Abstract
Background
Cholangiocarcinoma (CCA) is a rare malignant tumor of the biliary system. The heterogeneity of CCA leads to the lack of effective targeted treatment for CCA subtypes. The molecular characteristic of hilar CCA (hCCA) is still unclear.
Methods
A total of 63 hCCA patients were enrolled from Shanghai Eastern Hepatobiliary Surgery Hospital. Formalin-fixed, paraffin-embedded tumor tissues, and matched blood were collected and deep sequencing targeting 450 cancer genes were performed. Tumor mutation burden (TMB) was measured by an algorithm developed in-house. Correlation analysis was performed by Fisher’s exact test.
Results
The most commonly mutated genes were TP53 (51.7%), NF1 and KRAS (20%, for both), SMAD4 (16.7%), FAT3 and FRS2 (13.3%, for both), NF1 (11.7%), and KMT2C, MDM2, and ATM (10%, for each) in hCCA. ARID1A, GATA6, and PREX2 mutations commonly occurred in female and KMT2C mutations mainly occurred in patients under 60 years old. Statistical analysis showed the association between ARID1A mutation and tumor stage (P = 0.041) and between NF1 mutation and high TMB (P = 0.0095). Furthermore, ARID1B mutation was identified to associate with the poor prognosis of Chinese hCCA patients (P = 0.004).
Conclusion
The mutational characterization of hCCA is different from both extrahepatic CCA and intrahepatic CCA. ARID1B is a potential biomarker for prognosis prediction of Chinese hCCA patients.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cholangiocarcinoma (CCA) is a rare malignant tumor of the biliary system, which seriously threatens the life of patients [1]. According to the location of the disease, CCA was classified into intrahepatic CCA which is located within the hepatic parenchyma and extrahepatic CCA which consisted of hilar CCA (hCCA) and distal CCA [2]. Surgery is still the effective treatment for early CCA, although only a small subset of patients could be diagnosis because of the unclear clinical symptoms of early CCA [3]. The insensitivity of CCA to radiotherapy and chemotherapy leads to poor prognosis [4, 5]. Targeted therapy and immunotherapy based on biomarkers are effective treatments for malignant tumors [6,7,8]. However, there are few effective biomarkers for CCA, which need to be developed and explored for early identification and diagnosis.
The location-based classification is helpful to determine the preoperative treatment in clinic. Anyway, the boundary between subtypes is still ambiguous [9]. The heterogeneity of CCA leads to the lack of effective targeted treatment for CCA subtypes differ in response to the treatment [10]. With the development of next-generation sequencing (NGS) technology, it is possible to discover the differences among subtypes at the molecular level. Recent studies have shown that there are different molecular characteristics between intrahepatic CCA and extrahepatic CCA [11, 12]. Comprehensive whole-exome and transcriptome sequencing in a large cohort of 260 patients also revealed potentially targetable genetic driver alterations [13]. For example, the specific common mutations in intrahepatic CCA were IDH1, MCL1, PBRM1, FGFR2, and FGFR 3/4/19, whereas FBXW7, ERBB2, and RBM10 in extrahepatic CCA [11,12,13]. NGS studies revealed the genomic heterogeneity of CCA subtypes potentially affecting the future therapy trials [11]. Although extrahepatic CCA can be divided into hCCA and distal CCA, the prognosis of them were different. Waseem et al. reported that the mean survival of hCCA was lower than distal CCA, but similar to intrahepatic CCA [14]. Until now, few studies isolated hCCA and focused on its genomic characteristics.
In this study, we enrolled 63 Chinese hCCA patients to characterize their comprehensive genomic profiling, and aimed to identify the potential biomarkers for prognosis and provide evidence for further targeted therapy and immunotherapy.
Patients and methods
Patient enrollment and sample collection
From 2012 to 2019, 63 hCCA patients were enrolled from Shanghai Eastern Hepatobiliary Surgery Hospital according to the tumor locations. Informed consent was obtained from all patients and this study was approved by the Institutional Ethics Committee of Shanghai Eastern Hepatobiliary Surgery Hospital. According to the results of computed tomography or magnetic resonance imaging, the patients were given the necessary jaundice-reducing treatment. After the total bilirubin was less than 5 times of normal, surgical resection was performed. The tumor tissue samples were fixed in formalin, and then were embedded in paraffin within 24 h. Meanwhile, matched blood samples were collected as control. Formalin-fixed, paraffin-embedded (FFPE) tumor tissues containing at least 20% of tumor cells are considered to be composed of tumor tissue and can be used for further NGS detection.
Identification of genomic alterations and tumor mutation burden
DNAs of both FFPE tumor tissues and matched blood were obtained using QIAamp DNA FFPE Tissue Kit and QIAamp DNA Blood Midi Kit (Qiagen, Hilden, Germany), respectively, and sequenced using the next-generation sequencing-based YuanSu450™ gene panel of OrigiMed (Shanghai, China), from where the laboratory was certified by College of American Pathologists (CAP) and Clinical Laboratory Improvement Amendments (CLIA). The genes were captured and sequenced with a mean depth of 800 × using Illumina Nova (Illumina, Inc., CA). Genomic alteration was identified as following [15]: single-nucleotide variants (SNVs) were identified by MuTect (v1.7). Insertion–deletions (Indels) were identified using PINDEL (V0.2.5). The functional impact of genomic alterations was annotated by SnpEff3.0. Copy-number variation (CNV) regions were identified by Control-FREEC (v9.7) with the following parameters: window = 50 000 and step = 10 000. Gene fusions were detected through an in-house developed pipeline. Gene rearrangements were assessed by Integrative Genomics Viewer (IGV). Tumor mutation burden (TMB) was calculated by counting the coding somatic mutations, including SNVs and Indels, per megabase of the sequence examined in each patient.
Statistical analysis
Statistical analyses were performed using SPSS version 22.0 (SPSS Inc., Chicago, IL, USA). The Kaplan–Meier method and Cox regression were used to analyze survival. Fisher’s exact test was used to analyze significant differences. P < 0.05 was considered statistically significant.
Results
Clinical characteristics of hCCA patients
A total of 63 hCCA patients with a median age of 59 years (range 38–85 years) were enrolled in this study. These samples consisted of 41 (65.1%) male and 22 (34.9%) female. According to the pathological examination records, the tumor of patients was classified into stage I (4/63, 6.4%), stage II (35/63, 55.6%), stage III (16/63, 25.4%), and stage IV (5/63, 7.9%). The tumor stage of 3 (4.8%) patients was unclear. Three of the 63 patients harbored hepatitis B virus and no one harbored hepatitis C virus. The 27% of patients were identified as lymph-node metastasis positive. 57 (90.5%) patients have had radical surgery, and 54 of them were followed up, including 33 patients received postoperative adjuvant chemotherapy, 15 patients did not receive postoperative adjuvant treatment, and 6 patients with unknown postoperative treatment. According to intraoperative exploration, 8 (12.7%) patients were diagnosed with vascular invasion, 54 (85.7%) had no vascular invasion, and 1 patient had unknown information. According to Bismuth–Corlette classification [16], 8 patients were type I, 9 patients were type II, 17 patients were type IIIa, 25 patients were type IIIb, 2 patients were type IV, and 2 patients with unclear Bismuth–Corlette type. Patients’ clinical or pathological information is summarized and shown in Table 1.
Genomic alterations in hCCA
Three of the 63 patients did not detect the effective alterations. A total of 545 clinically relevant genomic alterations in 263 genes were identified in 60 hCCA patients. All these alterations included 331 (60.7%) substitution/Indels, 102 (18.7%) truncations, 89 (16.3%) gene amplifications, 21 (3.85%) fusion/rearrangement, and 2 (0.37%) gene homozygous deletions (Table S1). The most commonly mutated genes were TP53 (51.7%, 31/60), NF1 and KRAS (20%, 12/60, for both), SMAD4 (16.7%, 10/60), FAT3 and FRS2 (13.3%, 8/60, for both), NF1 (11.7%, 7/60), and KMT2C, MDM2, and ATM (10%, 6/60, for each) (Fig. 1). The most common mutations of ARID1A and SMAD4 were truncation mutant (10/12 and 7/10, respectively). The most common mutations of FRS2 and MDM2 were gene amplification (7/8 and 6/6). Notably, FRS2 and MDM2 amplifications were occurred simultaneously in 6 patients (Fig. 1).
Mutational landscape of 60 Chinese hCCA patients. The X-axis shows each case sample and the Y-axis shows each mutated gene. The bar graph upside shows the TMB value of the patients. The bar graph on the right shows the numbers of each mutated gene. Green represents substitution/indel mutations, red represents gene amplification mutations, blue represents gene homozygous deletion mutations, yellow represents fusion/rearrangement mutations, and purple represents truncation mutations
Correlations between mutated genes and the clinical characteristics of Chinese hCCA patients
To explore the potential biomarker, we performed association analyses between mutated genes and clinical characteristics such as gender and age. The most frequent mutated genes were TP53 (48.8%, 20/41), KRAS (17.07%, 7/41), SMAD4 (14.6%, 6/41), ATM and FAT3 (12.2%, 5/41, for both) in male, while TP53 (50%, 11/22), ARID1A (36.4%, 8/22), KRAS (22.7%, 5/22), FRS2, KMT2C, NF1, and SMAD4 (18.2%, 4/22, for each) in female. Statistical analysis showed that the mutational frequencies of ARID1A (P = 0.017), GATA6 (P = 0.039), and PREX2 (P = 0.039) were significantly higher in female than in male patients (Fig. 2a).
Based on tumor stage, we classified stage I and II into a group, and stage III and IV into another group, and found that ARID1A mutations were mainly occurred in stage I/II group. Statistical analysis showed a significantly association between ARID1A mutations and tumor stage I/II (P = 0.041) (Fig. 2b). In this study, there were 8 patients with vascular invasion. Statistical analysis showed that there was an association between KRAS mutation and vascular invasion (P = 0.043) (Fig. 2c).
In this cohort, most of the patients were over 40 years old, including 9 patients under 50 years old (1 of them was 38 years old), 14 patients between 50 and 59 years old, 29 patients between 60 and 69 years old, and 11 patients over 70 years old. Based on genomic alterations, we found the mutation of KMT2C mainly occurred in the patients under 60 years old. Statistical analysis also showed a significant association between age and the mutation of KMT2C (P = 0.002) (Fig. 2d). We also analyzed the clinical characteristic of lymph-node metastasis and no significantly associated gene mutations were detected.
We identified the TMB value of 60 patients with clinically relevant genomic alterations. The median TMB value was 3.8 mutations/Mb, ranged from 0 to 49.5 mutations/Mb. To explore TMB-related mutations, we divided patients into mutant and wild-type groups for each mutated gene. Our results showed that patients with NF1 mutation had a significant higher TMB than those without NF1 mutations (P = 0.0095) (Fig. 2e).
Bismuth–Corlette IIIa tumors located at the confluence of left and right hepatic ducts and invaded right hepatic ducts and Bismuth–Corlette Type IIIb tumors located at confluence of left and right hepatic ducts and invaded left hepatic ducts. We also analyze the association between mutated genes and Bismuth–Corlette subtype IIIa and IIIb. However, there were not any mutated genes associated with the invasion direction of the third subtype tumor was detected. Interestingly, a significant association between gender and invasion directions of Bismuth–Corlette subtype III tumor were identified (Fig. 2f).
ARID1B and RBM10 mutations were associated with the disease-free survival
Fifty-four patients with radical surgery were followed up and 51 of them were detected the effect alterations. The median disease-free survival (DFS) was 16 months (ranged from 1 to 54 months). Taking DFS as a continuous variable, we found the association between ARID1B and RBM10 mutations and DFS. Survival curve analysis showed that patients harboring ARID1B and RBM10 mutations had a shorter DFS time than those without mutations (Fig. 3a). To further confirm this result, gender, age, TMB, and other clinical types of patients were considered, and a multivariate cox regression analysis was performed. The results showed that ARID1B (P = 0.004) mutations were still significantly associated with shorter DFS, while RBM10 did not associate with DFS any more (Fig. 3b). This result indicated that the association between RBM10 and DFS was easily neutralized by other clinical characters. Meanwhile, multivariate cox regression analysis also showed that gender (P = 0.023) might be a potential factor in response to the correlation between these mutated genes and DFS.
Correlation analysis between mutated genes and disease-free survival (DFS). a Kaplan–Meier curves of the DFS in patients with (red)/without (blue) ARID1B and RBM10 mutations. b Multivariate cox regression analysis to confirm the correlation between DFS and ARID1B and RBM10 mutations. Forest plot showed the risk of DFS in various subgroups of patients such as gender, age, and tumor grade
Actionable target mutations of hCCA
Actionable alterations in various types of cancers were collected and summarized by the OncoKB team, and 17 clinically relevant genes with 26 potential therapies for CCA [17], such as cobimetinib/binimetinib/trametinib were potential target drug for KRAS mutations, debio1347/BGJ398/erdafitinib/AZD4547 were potential target drug for FGFR mutations, and trametinib/cobimetinib was potential target drug for NF1 mutations. In this cohort, there were 12 actionable mutated genes in 34 (54%) hCCA patients. The most common drug target mutations were KRAS (19.05%), FGFR (15.87%), and NF1 (11.11%) (Table 2). Interestingly, cobimetinib is the potential target drug for both KRAS mutation and NF1 mutation, and our data showed that nearly 30.16% (19/63) of hCCA patients harboring KRAS mutations or NF1 mutations may potentially benefit from it.
Discussion
CCA is a tumor with high heterogeneity, which occurred in the locations of intrahepatic, hilar, and distal common bile duct. Previous studies have suggested that hCCA and distal CCA are included in extrahepatic CCA, thus distinguishing them from intrahepatic CCA [18]. However, some intrahepatic CCA is the invasion of hCCA [18]. Akita et al. divided the intrahepatic CCA into perihilar CCA and peripheral CCA based on histologic [19]. Particularly, hCCA have been variably and inconsistently coded as either intrahepatic CCA or distal CCA. Although these three types of CCA are distinct in their presentation and natural history, as well as the approach to diagnosis and management [20, 21], few hCCA molecular characteristics have been reported. Here, we enrolled 63 hCCA patients and identified the mutational profile. In addition to the most common mutations of TP53, KRAS, SMAD4, ARID1A, and CDKN2A/B in CCA [20,21,22], the high mutation frequencies of MDM2 and FRS2 were detected in hCCA.
MDM2 and FRS2 are located at 12q13-15 chromosomal band and they are close to each other. This may be the main reason for the co-amplification of MDM2 and FRS2 in this cohort. Amplification of 12q13-15 region often occurred in liposarcoma tumors and low-grade osteosarcoma [23]. Previous studies showed that FRS2 and MDM2 amplification associated with the differentiation of liposarcoma [24]. FRS2 is a downstream binding protein of tyrosine kinase receptor and involved in the process of cell differentiation, proliferation, and tumorigenesis [25]. FRS2 can be phosphorylated by FGFRs to activate downstream pathways, such as MAPK and PI3K/Akt/mTOR pathways, so as to make tumor progress [25, 26]. In breast cancer, FRS2 is a biomarker with high risk of tamoxifen adjuvant therapy [27]. The high frequency of MDM2 and FRS2 amplification in hCCA supported the specific molecular mutational feature of hCCA, which may provide evidence for further precision medicine of hCCA.
KMT2C is a tumor suppressor due to its frequent mutations in multiple types of tumors [28,29,30]. KMT2C is associated with the poor prognosis in acute myeloid leukemia [31]. While in breast cancer, the association between KMT2C mutation and prognosis is controversial [32, 33]. Wang et al. reported that KMT2C mutations were more frequently occurred in patients over 50 years [33]. While in this study, the patients with KMT2C mutation were all under 60 years old, which indicated the association between KMT2C and age in hCCA. However, the function of KMT2C mutations is limited [33, 34].
NF1 encodes a GTPase activating protein and functions as a tumor suppressor gene in immature myeloid [35, 36]. Mutations of NF1 may lead to increased proliferation and tumorigenesis [37]. In CCA, low frequency of NF1 mutation was detected [12, 20,21,22]. A similar mutation frequency of NF1 in hCCA to intrahepatic CCA was detected in this study. Also, we first identified the association between NF1 mutation and high TMB in CCA. High TMB means to have more potential opportunity to benefit from immunotherapies [38, 39]. PD-L1 expression is also a biomarker for immunotherapy prediction [40]. Mou et al. reported intrahepatic CCA patients with high TMB and PD-L1-positive which exhibited a successful response to the combination of immunotherapy and chemotherapy [41]. Wang et al. also showed increased expression of PD-L1 on NF-associated tumors [42]. Together, our result implied that patients with NF1 mutation may have potential opportunity to be benefit from Immunotherapy.
ARID1A mutation is a frequent event in endometriosis-related ovarian carcinomas [43]. Low expression of ARID1A correlates with poor prognosis in intrahepatic CCA [44]. Although we did not detect the correlation between ARID1A mutation and DFS in this study, we found the significant association between ARID1A mutation and early tumor. ARID1A mutation and GATA 6 mutation were associated with gender in this study. These results are similar with the previous study in kinds of cancers [45, 46]. The previous study showed that GATA6 was a new predictor for poor prognosis of ovarian cancer [47]. Interestingly, our results also showed that gender may be a potential factor associated with DFS. These results implied the possible association between ARID1A and GATA6 mutations and the prognosis of female hCCA patients. However, further confirmation is still needed.
Vascular invasion is associated with high tumor grade [48]. There were 8 patients with vascular invasion in this study and all of them were of high tumor stage (III/IV). Interestingly, the association between KRAS mutation and vascular invasion were identified. KRAS mutation is a predictor for poor prognosis in many cancers [49, 50]. Similarly, although patients with KRAS mutation may benefit from cobimetinib/binimetinib/trametinib, our results also support that patients with KRAS mutations may have a higher risk of vascular invasion and poor prognosis.
Mutations in the ARID1B gene, which shares approximately 60% similarities in amino acid sequence with ARID1A, are a component of SWI/SNF chromatin remodeling complex and may play a role in cell cycle activation [51]. It is reported that the low expression of ARID1B is associated with the poor prognosis in bladder urothelial carcinoma and ovarian clear cell carcinoma [52, 53]. RBM10 is involved in the tissue damage repair and plays an important role in tumor progression in many cancer types [54,55,56,57]. The mutation of RBM10 was associated with the poor prognosis in lung adenocarcinoma [58]. In this study, our results showed the significant association between the mutations of ARID1B and RBM10 and short DFS. Further multivariate cox regression analysis confirms the associations between ARID1B and DFS, but not support the association between RBM10 and DFS. This may be due to the small cohort in this study. However, our results supported that Chinese hCCA patients with ARID1B mutation may have a poor prognosis. In total, we first reported the association between the mutation of ARID1B and short DFS, and suggested that ARID1B may be a potential prognosis biomarker for hCCA.
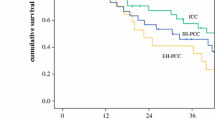
So far, there have been many studies on the mutation characteristics of CCA [21]. Previous studies have shown that the molecular characteristics of patients from different regions are different [12, 20]. All cases come from a single case center is a deficiency of this study. It is possible that the mutation characteristics of hCCA patients in this study may be different from those in other parts. To avoid the differences caused by regions, we compared the mutational characteristics of Chinese hCCA, intrahepatic CCA [22], and extrahepatic CCA [20]. The most common mutations of intrahepatic CCA were TP53, ARID1A, CDKN2A/B, TERT, IDH1/2, FGFR1/2/3/4, PBRM1, and SMAD4 [22]. While the most common mutations of extrahepatic CCA were TP53, KRAS, SMAD4, ARID1A, CDKN2A/B, TERT, and RBM10 [20]. In this study, the high-frequency mutations were TP53, KRAS, ARID1A, SMAD4, FGFR1/2/3/4, FRS2, CDKN2A/B, and MDM2. Compared with the most common mutated genes from intrahepatic CCA and extrahepatic CCA, we described the molecular characteristic of hCCA as follows: (I) Gene mutations similar to those in intrahepatic CCA, including TP53, KRAS, FGFR, PBRM1, and NF1. The mutational frequencies of TP53 and KRAS were significantly lower in hCCA and intrahepatic CCA than in extrahepatic CCA. Mutation information of FGFR, PBRM1, and NF1 from extrahepatic CCA was not available. (II) The mutational frequency of IDH1 was significantly lower in hCCA than in intrahepatic CCA, but similar to that of extrahepatic CCA. (III) Genes with higher mutational frequency in hCCA than in intrahepatic CCA and extrahepatic CCA, such as MDM2 and FRS2. (IV) Mutations in hCCA are similar to those in intrahepatic CCA and extrahepatic CCA, such as SMAD4, ARID1A, CDKN2A/B, TERT, and RBM10. Interestingly, there was a significant difference in SMAD4 mutation frequency between intrahepatic CCA and extrahepatic CCA, but no significant difference between hCCA and both intrahepatic CCA and extrahepatic CCA (Fig. 4). Meanwhile, lower frequency of KRAS, CDKN2A, BRAF, and IDH1, and higher frequency of ATM and PTEN were in hCCA. These results indicated less opportunity to benefit from the therapy of cobimetinib/binimetinib/trametinib (KRAS), abemaciclib/palbociclib/ribociclib (CDKN2A), PLX8394 (BRAF), and ivosidenib (IDH1), and more opportunity to benefit from the therapy of olaparib (ATM) and AZD8186/GSK2636771 (PTEN) in hCCA. Although no available alteration mutations in extrahepatic CCA, the frequency of NF1 was higher in hCCA than in intrahepatic CCA. This indicated the more opportunity to benefit from trametinib and cobimetinib in hCCA. In general, the opportunity to benefit from target drug of hCCA patients were different from those of intrahepatic CCA and extrahepatic CCA. Few studies reported the mutation characteristic of distal CCA. Although we failed to compare the molecular characteristics between hCCA and distal CCA, our results supported that hCCA is different from the previously reported intrahepatic CCA and extrahepatic CCA. The specific molecular feature of hCCA is of great significance in guiding the target drug treatment and further precision therapy of CCA.
Comparative analysis of high frequently mutated genes in hCCA (blue), intrahepatic CCA (red), and extrahepatic CCA (green). The X-axis represents the most mutated genes and the Y-axis represents the mutation frequency of each gene in different CCA subtypes. The significant differences were marked with * for P < 0.05, ** for P < 0.01, and *** for P < 0.001, NA not available
In conclusion, we firstly identified mutational landscape of hCCA and detected the correlation between mutated gene and clinical characteristics. Our results suggested potential biomarkers such as ARID1B, for potential therapy and prognosis of Chinese hCCA. Objectively, the single sampling and the small number of samples are shortcomings of this study. Further study with the expanded number of samples is still needed to confirm and supplement our results here. However, our research provided the molecular evidence that hCCA differs from intrahepatic CCA and extrahepatic CCA, and provided the evidence for guiding precise therapeutic strategies of Chinese hCCA.
Data availability
The datasets used and analyzed in this study are available from the corresponding author upon reasonable request.
References
Chen P, Li B, Zhu Y et al (2016) Establishment and validation of a prognostic nomogram for patients with resectable perihilar cholangiocarcinoma. Oncotarget 7(24):37319–37330. https://doi.org/10.18632/oncotarget.9104
Rizvi S, Gores GJ (2013) Pathogenesis, diagnosis, and management of cholangiocarcinoma. Gastroenterology 145(6):1215–1229. https://doi.org/10.1053/j.gastro.2013.10.013
Jarnagin WR, Fong Y, DeMatteo RP et al (2001) Staging, resectability, and outcome in 225 patients with hilar cholangiocarcinoma. Ann Surg 234(4):507–517. https://doi.org/10.1097/00000658-200110000-00010
Hidalgo E, Asthana S, Nishio H et al (2008) Surgery for hilar cholangiocarcinoma: the Leeds experience. Eur J Surg Oncol 34(7):787–794. https://doi.org/10.1016/j.ejso.2007.10.005
Yoo T, Park SJ, Han SS et al (2018) Proximal resection margins: more prognostic than distal resection margins in patients undergoing hilar cholangiocarcinoma resection. Cancer Res Treat 50(4):1106–1113. https://doi.org/10.4143/crt.2017.320
Schacke G (1985) Danger to pregnant women at the work site. Disagreement between employers and trade unions in the USA: sterilization or employment termination. Fortschr der Medizin 103(47–48):52–53
Du J, Zhou XJ (2017) Precise diagnosis and treatment of thymic epithelial tumors based on molecular biomarkers. Crit Rev Oncog 22(5–6):507–514. https://doi.org/10.1615/CritRevOncog.2017020577
Hainsworth JD, Meric-Bernstam F, Swanton C et al (2018) Targeted therapy for advanced solid tumors on the basis of molecular profiles: results from mypathway, an open-label, phase IIa multiple basket study. J Clin Oncol 36(6):536–542. https://doi.org/10.1200/jco.2017.75.3780
Miyazaki M, Ohtsuka M, Miyakawa S et al (2015) Classification of biliary tract cancers established by the Japanese Society of Hepato-Biliary-Pancreatic Surgery. J Hepato-Biliary-Pancreat Sci 22(3):181–196. https://doi.org/10.1002/jhbp.211
Huggett MT, Passant H, Hurt C et al (2014) Outcome and patterns of care in advanced biliary tract carcinoma (ABC): experience from two tertiary institutions in the United Kingdom. Tumori 100(2):219–224. https://doi.org/10.1700/1491.16421
Churi CR, Shroff R, Wang Y et al (2014) Mutation profiling in cholangiocarcinoma: prognostic and therapeutic implications. PLoS ONE 9(12):e115383. https://doi.org/10.1371/journal.pone.0115383
Tian W, Hu W, Shi X et al (2020) Comprehensive genomic profile of cholangiocarcinomas in China. Oncol Lett 19(4):3101–3110. https://doi.org/10.3892/ol.2020.11429
Nakamura H, Arai Y, Totoki Y et al (2015) Genomic spectra of biliary tract cancer. Nat Genet 47(9):1003–1010. https://doi.org/10.1038/ng.3375
Waseem D, Tushar P (2017) Intrahepatic, perihilar and distal cholangiocarcinoma: Management and outcomes. Ann Hepatol 16(1):133–139. https://doi.org/10.5604/16652681.1226927
Cao J, Chen L, Li H et al (2019) An accurate and comprehensive clinical sequencing assay for cancer targeted and immunotherapies. Oncologist 24(12):e1294–e1302. https://doi.org/10.1634/theoncologist.2019-0236
Bismuth H, Corlette MB (1975) Intrahepatic cholangioenteric anastomosis in carcinoma of the hilus of the liver. Surg Gynecol Obstetr 140(2):170–178
Chakravarty D, Gao J, Phillips SM et al (2017) OncoKB: a precision oncology knowledge base. JCO Precis Oncol. https://doi.org/10.1200/po.17.00011
Esnaola NF, Meyer JE, Karachristos A et al (2016) Evaluation and management of intrahepatic and extrahepatic cholangiocarcinoma. Cancer 122(9):1349–1369. https://doi.org/10.1002/cncr.29692
Akita M, Fujikura K, Ajiki T et al (2017) Dichotomy in intrahepatic cholangiocarcinomas based on histologic similarities to hilar cholangiocarcinomas. Mod Pathol 30(7):986–997. https://doi.org/10.1038/modpathol.2017.22
Xue L, Guo C, Zhang K et al (2019) Comprehensive molecular profiling of extrahepatic cholangiocarcinoma in Chinese population and potential targets for clinical practice. Hepatobiliary Surg Nutr 8(6):615–622. https://doi.org/10.21037/hbsn.2019.08.05
Lowery MA, Ptashkin R, Jordan E et al (2018) Comprehensive molecular profiling of intrahepatic and extrahepatic cholangiocarcinomas: potential targets for intervention. Clin Cancer Res 24(17):4154–4161. https://doi.org/10.1158/1078-0432.ccr-18-0078
Abdel-Wahab R, Liu S, Cao J et al (2019) AB045. P-13. genomic heterogeneity between Asian and Western intrahepatic cholangiocarcinoma. Hepatobiliary Surg Nutr 8:45–45. https://doi.org/10.21037/hbsn.2019.AB045
He X, Pang Z, Zhang X et al (2018) Consistent amplification of FRS2 and MDM2 in low-grade osteosarcoma: a genetic study of 22 cases with clinicopathologic analysis. Am J Surg Pathol 42(9):1143–1155. https://doi.org/10.1097/pas.0000000000001125
Ware PL, Snow AN, Gvalani M et al (2014) MDM2 copy numbers in well-differentiated and dedifferentiated liposarcoma: characterizing progression to high-grade tumors. Am J Clin Pathol 141(3):334–341. https://doi.org/10.1309/ajcplyu89xhsnhqo
Gotoh N (2008) Regulation of growth factor signaling by FRS2 family docking/scaffold adaptor proteins. Cancer Sci 99(7):1319–1325. https://doi.org/10.1111/j.1349-7006.2008.00840.x
Manuvakhova M, Thottassery JV, Hays S et al (2006) Expression of the SNT-1/FRS2 phosphotyrosine binding domain inhibits activation of MAP kinase and PI3-kinase pathways and antiestrogen resistant growth induced by FGF-1 in human breast carcinoma cells. Oncogene 25(44):6003–6014. https://doi.org/10.1038/sj.onc.1209592
Zhong X, Xie G, Zhang Z et al (2016) MiR-4653-3p and its target gene FRS2 are prognostic biomarkers for hormone receptor positive breast cancer patients receiving tamoxifen as adjuvant endocrine therapy. Oncotarget 7(38):61166–61182. https://doi.org/10.18632/oncotarget.11278
Fujimoto A, Totoki Y, Abe T et al (2012) Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat Genet 44(7):760–764. https://doi.org/10.1038/ng.2291
Zang ZJ, Cutcutache I, Poon SL et al (2012) Exome sequencing of gastric adenocarcinoma identifies recurrent somatic mutations in cell adhesion and chromatin remodeling genes. Nat Genet 44(5):570–574. https://doi.org/10.1038/ng.2246
Li WD, Li QR, Xu SN et al (2013) Exome sequencing identifies an MLL3 gene germ line mutation in a pedigree of colorectal cancer and acute myeloid leukemia. Blood 121(8):1478–1479. https://doi.org/10.1182/blood-2012-12-470559
Chen C, Liu Y, Rappaport RA et al (2014) MLL3 is a haploinsufficient 7q tumor suppressor in acute myeloid leukemia. Cancer Cell 25(5):652–665. https://doi.org/10.1016/j.ccr.2014.03.016
Sato K, Akimoto K (2017) Expression levels of KMT2C and SLC20A1 identified by information-theoretical analysis are powerful prognostic biomarkers in estrogen receptor-positive breast cancer. Clin Breast Cancer 17(3):e135–e142. https://doi.org/10.1016/j.clbc.2016.11.005
Wang Y (2019) Association between histone lysine methyltransferase KMT2C mutation and clinicopathological factors in breast cancer. Biomed Pharmacother. https://doi.org/10.1016/j.biopha.2019.108997
Rao RC, Dou Y (2015) Hijacked in cancer: the KMT2 (MLL) family of methyltransferases. Nat Rev Cancer 15(6):334–346. https://doi.org/10.1038/nrc3929
Hr B, Da S, Frank M (1990) The GTPase superfamily: a conserved switch for diverse cell functions. Nature 348(6297):125–132. https://doi.org/10.1038/348125a0
Side L, Taylor B, Cayouette M et al (1997) Homozygous inactivation of the NF1 gene in bone marrow cells from children with neurofibromatosis type 1 and malignant myeloid disorders. N Engl J Med 336(24):1713–1720. https://doi.org/10.1056/nejm199706123362404
Basu TN, Gutmann DH, Fletcher JA et al (1992) Aberrant regulation of ras proteins in malignant tumour cells from type 1 neurofibromatosis patients. Nature 356(6371):713–715. https://doi.org/10.1038/356713a0
High TMB predicts immunotherapy benefit (2018). Cancer Discov 8(6):668. https://doi.org/10.1158/2159-8290.cd-nb2018-048
Chalmers ZR, Connelly CF, Fabrizio D et al (2017) Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med 9(1):34. https://doi.org/10.1186/s13073-017-0424-2
Patel SP, Kurzrock R (2015) PD-L1 expression as a predictive biomarker in cancer immunotherapy. Mol Cancer Ther 14(4):847–856. https://doi.org/10.1158/1535-7163.mct-14-0983
Mou H, Yu L, Liao Q et al (2018) Successful response to the combination of immunotherapy and chemotherapy in cholangiocarcinoma with high tumour mutational burden and PD-L1 expression: a case report. BMC Cancer 18(1):1105. https://doi.org/10.1186/s12885-018-5021-2
Wang S, Liechty B, Patel S et al (2018) Programmed death ligand 1 expression and tumor infiltrating lymphocytes in neurofibromatosis type 1 and 2 associated tumors. J Neurooncol 138(1):183–190. https://doi.org/10.1007/s11060-018-2788-6
Kim T, Yoo J, Wang Z et al (2015) ARID1A Is essential for endometrial function during early pregnancy. PLoS Genet 11(9):e1005537. https://doi.org/10.1371/journal.pgen.1005537
Yang S, Wang A, Du J et al (2016) Low expression of ARID1A correlates with poor prognosis in intrahepatic cholangiocarcinoma. World J Gastroenterol 22(25):5814–5821. https://doi.org/10.3748/wjg.v22.i25.5814
Zhang L, Jia CW, Lu ZH et al (2016) ARID1A expression of SWI/SNF remodeling complex in pancreatic neuroendocrine tumor. Chin J Pathol 45(8):571–574. https://doi.org/10.3760/cma.j.issn.0529-5807.2016.08.015
Sasaki M, Sato Y, Nakanuma Y (2017) Mutational landscape of combined hepatocellular carcinoma and cholangiocarcinoma, and its clinicopathological significance. Histopathology 70(3):423–434. https://doi.org/10.1111/his.13084
Shen W, Niu N, Lawson B et al (2019) GATA6: a new predictor for prognosis in ovarian cancer. Hum Pathol 86:163–169. https://doi.org/10.1016/j.humpath.2019.01.001
Spolverato G, Ejaz A, Kim Y et al (2014) Tumor size predicts vascular invasion and histologic grade among patients undergoing resection of intrahepatic cholangiocarcinoma. J Gastrointest Surg 18(7):1284–1291. https://doi.org/10.1007/s11605-014-2533-1
Guan JL, Zhong WZ, An SJ et al (2013) KRAS mutation in patients with lung cancer: a predictor for poor prognosis but not for EGFR-TKIs or chemotherapy. Ann Surg Oncol 20(4):1381–1388. https://doi.org/10.1245/s10434-012-2754-z
Lin G, Zheng XW, Li C et al (2012) KRAS mutation and NF-κB activation indicates tolerance of chemotherapy and poor prognosis in colorectal cancer. Dig Dis Sci 57(9):2325–2333. https://doi.org/10.1007/s10620-012-2172-x
Khursheed M, Kolla J, Kotapalli V et al (2013) ARID1B, a member of the human SWI/SNF chromatin remodeling complex, exhibits tumour-suppressor activities in pancreatic cancer cell lines. Br J Cancer 108(10):2056–2062. https://doi.org/10.1038/bjc.2013.200
Wang B, Xie H, Ma C et al (2017) Expression of ARID1B Is associated with poor outcomes and predicts the benefit from adjuvant chemotherapy in bladder urothelial carcinoma. J Cancer 8(17):3490–3497. https://doi.org/10.7150/jca.19109
Wang M, Fan W, Ye M et al (2018) Molecular profiles and tumor mutational burden analysis in Chinese patients with gynecologic cancers. Sci Rep 8(1):8990. https://doi.org/10.1038/s41598-018-25583-6
Zhao J, Sun Y, Huang Y et al (2017) Functional analysis reveals that RBM10 mutations contribute to lung adenocarcinoma pathogenesis by deregulating splicing. Sci Rep 7:40488. https://doi.org/10.1038/srep40488
Jackson T, Du L, Janesko-Feldman K et al (2015) The nuclear splicing factor RNA binding motif 5 promotes caspase activation in human neuronal cells, and increases after traumatic brain injury in mice. J Cereb Blood Flow Metab 35(4):655–666. https://doi.org/10.1038/jcbfm.2014.242
Giannakis M, Mu X, Shukla S et al (2016) Genomic correlates of immune-cell infiltrates in colorectal carcinoma. Cell Rep 15(4):857–865. https://doi.org/10.1016/j.celrep.2016.03.075
Loiselle JJ, Roy JG, Sutherland LC (2017) RBM10 promotes transformation-associated processes in small cell lung cancer and is directly regulated by RBM5. PLoS ONE 12(6):e0180258. https://doi.org/10.1371/journal.pone.0180258
Yin LL, Wen XM, Li M et al (2018) A gene mutation in RNA-binding protein 10 is associated with lung adenocarcinoma progression and poor prognosis. Oncol Lett 16(5):6283–6292. https://doi.org/10.3892/ol.2018.9496
Funding
This work was supported by the Shanghai Municipal Health Commission Integrative Innovation Project [2019CXJQ03] and Key project of Jiading District Health Construction Commission of Shanghai (No. 2020-ZD-01).
Author information
Authors and Affiliations
Contributions
FF, XW, QG, YW, YY, QC, BL, BY, and CL collected patients’ consents and samples, and analyzed data; XS, QH, LZ, CG, and XJ wrote the manuscript; XJ and CG designed and supervised the study.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
The project was approved by the Ethics Committee of Shanghai Eastern Hepatobiliary Surgery Hospital.
Informed consent
Informed consent for participation was obtained from all subjects.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
About this article
Cite this article
Feng, F., Wu, X., Shi, X. et al. Comprehensive analysis of genomic alterations of Chinese hilar cholangiocarcinoma patients. Int J Clin Oncol 26, 717–727 (2021). https://doi.org/10.1007/s10147-020-01846-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10147-020-01846-z