Abstract
Lichenysin is categorized into the family of lipopeptide biosurfactants and has a variety of applications in the petroleum industry, bioremediation, pharmaceuticals, and the food industry. Currently, large-scale production is limited due to the low yield. This study found that lichenysin production was repressed by supplementation of extracellular amino acids. The global transcriptional factor CodY was hypothesized to prevent lichenysin biosynthesis under an amino acid-rich condition in Bacillus licheniformis. Thus, the codY null strain was constructed, and lichenysin production was increased by 31.0% to 2356 mg/L with the addition of precursor amino acids, and the lichenysin production efficiency was improved by 42.8% to 98.2 mg/L• h. Correspondingly, the transcription levels of the lichenysin synthetase gene lchAA, and its corresponding regulator genes comA, degQ, and degU, were upregulated. Also, the codY deletion enhanced biosynthesis of lichenysin precursor amino acids (Gln, Ile, Leu, and Val) and reduced the formation of byproducts, acetate, acetoin, and 2,3-butanediol. This study firstly reported that lichenysin biosynthesis was negatively regulated by CodY and lichenysin production could be further improved with the precursor amino acid amendment in the codY null strain.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Lichenysin, a lipopeptide biosurfactant produced by Bacillus licheniformis strains, is generally composed of a heptapeptide head group with a sequence of l-Gln1 → d-Leu2 → l-Leu3 → l-Val4 → l-Asp5 → d-Leu6 → l-Ile7 linked to a lactone ring by a C13–15 β-hydroxy fatty acid (Nerurkar 2010). Among the lipopeptide family, surfactin has been reported as the most powerful and the best characterized biosurfactant. However, lichenysin has higher surface activity, chelating activity, and hemolytic activity than surfactin (Grangemard et al. 2001), and lichenysin has potential applications in oil exploitation, detergents, environmental remediation, biological control of agriculture, and so on (Nerurkar 2010; Wu et al. 2015). However, the large-scale industrial production and commercialization of lichenysin are limited by its high production cost and low yield.
Lichenysin is synthesized by a multi-enzyme peptide synthetase complex called nonribosomal peptide synthetases (NRPSs), including four peptide synthetase genes lchAA, lchAB, lchAC, and lchA-TE (Nerurkar 2010; Madslien et al. 2013). It has been reported that the transcription of lichenysin is activated by the phosphorylated form of ComA, and this DNA-binding activator protein recognized the promoter region of lichenysin A synthetase operon (Yakimov and Golyshin 1997). However, little is known about the biosynthetic regulation mechanism of lichenysin, which limits further development of lichenysin production at an industrial scale.
The peptide of lichenysin is composed of aspartate, glutamine, leucine, isoleucine, and valine. Lichenysin fermentation generally adopts a synthetic medium, which is important to add precursor amino acids (the component units of lichenysin) in the culture medium for enhancing the lichenysin production. Adding l-glutamate and l-aspartate to the culture of B. licheniformis BAS50 increased the lichenysin A yield by two- and fourfold, respectively (Yakimov et al. 1996). Hashizume et al. (2008) discovered a new type of tripropeptin, consisting of a cyclic octapeptide core and a fatty acyl side chain, produced by Lysobacter sp. BMK333-48F3 with medium containing isoleucine, and the yield of this tripropeptin depended on the quantity of isoleucine. The addition of different amino acids significantly influenced the proportion of surfactin variants with different fatty acids (Liu et al. 2012). These studies suggested that addition of precursor amino acids might play an important role in the lipopeptide biosynthesis. Thus, it is possible to improve the yield of lichenysin by adding precursor amino acids.
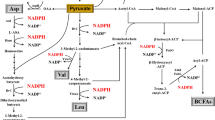
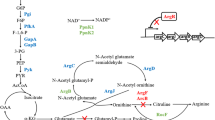
CodY is a global transcriptional regulator in many Gram-positive bacteria that controls the expression of many late-exponential-phase and early-stationary-phase genes (Sonenshein 2005). In Bacillus subtilis, the DNA-binding activity of CodY is enhanced by its interactions with two cofactors, GTP and branched-chain amino acids (BCAAs), which independently and additively increase the affinity of CodY for its target sites (Brinsmade et al. 2014; Belitsky 2015). CodY controls transcription by at least four different mechanisms: negative or positive regulation by binding a promoter site, negative regulation by interfering with the binding of a positive regulator, and negative regulation by acting as a roadblock to RNA polymerase (Belitsky and Sonenshein 2013). B. subtilis CodY inhibited the synthesis of BCAAs under amino acid-rich growth conditions (Fujita et al. 2014). BCAAs are the precursor amino acids of lichenysin biosynthesis. Hence, CodY is likely to influence the biosynthesis of lichenysin in B. licheniformis.
B. licheniformis WX02-Psrflch was constructed by our group to improve lichenysin production by promoter replacement (Qiu et al. 2014). This study aims to further increase the lichenysin production by the addition of extracellular amino acids in WX02-Psrflch and understand the regulation mechanism of lichenysin biosynthesis. It is hypothesized that CodY might negatively regulate biosynthesis of lichenysin in B. licheniformis. To test this hypothesis, we constructed the codY knockout strain, WX02-PsrflchΔcodY, and analyzed the expression levels of key genes involved in the biosynthesis of lichenysin, precursor amino acids, and the overflow metabolites with the amendment of the precursor amino acids.
Materials and methods
Bacterial strains, plasmids, primers, and growth media
Experiments were performed with the strains and plasmids listed in Table 1. The oligonucleotide primers listed in Table S1 and Table S2 were designed on the basis of the B. licheniformis WX-02 (B. licheniformis CCTCC M208065) genome sequence [GenBank: AHIF00000000] (Yangtse et al. 2012). B. licheniformis WX02-Psrflch was used as the starting strain for transformation. The Escherichia coli DH5α and B. licheniformis were grown in Luria-Bertani (LB) medium containing 10 g/L tryptone, 5 g/L yeast extract, and 10 g/L NaCl at 37 °C for 12 h. The seed culture (3% inoculum size) was transferred into 250-mL Erlenmeyer flasks containing 50 mL lichenysin fermentation medium (Qiu et al. 2014) with slight modifications comprising 30 g/L glucose, 5 g/L NH4NO3, 60 mM KH2PO4, 80 mM Na2HPO4, 0.4 mM FeSO4·7H2O, 0.4 mM MnSO4·H2O, 0.8 mM MgSO4·7H2O, 7 μM CaCl2, and 4 μM Na2-EDTA at an initial pH 7.0, and incubated at 37 °C for 36 h.
Optimization of precursor amino acid supplementation
Precursor amino acids (the component units of lichenysin) used in this experiment were l-aspartate (Asp), l-glutamate (Glu), l-isoleucine (Ile), l-leucine (Leu), and l-valine (Val). Two strategies were adopted for addition of precursor amino acids to the lichenysin fermentation medium: one was the addition of an individual amino acid at different concentrations (0.2, 0.5, and 2 g/L), and the other was the addition of an equally compounded amino acid mix at different concentrations (0.05, 0.2, 0.5, and 1 g/L). The medium without precursor amino acids added served as the control.
Construction of plasmids
The gene codY was deleted by the double-crossover homologous recombination method with the primers listed in Table S1 (Liang et al. 2015). First, two homologous arms (homologous to the 5′ and 3′ coding regions of the codY gene) of approximately 500 bp were amplified by PCR from the genomic DNA of B. licheniformis WX-02 by primers of codYKF1 and codYKR1, and codYKF2 and codYKR2, respectively. These two homologous arms were ligated by splicing with overlapping extension PCR (SOE-PCR) with primers of codYKF1 and codYKR2. The DNA fragment was sub-cloned in a vector T2(2)-ori joined by Sac I and Xba I restriction sites. The resulting plasmid was further verified by sequencing. A recombinant vector for codY deletion was designated as T2(2)-oriΔcodY (Fig. S1A).
The fusion of the P43 promoter of B. subtilis 168, codY gene, and terminator amyL gene of B. licheniformis WX-02 was amplified by PCR with primers of P43-F and P43Cy-R, Cy43-F and Cy-R, and TLCy-F and TL-R (Table S1), respectively, with the templates of genomic DNA from B. licheniformis WX-02 or B. subtilis 168. Then, the DNA fragment amplified by SOE-PCR was cloned into the plasmid of pHY300PLK joined by the EcoR I and Xba I restriction sites. The resulting plasmid was verified by sequencing. A recombinant vector for expression of codY was designated as pHYcodY (Fig. S1B).
Construction of the codY knocked-out strain of WX02-Psrflch
The codY deletion from B. licheniformis WX02-Psrflch was done by our previously established methods (Qi et al. 2014). Briefly, WX02-Psrflch was electrotransformed with T2(2)-oriΔcodY, and the transformants were then selected by kanamycin resistance (20 μg/mL) followed by verification with PCR using the primers codYKF1 and codYKR2 (Table S1). The selected positive transformants were cultured in the LB medium containing kanamycin (20 μg/mL) at 45 °C for 8 h to promote the first crossover in the cells. The selected colonies with single crossover were repeatedly cultured in the LB medium at 37 °C for 8 h. After serial transfers without antibiotics, cells were plated on LB agar plates for selecting kanamycin-sensitive colonies. The codY knockout strain was selected by the second crossover and confirmed by PCR with primers of codYYF and codYYR (Table S1) for WX02-PsrflchΔcodY (Fig. S2A).
Construction of the codY overexpression strain of WX02-Psrflch
The codY expression plasmid pHYcodY vector and noncarrier plasmid pHY300PLK were transformed into B. licheniformis WX02-Psrflch cells according to the method described previously (Tian et al. 2014), and the transformants were first selected by LB agar plates with 20 μg/mL tetracycline. The recombinant strains were named as WX02-Psrflch/pHYcodY and WX02-Psrflch/pHY300, respectively (Fig. S2B).
Extraction and determination of lichenysin
The extraction of lichenysin was performed according to our previously reported method (Qiu et al. 2014). The cell cultures were centrifuged at 8000×g for 15 min to remove the cells, and the supernatant was adjusted to pH 2.0. The acidified supernatant was stored at 4 °C overnight to precipitate the crude lichenysin. The lichenysin-containing precipitates were collected by centrifugation at 8000×g for 15 min at 4 °C and then dissolved with methanol. This solution was further centrifuged at 8000×g for 15 min for collecting the lichenysin-containing supernatant. The supernatant was then filtered through a 0.22-μm filter to determine the concentration of lichenysin by HPLC with an Agilent TC-C18 column (250 mm × 4.6 mm, 5 μm) (Qiu et al. 2014). The mobile phase consisted of 80% acetonitrile as solvent A and 100% acetonitrile as solvent B with the addition of 3.8 mM trifluoroacetic acid. The elution conditions were as follows: solvent A 50% at 0–5 min, then from 50 to 30% at 25 min, 30% at 30 min, and 50% at 31–33 min. The flow rate was 0.9 mL/min at 25 °C and detected by UV absorption at 210 nm.
Transcript analysis
The total RNA was extracted from B. licheniformis WX02-Psrflch and codY knocked-out strain at the late exponential phase using the TRIzol® Reagent (Invitrogen, USA). DNase I-treated RNA was used as the template to synthesize complementary DNA (cDNA) at 42 °C with the Thermo Scientific RevertAid First-Strand cDNA Synthesis Kit with Random Primers. Quantitative PCR analysis was performed with the Maxima® SYBR Green/ROX qPCR Master Mix (Thermo) with the manufacturer’s instructions. For analysis of the transcriptional level, resulting cDNA was used for PCR amplification with primers (Table S2) under the following conditions: 95 °C for 10 min, then 40 cycles of 95 °C (15 s), 58 °C (15 s), and 72 °C (30 s) with a single fluorescence measurement, followed with a melting curve program (a continuous fluorescence measurement from 60 to 95 °C with a slow heating unit). The 16S rRNA sequence served as the positive internal control for RT-PCR assay. All assays were performed in triplicate, and the reaction without template was used as the negative control.
Biomass and metabolite analysis
The biomass was determined by cell dry weight (CDW) and expressed as grams per liter of CDW. Residual glucose concentration was determined through a SBA-40E biosensor analyzer (Institute of Biology, Shandong Academy of Sciences, China). The production of acetoin, 2,3-butanediol, and acetate in the broth was analyzed, respectively, by GC (Qi et al. 2014) and HPLC (Santos et al. 2014).
Statistics
Data were analyzed using analysis of variance. The t test was used to determine statistical difference for the paired comparisons. All samples were run in triplicate.
Results
Effects of precursor amino acids addition on lichenysin production by B. licheniformis WX02-Psrflch
In order to improve lichenysin production, individual precursor amino acid or the amino acid mixtures (Asp, Glu, Ile, Leu, and Val) were added into the media to investigate their effects on lichenysin production in B. licheniformis WX02-Psrflch. It was found that the yield of lichenysin decreased with the addition of individual amino acid (Fig. 1a) or a mixture of amino acids (Fig. 1b) into the fermentation medium. Compared to the control, the lichenysin production decreased by 97.3% (Fig. 1a) and 26.3% (Fig. 1b), when Val was added at 2 g/L and the amino acid mixture at 1 g/L in the lichenysin fermentation medium, respectively.
Effects of various precursor amino acid concentrations on the yield of lichenysin in B. licheniformis WX02-Psrflch: a an individual precursor amino acid addition, p < 0.05 at 2 g/L versus the control, and b equally compounded precursor amino acid mixture addition, p < 0.05 at 1 g/L versus the control. All experiments in triplicate
Regulation of lichenysin production by CodY with the addition of precursor amino acids
For the codY knocked-out strain, we tested different concentrations of the precursor amino acid mixtures. The results showed that the addition of 0.5 g/L precursor amino acids led to the highest yield of lichenysin at 2527 mg/L, increased by 25% compared with the control (Fig. 2a). Therefore, the culture medium, containing 0.5 g/L of Asp, Glu, Ile, Leu, and Val, was chosen as the lichenysin fermentation medium for the subsequent experiments.
Effects of precursor amino acids on the yield of lichenysin: a equally compounded amino acid mixture addition with different concentrations in codY knocked-out strain, p < 0.05 at 0.5 g/L versus the control; b effects of codY knocked-out (p < 0.05 versus WX02-Psrflch) and overexpression (p < 0.05 versus WX02-Psrflch) mutants on the lichenysin production. All experiments in triplicate
Effects of codY on the lichenysin production with the presence of precursor amino acids were investigated in the codY deletion and overexpression strains. The lichenysin production increased by 25.7% in the codY knockout strain WX02-PsrflchΔcodY strain (2301 mg/L) and decreased by 94.5% in the codY overexpressed strain WX02-Psrflch/pHYcodY strain (100 mg/L) compared to the control strain WX02-Psrflch (1831 mg/L) in Fig. 2b. Also, the yield of lichenysin at 1619 mg/L in noncarrier control strain WX02-Psrflch/pHY300 decreased slightly compared to WX02-Psrflch. These results indicated that the yield of lichenysin was improved in the codY deletion mutant under the precursor amino acid amendment, and suggested that the CodY transcription factor indeed negatively regulated biosynthesis of lichenysin.
Effects of codY deletion on cell growth and lichenysin biosynthesis
Since the codY null strain, WX02-PsrflchΔcodY, was confirmed to produce the highest level of lichenysin in Fig. 2, it is worthwhile to further study how CodY regulates lichenysin biosynthesis. As shown in Fig. 3a, both WX02-PsrflchΔcodY and the control WX02-Psrflch showed strong growth in the first 10 h, concomitantly with the glucose depletion. WX02-PsrflchΔcodY, however, reached a slightly higher biomass at 5.3 g/L, increased by 12.8%.
Effects of codY deletion on the fermentation of lichenysin under the addition of precursor amino acids: a time profiles of residual glucose, cell growth, and lichenysin production; b transcriptional levels of the key genes involved in lichenysin biosynthesis. p < 0.05 for lchAA, comA, and degQ, respectively, in WX02-Psrflch versus WX02-Psrflch△codY. All experiments in triplicate
The maximum yield of lichenysin reached 2356 mg/L, 31.0% higher than the control at 1798 mg/L. Moreover, the lichenysin fermentation period, defined as the duration prior to the maximum yield, of codY knocked-out strain was cut short from 32 to 24 h, which means fermentation efficiency was greatly improved by 42.8% from 56.2 to 98.2 mg/L• h. These results indicated that codY deletion mutant helped to improve not only the lichenysin yield but also the production efficiency.
To further understand the regulation mechanism for enhanced lichenysin production by codY deletion under precursor amino acid amendment, the transcriptional levels of several genes involved in lichenysin biosynthesis were analyzed. lchAA is the one of lichenysin synthetase genes, and the result showed that lchAA transcriptional level in the codY knockout strain was increased by 1.8-fold compared with the control (WX02-Psrflch) (Fig. 3b). Meanwhile, the transcriptional levels of the regulators comA, degQ, and degU were also measured, and their transcriptional levels were upregulated by 1.9-fold, 3.2-fold, and 1.2-fold, respectively, in the codY deletion mutant (Fig. 3b). These results suggested that codY deletion stimulated the expression of ComA and DegU, which further activated lchAA gene to enhance lichenysin production.
Effects of codY deletion on biosynthesis of precursor amino acids
Asp, Gln, Ile, Leu, and Val were the precursor amino acids for biosynthesis of lichenysin, but it is difficult to determine their in vivo concentrations due to their transient transformation to lichenysin. Therefore, the genes responsible for precursor amino acid biosynthesis were investigated, instead of the amino acids. The results showed that the transcriptional levels of key genes involved in branched-chain amino acid biosynthesis, ilvA, ilvB, leuA, bcd, and alsS, were all upregulated by 2.3-fold, 1.2-fold, 2.5-fold, 7.5-fold, and 12.4-fold, respectively, in the codY deletion mutant, compared to the control (WX02-Psrflch) (Fig. 4). The expression of Gln and Glu biosynthesis genes glnA and rocG was upregulated by 3.1-fold and 2.5-fold, respectively, and the transcription of the Asp biosynthesis gene aspB remained unchanged. These results indicated that codY deletion enhanced the biosynthesis of intracellular Gln, Ile, Leu, and Val, which may further improve the lichenysin production.
Effects of codY deletion on the overflow metabolites
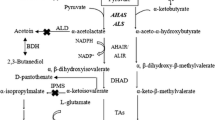
In addition, in vitro overflow metabolites were analyzed, and acetate, acetoin, and 2,3-butanediol were found to be major overflow metabolites affecting lichenysin production. The maximum yields of acetate (2.8 g/L) in Fig. 5a, acetoin (1.7 g/L) in Fig. 5b, and 2,3-butanediol (2 g/L) in Fig. 5c were sharply decreased by 50.9, 41.4, and 25.9% in the codY deletion mutant, respectively. These results showed that the codY deletion reduced the overflow metabolites during lichenysin fermentation, which could be one reason for the enhanced lichenysin production.
Effects of codY deletion on the overflow metabolites during lichenysin fermentation: a the changes of acetate concentration; b the changes of acetoin concentration; c the changes of 2,3-butanediol concentration; d transcriptional levels of the key genes involved in biosynthesis of overflow metabolites. p < 0.05 for lchAA and ackA, respectively, in WX02-Psrflch versus WX02-Psrflch△codY. All experiments in triplicate
Also, the gene expression levels of these key overflow metabolites were further analyzed. Acetate is synthesized from pyruvate via acetate kinase encoded by ackA and acetoin from pyruvate via -acetolactate decarboxylase encoded by alsD. Subsequently, 2,3-butanediol is biosynthesized from acetoin by butanediol dehydrogenase. As shown in Fig. 5d, the transcriptional levels of ackA and alsD were reduced by 50.0 and 12.5% compared to the control strain WX02-Psrflch. Thus, the transcriptional levels of ackA and alsD elucidated the possible mechanism for the reduced overflow metabolites under codY deletion.
Discussion
Synthetic chemical surfactants have been widely used in detergents, functional food additives, oil recovery, and impose environmental problems after discharge into the surroundings. Hence, biosurfactants, a class of biodegradable and eco-friendly compounds, are attracting growing attention. Lichenysin exhibits high surface activity, low toxicity, and antimicrobial activity. Low yield leads to the bottleneck for the wide applications of biosurfactants.
B. licheniformis WX02-Psrflch was previously constructed in our group by promoter replacement for enhanced lichenysin biosynthesis (Qiu et al. 2014), which is one common strategy applied for production improvement of many microbial metabolites (Jiao et al. 2017; Dangel et al. 2010; Watanabe et al. 2016). The addition of precursor amino acids was reported to increase the biosurfactant yields (Yakimov et al. 1996; Hashizume et al. 2008; Liu et al. 2012).
In order to further improve lichenysin production, individual precursor amino acid or amino acid mixtures were added into the media to investigate their effects on lichenysin production. Our results, however, indicated that the addition of precursor amino acids suppressed the lichenysin production. In B. subtilis, the activation of ilv-leu expression was negatively regulated through CodY binding to the promoter region under an amino acid-rich growth condition, which further inhibited the biosynthesis of BCAAs (Fujita et al. 2014). Therefore, we hypothesized that the addition of precursor amino acids activated the CodY-dependent repression of the BCAA biosynthesis during lichenysin fermentation in B. licheniformis WX02-Psrflch, which subsequently affected the supply of precursor amino acids for lichenysin biosynthesis, and further decreased the lichenysin production. In other words, CodY binding affinity to the promoter region of ilv-leu operon is increased with the cofactor branched-chain amino acids (BCAAs), which negates the biosynthesis of BCAAs and subsequently reduces the supplies of these precursor amino acids for lichenysin synthesis.
The codY knockout strain was constructed to validate this hypothesis, and lichenysin production was indeed improved by 31% with the addition of precursor amino acids. In order to investigate the regulation mechanism of CodY on lichenysin production, gene expression levels were analyzed according to the lichenysin biosynthetic pathway. lchAA was one of lichenysin synthetase genes and mainly responsible for the biosynthesis of lichenysin directly (Qiu et al. 2014). In B. subtilis, the regulators ComA and DegU positively regulated srfA operon for surfactin production (Duitman et al. 2007; Roongsawang et al. 2010), and DegQ promoted the phosphorylation of DegU (Kobayashi 2007). As the global transcriptional factor, CodY negatively regulated the lchAA and its regulator genes comA and degU, as well as the genes involving BCAA biosynthesis, with the addition of precursor amino acids in B. licheniformis. In short, CodY negatively regulates the regulator genes comA and degU, which positively regulate lichenysin synthetase genes. As the cascade regulation, CodY eventually represses lichenysin biosynthesis.
Since CodY was discovered to repress BCAA biosynthesis (Fujita et al. 2014) and lichenysin biosynthesis in this study, these results well explained that the addition of precursor amino acids decreased lichenysin yield in B. licheniformis. It is noteworthy that CodY also positively regulated production of overflow metabolites, acetate, acetoin, and 2,3-butanediol. Acetate is produced from pyruvate via acetate kinase encoded by ackA in B. subtilis (Shivers et al. 2006). Acetoin is produced from pyruvate via -acetolactate decarboxylase encoded by alsD, and 2,3-butanediol is subsequently synthesized from acetoin by butanediol dehydrogenase (Wang et al. 2016). Biosynthesis of these metabolites was deactivated by knocking out codY, which could be another explanation for enhanced lichenysin production by potentially lowering the metabolic flux of the byproducts. In a word, CodY positively regulates biosynthesis of acetate, acetoin, and 2,3-butanediol. Therefore, knocking out codY represses biosynthesis of these byproducts, which helps the metabolic flux towards lichenysin production.
References
Belitsky BR (2015) Role of branched-chain amino acid transport in Bacillus subtilis CodY activity. J Bacteriol 197:1330–1338
Belitsky BR, Sonenshein AL (2013) Genome-wide identification of Bacillus subtilis CodY-binding sites at single-nucleotide resolution. Proc Natl Acad Sci 110:7026–7031
Brinsmade SR, Alexander EL, Livny J, Stettner AI, Segrè D, Rhee KY, Sonenshein AL (2014) Hierarchical expression of genes controlled by the Bacillus subtilis global regulatory protein CodY. Proc Natl Acad Sci 111:8227–8232
Dangel V, Westrich L, Smith MCM, Heide L, Gust B (2010) Use of an inducible promoter for antibiotic production in a heterologous host. Appl Microbiol Biotechnol 87:261–269
Duitman EH, Wyczawski D, Boven LG, Venema G, Kuipers OP, Hamoen LW (2007) Novel methods for genetic transformation of natural Bacillus subtilis isolates used to study the regulation of the mycosubtilin and surfactin synthetases. Appl Environ Microbiol 73:3490–3496
Fujita Y, Satomura T, Tojo S, Hirooka K (2014) CcpA-mediated catabolite activation of the Bacillus subtilis ilv-leu operon and its negation by either CodY- or TnrA-mediated negative regulation. J Bacteriol 196:3793–3806
Grangemard I, Wallach J, Maget-Dana R, Peypoux F (2001) Lichenysin: a more efficient cation chelator than surfactin. Appl Biochem Biotechnol 90:199–210
Hashizume H, Igarashi M, Sawa R, Adachi H, Nishimura Y, Akamatsu Y (2008) A new type of tripropeptin with anteiso-branched chain fatty acid from Lysobacter sp. BMK333-48F3. J Antibiot 61:577–582
Jiao S, Li X, Yu HM, Yang H, Li X, Shen ZY (2017) In situ enhancement of surfactin biosynthesis in Bacillus subtilis using novel artificial inducible promoters. Biotechnol Bioeng 114(4):832–842
Kobayashi K (2007) Gradual activation of the response regulator DegU controls serial expression of genes for flagellum formation and biofilm formation in Bacillus subtilis. Mol Microbiol 66:395–409
Liang C, Huo Y, Qi G, Wei X, Wang Q, Chen S (2015) Enhancement of L-valine production in Bacillus licheniformis by blocking three branched pathways. Biotechnol Lett 37:1243–1248
Liu JF, Yang J, Yang SZ, Ye RQ, Mu BZ (2012) Effects of different amino acids in culture media on surfactin variants produced by Bacillus subtilis TD7. Appl Biochem Biotechnol 166:2091–2100
Madslien EH, Ronning HT, Lindback T, Hassel B, Andersson MA, Granum PE (2013) Lichenysin is produced by most Bacillus licheniformis strains. J Appl Microbiol 115:1068–1080
Nerurkar AS (2010) Structural and molecular characteristics of lichenysin and its relationship with surface activity. Biosurfactants 672:304–315
Qi GF, Kang YF, Li L, Xiao AF, Zhang SM, Wen ZY, Xu DH, Chen SW (2014) Deletion of meso-2,3-butanediol dehydrogenase gene budC for enhanced D-2,3-butanediol production in Bacillus licheniformis. Biotechnol Biofuels 7:6–12
Qiu Y, Xiao F, Wei X, Wen Z, Chen S (2014) Improvement of lichenysin production in Bacillus licheniformis by replacement of native promoter of lichenysin biosynthesis operon and medium optimization. Appl Microbiol Biotechnol 98:8895–8903
Roongsawang N, Washio K, Morikawa M (2010) Diversity of nonribosomal peptide synthetases involved in the biosynthesis of lipopeptide biosurfactants. Int J Mol Sci 12:141–172
Santos CC, Libeck BS, Schwan RF (2014) Co-culture fermentation of peanut-soy milk for the development of a novel functional beverage. Int J Food Microbiol 186:32–41
Shivers RP, Dineen SS, Sonenshein AL (2006) Positive regulation of Bacillus subtilis ackA by CodY and CcpA: establishing a potential hierarchy in carbon flow. Mol Microbiol 62:811–822
Sonenshein AL (2005) CodY, a global regulator of stationary phase and virulence in Gram-positive bacteria. Curr Opin Microbiol 8:203–207
Tian G, Fu J, Wei X, Ji Z, Ma X, Qi G, Chen S (2014) Enhanced expression of pgdS gene for high production of poly-γ-glutamic aicd with lower molecular weight in Bacillus licheniformis WX-02. J Chem Technol Biotechnol 89:1825–1832
Wang J, Yuan H, Wei X, Chen J, Chen S (2016) Enhancement of poly-γ-glutamic acid production by alkaline pH stress treatment in Bacillus licheniformis WX-02. J Chem Technol Biotechnol 91(9):2399–2403
Watanabe T, Morita T, Koike H, Yarimizu T, Shinozaki Y, Sameshima-Yamashita Y, Yoshida S, Koitabashi M, Kitamoto H (2016) High-level recombinant protein production by the basidiomycetous yeast Pseudozyma antarctica under a xylose-inducible xylanase promoter. Appl Microbiol Biotechnol 100(7):3207–3217
Wu Q, Zhang R, Peng S, Xu Y (2015) Transcriptional characteristics associated with lichenysin biosynthesis in Bacillus licheniformis from Chinese Maotai-flavor liquor making. J Agr Food Chem 63:888–893
Yakimov MM, Golyshin PN (1997) ComA-dependent transcriptional activation of lichenysin A synthetase promoter in Bacillus subtilis cells. Biotechnol Prog 13:757–761
Yakimov MM, Fredrickson HI, Timmis KN (1996) Effect of heterogeneity of hydrophobic moieties on surface activity of lichenysin A, a lipopeptide biosurfactant from Bacillus licheniformis BAS50. Biotechnol Appl Biochem 23:13–18
Yangtse WM, Zhou YH, Lei Y, Qiu YM, Wei XT, Ji ZX, Qi GF, Yong YC, Chen LL, Chen SW (2012) Genome sequence of Bacillus licheniformis WX-02. J Bacteriol 194:3561–3562
Acknowledgements
This work was funded by the National Science & Technology Pillar Program during the Twelfth Five-Year Plan Period (2013AA102801-52), the National Program on Key Basic Research Project (973 Program, No. 2015CB150505), and the Science and Technology Program of Wuhan (20160201010086).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
This paper does not contain any studies with human participants or animals performed by any of the authors.
Electronic supplementary material
ESM 1
(PDF 301 kb)
Rights and permissions
About this article
Cite this article
Zhu, C., Xiao, F., Qiu, Y. et al. Lichenysin production is improved in codY null Bacillus licheniformis by addition of precursor amino acids. Appl Microbiol Biotechnol 101, 6375–6383 (2017). https://doi.org/10.1007/s00253-017-8352-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-017-8352-z