 36(3):46–60. https://doi.org/10.1139/cjm-2016-0288
Fischbach MA, Lai JR, Roche ED, Walsh CT, Liu DR (2007) Directed evolution can rapidly improve the activity of chimeric assembly-line enzymes. Proc Natl Acad Sci U S A 104(29):11951–11956. https://doi.org/10.1073/pnas.0705348104
Galea CA, Han M, Zhu Y, Roberts K, Wang J, ThompsonL PEJ, Velkov T (2017) Characterization of the Polymyxin D Synthetase Biosynthetic Cluster and Product Profile of Paenibacillus polymyxa ATCC 10401. J Nat Prod 80(5):1264–1274. https://doi.org/10.1021/acs.jnatprod.6b00807
Galea CA, Roberts KD, Zhu Y, Thompson PE, Li J, Velkov T (2017) Functional characterization of the unique terminal thioesterase domain from polymyxin synthetase. Biochemistry 56:657–668. https://doi.org/10.1021/acs.biochem.6b01139
Gao L, Guo J, Fan Y, Ma Z, Lu Z, Zhang C, Zhao H, Bie X (2018) Module and individual domain deletions of NRPS to produce plipastatin derivatives in Bacillus subtilis. Microb Cell Fact 17(1):84. https://doi.org/10.1186/s12934-018-0929-4
Han JW, Kim EY, Lee JM, Yun SK, Bang E, Kim BS (2012) Site-directed modification of the adenylation domain of the fusaricidin nonribosomal peptide synthetase for enhanced production of fusaricidin analogs. Biotechnol Lett 34(7):1327–1334. https://doi.org/10.1007/s10529-012-0913-8
Hee KH, Yee K, Leaw J, Ong JL, Lee LS (2017) Development and validation of liquid chromatography tandem mass spectrometry method quantitative determination of polymyxin B1, polymyxin B2, polymyxin B3 and isoleucine-polymyxin B1 in human plasma and its application in clinical studies. J Pharm Biomed Anal 140:91–97. https://doi.org/10.1016/j.jpba.2017.03.018
Hmidet N, Ben AH, Jacques P (2017) Nasri M (2017) Enhancement of Surfactin and Fengycin Production by Bacillus mojavensis A21: Application for Diesel Biodegradation. Biomed Res Int 3:1–8. https://doi.org/10.1155/2017/5893123
Jenny T, Richard L, Laura N, Majid AN, Matthew S, Anna-Winona S, Smith CP, Jason M (2012) Introduction of a non-natural amino acid into a nonribosomal peptide antibiotic by modification of adenylation domain specificity. Angew Chem Int Ed Engl 51(29):7181–7184. https://doi.org/10.1002/anie.201202043
Jiang J, Gao L, Bie X, Lu Z, Liu H, Zhang C, Lu F, Zhao H (2016) Identification of novel surfactin derivatives from NRPS modification of Bacillus subtilis and its antifungal activity against Fusarium moniliforme. BMC Microbiol 16:31–44. https://doi.org/10.1186/s12866-016-0645-3
Jie H, Ledesma KR, Wai-Ying L, Figueroa DA, Tze-Peng L, Diana S-LC, Tam VH (2010) Variability of polymyxin B major components in commercial formulations. Int J Antimicrob Agents 35:308–310. https://doi.org/10.1016/j.ijantimicag.2009.11.005
Junqiang W, Rongjun G, Wenchao W, Guizhen M, Shidong L (2018) Insight into the surfactin production of Bacillus velezensis B006 through metabolomics analysis. J Ind Microbiol Biotechno 45:1033–1044. https://doi.org/10.1007/s10295-018-2076-7
Kim SY, Park SY, Choi SK, Park SH (2015) Biosynthesis of polymyxins B, E, and P using genetically engineered polymyxin synthetases in the surrogate host Bacillus subtilis. J Microbiol Biotechnol. https://doi.org/10.4014/jmb.1505.05036
Knight-Connoni V, Mascio C, Chesnel L, Silverman J (2016) Discovery and development of surotomycin for the treatment of Clostridium difficile. J Ind Microbiol Biotechnol 43:195–204. https://doi.org/10.1007/s10295-015-1714-6
Koh JJ, Lin S, Beuerman RW, Liu S (2017) Recent advances in synthetic lipopeptides as anti-microbial agents: designs and synthetic approaches. Amino Acids 49(12):1–25. https://doi.org/10.1007/s00726-017-2476-4
Kraas FI, Helmetag V, Wittmann M, Strieker M, Marahiel MA (2010) Functional dissection of surfactin synthetase initiation module reveals insights into the mechanism of lipoinitiation. Chem Biol 17(8):872–880. https://doi.org/10.1016/j.chembiol.2010.06.015
Kries H, Niquille DL, Hilvert D (2015) A subdomain swap strategy for reengineering nonribosomal peptides. Chem Biol 22(5):640–648. https://doi.org/10.1016/j.chembiol.2015.04.015
Kudo F, Miyanaga A, Eguchi T (2019) Structural basis of the nonribosomal codes for nonproteinogenic amino acid selective adenylation enzymes in the biosynthesis of natural products. J Ind Microbiol Biotechnol 46:515–536. https://doi.org/10.1007/s10295-018-2084-7
Kuratsu Y, Arai Y, Inuzuka K, Suzuki T (1983) Stimulatory effect of aspartic acid on colistin production by Bacillus polymyxa. Agricult Biol Chem 47(11):2607–2612. https://doi.org/10.1080/00021369.1983.10865997
Landman D, Georgescu C, Martin DA, Quale J (2008) Polymyxins revisited. Clin Microbiol Rev 21(3):449. https://doi.org/10.1128/CMR.00006-08
Liu L, Xu QM, Chen T, Cheng JS, Yuan YJ (2017) Artificial consortium that produces riboflavin regulates distribution of acetoin and 2, 3-butanediol by Paenibacillus polymyxa CJX518. Eng Life Sci 17(9):1039–1049. https://doi.org/10.1002/elsc.201600239
Liu Q, Fan W, Zhao Y, Deng Z, Feng Y (2019) Probing and engineering the fatty acyl substrate selectivity of starter condensation domains of nonribosomal peptide synthetases in lipopeptide biosynthesis. Biotechnol J. https://doi.org/10.1002/biot.201900175
Luo S, Chen XA, Mao XM, Li YQ (2018) Regulatory and biosynthetic effects of the bkd gene clusters on the production of daptomycin and its analogs A21978C1–3. J Ind Microbiol Biotechnol 45:271–279. https://doi.org/10.1007/s10295-018-2011-y
Manchandani P, Dubrovskaya Y, Gao S, Tam VH (2016) Comparative pharmacokinetic profiling of different polymyxin B components. Antimicrob Agents Chemother 60(11):6980–6982. https://doi.org/10.1128/AAC.00702-16
McErlean M, Overbay J, Van Lanen S (2019) Refining and expanding nonribosomal peptide synthetase function and mechanism. J Ind Microbiol Biotechnol 46:493–513. https://doi.org/10.1007/s10295-018-02130-w
Mohd Zeeshan A, Gitanjali Y, Gokhale RS, Debasisa M (2004) NRPS-PKS: a knowledge-based resource for analysis of NRPS/PKS megasynthases. Nucleic Acids Res 32(2):W405–W413. https://doi.org/10.1093/nar/gkh359
Niu B, Vater J, Rueckert C, Blom J, Lehmann M, Ru JJ, Chen XH, Wang Q, Borriss R (2013) Polymyxin P is the active principle in suppressing phytopathogenic Erwinia spp. by the biocontrol rhizobacterium Paenibacillus polymyxa M-1. BMC Microbiol 13(1):137–137. https://doi.org/10.1186/1471-2180-13-137
Olishevska S, Nickzad A, Déziel E (2019) Bacillus and Paenibacillus secreted polyketides and peptides involved in controlling human and plant pathogens. Appl Microbiol Biotechnol 103(1):1–27. https://doi.org/10.1007/s00253-018-9541-0
Pogue JM, Ortwine JK, Kaye KS (2015) Editorial commentary: Optimal usage of colistin: Are we any closer? Clin Infect Dis 4(12):847–847. https://doi.org/10.1093/cid/civ723
Roberts KD, Azad MA, Wang J, Horne AS, Thompson PE, Nation RL, Velkov T, Li J (2015) Antimicrobial activity and toxicity of the major lipopeptide components of polymyxin B and colistin: Last-line antibiotics against multidrug-resistant Gram-negative bacteria. ACS Infect Dis 1(11):568–575. https://doi.org/10.1021/acsinfecdis.5b00085
Sandri AM, Landersdorfer CB, Jacob J, Boniatti MM, Dalarosa MG, Falci DR, Behle TF, Bordinhão RC, Wang J, Forrest A (2013) Population pharmacokinetics of intravenous polymyxin B in critically ill patients: implications for selection of dosage regimens. Clin Infect Dis 57(4):524–531. https://doi.org/10.1093/cid/cit334
Sen S, Borah SN, Bora A, Deka S (2017) Production, characterization, and antifungal activity of a biosurfactant produced by Rhodotorula babjevae YS3. Microb Cell Fact 16(1):95. https://doi.org/10.1186/s12934-017-0711-z
Shen Y, Zhou H, Jiao X, Wang Y, Zhang Q, Walsh TR, Bing S, Wu C, Hu Y, Lu Y (2018) Anthropogenic and environmental factors associated with high incidence of mcr-1 carriage in humans across China. Nat Microbiol 3(9): 1054-1062. https://doi.org/10.1038/s41564-018-0205-8
Sieber SA, Marahiel MA (2005) Molecular mechanisms underlying nonribosomal peptide synthesis: approaches to new antibiotics. Chem Rev 105(2):715–738. https://doi.org/10.1021/cr0301191
Sivanesan S, Roberts K, Wang J, Chea SE, Thompson PE, Li J, Nation RL, Velkov T (2017) Pharmacokinetics of the individual major components of polymyxin B and colistin in rats. J Nat Prod 80(1):225–229. https://doi.org/10.1021/acs.jnatprod.6b01176
Song CW, Park JM, Chung SC, Lee SY, Song H (2019) Microbial production of 2,3-butanediol for industrial applications. J Ind Microbiol Biotechnol 46:1583–1601. https://doi.org/10.1007/s10295-019-02231-0
Soo-Keun C, Soo-Young P, Rumi K, Seong-Bin K, Choong-Hwan L, Kim JF, Seung-Hwan P (2009) Identification of a polymyxin synthetase gene cluster of Paenibacillus polymyxa and heterologous expression of the gene in Bacillus subtilis. J Bacteriol 191(10):3350–3358. https://doi.org/10.1128/JB.01728-08
Soo-Young P, Soo-Keun C, Jihoon K, Tae-Kwang O, Seung-Hwan P (2012) Efficient production of polymyxin in the surrogate host Bacillus subtilis by introducing a foreign ectB gene and disrupting the abrB gene. Appl Environ Microbiol 78(12):4194–4199. https://doi.org/10.1128/AEM.07912-11
Vater J, Herfort S, Doellinger J, Weydmann M, Lasch P, Borriss R (2018) Genome mining of lipopeptide biosynthesis of Paenibacillus polymyxa E681 in combination with mass spectrometry discovery of the lipoheptapeptide paenilipoheptin. Chembiochem 19(7):744–753. https://doi.org/10.1002/biot.201400577
Velkov T, Roberts KD, Thompson PE, Li J (2016) Polymyxins: a new hope in combating Gram-negative superbugs? Future Med Chem 8(10):1017–1025. https://doi.org/10.4155/fmc-2016-0091
Wang C, Cao Y, Wang Y, Sun L, Song H (2019) Enhancing surfactin production by using systematic CRISPRi repression to screen amino acid biosynthesis genes in Bacillus subtilis. Microb Cell Fact 18(1):90. https://doi.org/10.1186/s12934-019-1139-4
Wenzler E, Bunnell KL, Danziger LH (2018) Clinical use of the polymyxins: the tale of the fox and the cat. Int J Antimicrob Agents 51(5):700–706. https://doi.org/10.1016/j.ijantimicag.2017.12.023
Wittmann M, Linne U, Pohlmann V, Marahiel MA (2010) Role of DptE and DptF in the lipidation reaction of daptomycin. FEBS J 275(21):5343–5354. https://doi.org/10.1111/j.1742-4658.2008.06664.x
Yu Z, Cai Y, Qin W, Lin J, Qiu J (2015) Polymyxin E induces rapid Paenibacillus polymyxa death by damaging cell membrane while Ca2+ can protect cells from damage. PLoS One 10(8):e0135198–e0135207. https://doi.org/10.1371/journal.pone.0135198
Yu Z, Guo C (2015) Qiu J (2015) Precursor amino acids inhibit polymyxin E biosynthesis in Paenibacillus polymyxa, probably by affecting the expression of polymyxin E biosynthesis-associated genes. Biomed Res Int 3:1–11. https://doi.org/10.1155/2015/690830
Zahra K, Prince RA, Danziger LH, Rotschafer JC, Rhomberg PR, Jones RN (2015) Microbiological Assessment of Polymyxin B Components Tested Alone and in Combination. Antimicrobial Agents Chemotherapy 59:7823–7825. https://doi.org/10.1128/AAC.01021-15
Zhang F, Wang Y, Jiang Q, Chen Q, Karthik L, Zhao Y-L, Li Z (2018) Substrate selection of adenylation domains for nonribosomal peptide synthetase (NRPS) in bacillamide C biosynthesis by marine Bacillus atrophaeus C89. J Ind Microbiol Biotechnol 45:335–344. https://doi.org/10.1007/s10295-018-2028-2
Zhang L, Cao C, Jiang R, Xu H, Xue F, Huang W, Ni H, Gao J (2018) Production of R, R-2,3-butanediol of ultra-high optical purity from Paenibacillus polymyxa ZJ-9 using homologous recombination. Bioresour Technol 261:272–278. https://doi.org/10.1016/j.biortech.2018.04.036
Zhao J, Zhao P, Quan C, Jin L, Zheng W, Fan S (2015) Comparative proteomic analysis of antagonistic Bacillus amyloliquefaciens Q-426 cultivated under different pH conditions. Biotechnol Appl Biochem 62:574–581. https://doi.org/10.1002/bab.1293
Zhi Y, Wu Q, Xu Y (2017) Genome and transcriptome analysis of surfactin biosynthesis in Bacillus amyloliquefaciens MT45. Sci Rep 7:40976. https://doi.org/10.1038/srep40976
Zhu C, Xiao F, Qiu Y, Wang Q, He Z, Chen S (2017) Lichenysin production is improved in codY null Bacillus licheniformis by addition of precursor amino acids. Appl Microbiol Biotechnol 101(4):1–9. https://doi.org/10.1007/s00253-017-8352-z
Acknowledgments
This study was supported by the National Key Research and Development Project of China (No. 2018YFA0902200), the National Natural Science Foundation of China (No. 21878224, 21576201).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yuan, Y., Xu, QM., Yu, SC. et al. Control of the polymyxin analog ratio by domain swapping in the nonribosomal peptide synthetase of Paenibacillus polymyxa. J Ind Microbiol Biotechnol 47, 551–562 (2020). https://doi.org/10.1007/s10295-020-02275-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-020-02275-7