Abstract
Peanut is one of the most valuable legumes, grown mainly in arid and semi-arid regions, where its production may be hindered by the lack of water. Therefore, breeding drought tolerant varieties is of great importance for peanut breeding programs around the world. Unlike cultivated peanuts, wild peanuts have greater genetic diversity and are an important source of alleles conferring tolerance/resistance to abiotic and biotic stresses. To decipher the transcriptome changes under drought stress, transcriptomics of roots of highly tolerant Arachis duranensis (ADU) and moderately susceptible A. stenosperma (AST) genotypes were performed. Transcriptome analysis revealed an aggregate of 1465 differentially expressed genes (DEGs), and among the identified DEGs, there were 366 single nucleotide polymorphisms (SNPs). Gene ontology and Mapman analyses revealed that the ADU genotype had a higher number of transcripts related to DNA methylation or demethylation, phytohormone signal transduction and flavonoid production, transcription factors, and responses to ethylene. The transcriptome analysis was endorsed by qRT-PCR, which showed a strong correlation value (R2 = 0.96). Physio-biochemical analysis showed that the drought-tolerant plants produced more osmolytes, ROS phagocytes, and sugars, but less MDA, thus attenuating the effects of drought stress. In addition, three SNPs of the gene encoding transcription factor NFAY (Aradu.YE2F8), expansin alpha (Aradu.78HGD), and cytokinin dehydrogenase 1-like (Aradu.U999X) exhibited polymorphism in selected different genotypes. Such SNPs could be useful for the selection of drought-tolerant genotypes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Peanut is one of the most important legumes grown all over the world, replacing soybean in terms of global production (Toomer 2018). Peanut cultivation is well suited to semi-tropical and semi-arid tropical climates. Peanut production covers about 27 million ha of land worldwide and yields up to 43 million t of pods per year (Launio et al. 2018). It is generally grown as a rain-fed crop. India is the second largest groundnut producer in the world, yielding 6.8 million ton 5.8 million ha of agricultural land (FAOSTAT 2019). Maintaining and even increasing groundnut yield to meet the mounting population demand while the environmental situations deteriorate is a major hurdle for the groundnut industry. Development of drought tolerant cultivars adapted to different types of drought stress is significant for many groundnut breeding programs (Oshunsanya et al. 2019). Drought causes production losses of about six million t of groundnut per year worldwide (Jiang et al. 2021). The global drought situation has worsened recently, showing a trend towards longer periods, more frequent occurrences and a wider range. Furthermore, the magnitude and intensity of global drought are expected to increase over the next 30–90 years (Balyan et al. 2020). Recent studies have shown that drought stress affects a variety of biological processes, including water physiology, membrane permeability, nutrient uptake and mobility, enzyme function, pigment content, photosynthesis, and assimilate transport (Jiang et al. 2020; Reddy et al. 2021; Wang et al. 2019).
Drought tolerance is a complex attribute, usually associated with many loci, each of which has only a minor effect. The wild legumes are very good materials for the study of drought tolerance as they usually grow in arid regions (Saxena et al. 2011). These plants have evolved complex mechanisms for sensing external signals that enable them to adapt to changing environmental conditions and thus mitigate the damage caused by various stressors. These pathways include drought stress perception, signal transduction, transcriptional activation of drought-responsive target genes, and synthesis of drought stress-related proteins and other molecules that together help the plant survive (Lamaoui et al. 2018). Changes in gene expression lead to a variety of visible symptoms, such as leaf curling, stomata closure, epicuticular wax deposition, ability to remain green, osmotic adjustment, increased root length leading to photochemical quenching, higher WUE, resistance to photoinhibition, and membrane stabilization, all of which are traits of drought tolerance (Joshi et al. 2016). There are also several phenotypic trade-offs such as delayed flowering, lower plant height, and lower growth rate (Kumar et al. 2021). In addition, deeper rooting and increased lateral roots for drought tolerance have been observed in grain legumes (Maga 2021).
When it comes to the most critical organs during drought, roots are often the first to detect water deficit and transmit it to shoots and leaves (Gui et al. 2021). Drought stress often restricts shoot development, while root growth is stimulated to accelerate the remobilization of photoassimilates from shoots to roots to cope with drought stress (Martignago et al. 2020). Plant root system architecture (RSA) is a critical developmental and agronomic feature that influences plant adaptation and production in water-scarce environments (Tamir, Xiong et al. 2021). It is well known that root traits are critical for drought resistance in legumes (Abdelrahman et al. 2018; Marshall et al. 2016; Polania et al. 2017). Phenotyping of roots of 270 chickpea genotypes revealed inherent genetic heterogeneity for root traits within the studied chickpea population and that the genotypes with deep roots had twice as long taproot and more root branching than the shallow rooted genotypes (Abdelrahman, et al. 2018). Root traits affect not only water uptake but also nutrient uptake (Borden et al. 2020); therefore, root traits may be useful in the production of novel cultivars with improved outcomes under drought and/or nutrient deficiencies (Wissuwa et al. 2016). On an accounting basis, 41 soybean accessions were tested for drought tolerance in the greenhouse (Yan et al. 2020). In this study, root metaxylem was found to play a major role in water use efficiency and stomatal conductance during the reproductive stage, and the amount of root metaxylem correlated with higher yields under drought stress in soybean (Yan, et al. 2020). In recent years, the use of the RNA-seq approach to comprehend how biotic and abiotic stress tolerance has increased in various crop species (Rathod et al. 2020a, b; Rathod et al. 2020a, b). It has been extensively utilized to assess the structures and expression patterns of numerous genes in response to stress conditions in a variety of plants, including rice, poplar, and chickpea (Che Omar et al. 2016; Garg et al. 2016; Lorrain et al. 2018). There are a few reports about transcriptomic studies related to molecular mechanisms during drought in peanuts. Shen et al. investigated transcriptomes of drought-stressed leaves of a drought-tolerant variety, FH1, and discovered transcriptional modifications after 7 days of drought treatment (Shen et al. 2015). Another research, conducted by Brasileiro et al., studied wild-peanut plant transcriptomes that have been subjected to drought stress for 11 days (Brasileiro et al. 2015). Zhao et al., on the other hand, have directly analyzed transcriptomic responses of peanut root tissues to shorter-drought (two-days) in J1, another well-known drought-tolerant peanut type (Zhao et al. 2018). Taking into account these drought-transcriptomic studies and other related studies, it can be concluded that drought stresses can trigger differential gene expression involved in different signaling pathways. Nonetheless, molecular research on drought-tolerance processes in peanut is still in its early stages, owing to its massive allotetraploid genome size. So, filling information gaps about molecular changes in drought responses during seedling stage would be favorable in developing drought tolerant genotypes in peanut. Thus, the current research uses physico-biochemical properties and RNA-seq to find genes and the molecular pathways involved in providing peanuts, tolerance to drought.
Material and methods
Plant materials and growth conditions
Experiments were conducted with two genotypes of wild peanut, Arachis duranensis (tolerant) and A. stenosperma (susceptible). For this study, a randomized complete block design with split plot was used, and seeds were sown at 10 seeds m−1 in a single row (15 × 120 cm) under a rain shelter at Junagadh Agricultural University (JAU), Gujarat, India, to create an artificial drought stress condition. Two rain shelters were established with two treatments, namely, well-watered and mid-season drought, each containing three blocks. Both treatments were irrigated immediately after sowing to promote uniform germination. The irrigated treatment (referred to as the WW) was irrigated throughout the growing season based on the evapotranspiration replacement described by Stansell et al. (1976). The drought-stressed variant (DS) was fully irrigated during the initial phase of the growing season until 42 days after planting. Thereafter, a water deficit was created by withholding water for up to 4 weeks, starting at a soil water potential of − 10 kPa at 20-cm depth and gradually increasing to − 700 kPa after treatment for 1 week and further increasing to − 1050 to − 1200 kPa the next week, which was maintained for another 2 weeks. Specific leaf area, 15 N, and 13C were measured following the protocol of Dang et al. (2012) to determine the physiological responses to drought stress. Apart from water treatment, all other agronomic managements were carried out in accordance with the best management practices for groundnut of the College of India. After drought period, random root samples were collected for each genotype and immediately frozen in liquid nitrogen before processing at − 80 °C. Three random root samples were pooled from each biological replicate and about 0.2 g was crushed in liquid nitrogen for RNA extraction (Eybishtz et al. 2009).
Physio-biochemical analysis of different parameters
The phenotype changes induced by drought were examined in wild variety, by randomly selecting the second-compound leaves from treatment, control, and re-watered seedlings, and then measuring the physiological indexes. Various physio-biochemical parameters like relative water content (RWC) (Barrs and Weatherley 1962), osmotic potential (OP) (Bhauso et al. 2014), electrolytic leakage (EL) (Wang et al. 2008), proline (Bates et al. 1973), total chlorophyll content (Hiscox and Israelstam 1979), and antioxidant enzymes such as catalase (CAT), glutathione peroxidase (GPX), ascorbate peroxidase (APX), superoxide dismutase (SOD), and peroxidase (POX) estimated. Three independent biological replicates were used for each measurement.
RNA isolation and sequencing
Total RNA was extracted from the control and treatment groups using the Xcelgen Plant RNA Kit according to the manufacturer’s instructions, and the purity and integrity of the extracted RNA were determined using a NanoDrop ND -1000 UV/Vis spectrophotometer (Thermo Scientific, Wilmington, DE) and an Agilent 2100 Bioanalyzer (Agilent). High-quality samples were submitted to the Illumina TruSeq Stranded Total RNA Library Construction Kit for pair-end library preparation. The average size of a library was 320 bp. Libraries were constructed using 1 g of total RNA with the Illumina TruSeq Stranded Total RNA Library Preparation Kit according to the manufacturer’s instructions, followed by ribo removal with rRNA removal beads prior to purification, fragmentation, and preparation for cDNA synthesis. The resulting RNA fragment was converted to first-strand cDNA and then to second-strand cDNA, which was then subjected to A-tailing, adapter index ligation, and PCR amplification. Quality and quantity controls were performed using a Bioanalyzer 2100 (Agilent Technologies) and a High Sensitivity (HS) DNA chip according to the manufacturer’s instructions.
Quality control, alignment with reference genome, sequence annotation, and (DEG) analysis
The RNA-seq reads quality was estimated using the Illumina TruSeq pipeline and FastQC software (Brown et al. 2017; Hamid et al. 2019). For the quality control of the sequences, low-quality data containing reads < 50 bp, adaptor dimers, homopolymers, and ambiguous bases (‘N’) were removed by filtering by the PRINSEQ-lite v0.20.4. Also, sequencing reads that had quality scores below 25, were removed by a downstream analysis (Schmieder and Edwards 2011). The GFF and fasta files of genome-A (Arachis duranensis) were retrieved from PeanutBase (http://peanutbase.org) and served as the reference for alignment (Rathod, et al. 2020a, 2020b). The STAR 2.5.1a software (https://github.com/alexdobin/STAR/releases/) was used for mapping the filtered reads against the reference genome (Dobin and Gingeras 2015). DEGs among treated, and well-watered (non-stressed) root samples were recognized by the Cuffdiff tool of the Cufflinks package (http://sihua.us/Cufflinks.htm) (Ghosh and Chan 2016; Trapnell et al. 2010). Moreover, the normalized transcripts were quantified by the same tool and denoted in fragments per kilo base per million mapped reads (FPKM) (Trapnell et al. 2009). A false discovery rate (FDR) of < 0.05% and < 0.01 p value with > 2 log2-fold change were used for the production of high-quality data for the statistical analysis of DEGs (Tulsani et al. 2020). The expressed sequences shared by various samples were depicted and compared using Venn diagrams using VENNY software (Oliveros 2007). The cleaned reads for each library are displayed in Table S1. All the raw sequences for the samples were compiled at https://www.ncbi.nlm.nih.gov/sra/PRJNA751721.
Gene Ontology and pathway analysis
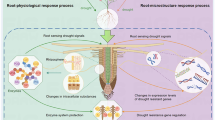
Gene Ontology (GO) and pathway analysis provide a limited vocabulary of specified words that describe the properties of gene products (Hamid et al. 2018). Gene Ontology analysis was performed using the transcripts ID’s of all differentially expressed genes. Blast2GO Command Line v1.4.1 commercial software was used for this purpose. KEGG analysis was used to get a better overview of significantly enriched pathways in DEGs. For both analyses, the resulting p value was adjusted using the FDR correction, with a threshold of 0.01 for the FDR. The overall bioinformatics workflow is graphically represented in Fig. 1.
SNP identification
SNPs were identified using SAMtools (Thakur and Randhawa 2018) and subsequently annotated using SnpEff50 software. For both ADU and AST, the BatchPrimer3 program (https://probes.pw.usda.gov/cgi-bin/batchprimer3/ batchprimer3.cgi) was used to generate allele-specific primers. qRT-PCR was used to validate SNPs. Homozygous and heterozygous alleles were identified based on differences in Ct value.
Validation of DEGs using qRT-PCR
To authenticate the fidelity of RNA-seq, 12 selected putative drought-tolerance-related DEGs and eight TFs (2 BHLH, 2 WRKY, 2 NAC, and 2 MYB) were used in qRT-PCR verification. Beacon Designer 7.0 program was used to design the qRT-PCR primers, and the specificity of the primers was assessed by aligning the primer sequences against the NCBI database (Hamid et al. 2020). To generate cDNA from whole RNA, a TaKaRa PrimeScript RT reagent kit (Perfect Real Time) was utilized. Reactions were carried out based on the SYBR Premix Ex Taq TM guideline using an Applied Biosystems 7500 Fast Real-Time PCR System with a 20-µL reaction mixture, which composed 10 µL 2 × SYBR Premix, 0.4 µL (200 nM) of each primer, and 1 µL of template cDNA. Temperature conditions for amplification were set according to the manufacturer’s instructions. Three biological replicates were used for each of the selected genes and the 2−ΔΔCT method was used to estimate relative gene expression levels. Actin 11 was used as an internal control for RT-qPCR (Rathod, et al. 2020a, 2020b).
Results
Physio-biochemical features
To understand the phenotypic changes and subsequent physiological responses between two peanut genotypes, various physio-biochemical parameters such as RWC, OP, EL, proline, total chlorophyll content and antioxidant enzymes such as SOD, APX, catalase, POX, and glutathione peroxidase (GPX) were studied in three independent biological replicates. It was observed that drought-tolerant groundnut ADU exhibited higher RWC (72.98%) than drought susceptible cultivar AST (57.40%) under stress conditions. RWC of ADU was also higher than AST after resumption of irrigation (Fig. 2a).
Changes in relative water content (a), Osmotic potential (b), Electrolytic leakage (c), Proline content (d), MDA (e), total chlorophyll content (f), SOD (g), APX (h), CAT (i), GPX, (j), and POX (k) under well-watered (WW), drought stress (DS) and re-watered (RW) conditions of ADU and AST genotypes. Bars with the same small letters do not statistically differ by the Tukey test at P ≤ 0.05
The osmotic potential was highest (1446 mM/kg) in ADU (DS) exposed to drought stress, while the lowest osmotic potential was observed in the control AST plants (533.00 mM/kg). There was a sharp increase (by 1446 mM/kg) in the accumulation of osmolytes in the ADU when exposed to drought, which helps the plants to survive in drought, while the increase in osmotic potential was lower in the drought susceptible AST (increase by 1426 mM/kg). After resumption of irrigation, the drought-tolerant plant showed a greater decrease in osmolyte accumulation (decreased by 143.33 mM/kg). The accumulation of osmolytes in the tolerant genotype showed a bell trend such that well-watered plants showed the least accumulation (533 mM/kg), while stressed plants showed a significant increase in osmolyte accumulation (increased by 1446 mM/kg), while after resumption of irrigation, the accumulation of these compounds decreased (decreased by 143.33 mM/kg) (Fig. 2b). In contrast, osmolyte concentration in the re-watered susceptible plants hardly decreased (decreased by 119.33 mM/kg) (Fig. 2b).
For electrolytic leakage (EL) ADU followed the same trend as OP; there was significant increase in the electrolytic leakage (EL) in both the varieties during drought stress while the level of electrolytic leakage reduced upon revival. The level of electrolytic leakage is highest in AST in drought stress (94.27%) and least in control plants of ADU (39.23%). Moreover, the increase in the EL was more in AST (46.98% increase) upon imposition of drought stress when compared to ADU (27.53% increase) (Fig. 2c). The proline content was highest in the ADU subjected to drought stress (27.53% increase) while the control ADU plants accumulated the lowest amount of proline (34.14 µg/g FW). There was a sharp increase (increased by 86.69 µg/g FW) in the concentration of proline in ADU, when subjected to drought, which helps the plants to survive in drought while the increase in the proline content in the drought susceptible, AST was lower (increased by 58.41 µg/g FW). Upon revival, the drought-tolerant plant showed more reduction in proline content (decreased by 58.90 µg/g FW) while the drought-stressed plant did not show much reduction (28.41 µg/g FW) (Fig. 2d).
Lipid peroxidation and H2O2 production are reliable biomarkers of cellular oxidative stress. We evaluated the variations in lipid hydroperoxide generation rates by measuring MDA concentration in stressed leaves and control environments. MDA concentration in peanut plant leaves increased significantly under drought stress conditions. However, AST showed significantly higher MDA concentrations during drought treatment compared with the ADU genotype (Fig. 2e), indicating less membrane damage in the ADU genotype. Chlorophyll content was highest in the well-watered ADU genotype (0.78 mg/g), while the lowest chlorophyll content was observed in AST under drought stress (0.11 mg/g). The chlorophyll content of AST decreased more under drought (by 0.55 mg/g), while the chlorophyll content of the drought-tolerant ADU was lower (by 0.35 mg/g) (Fig. 2f).
Drought causes oxidative stress in plant cells. However, considerable stress tolerance is achieved by scavenging reactive oxygen species (ROS). Therefore, we determined the ROS-dependent enzyme level in each of the experimental conditions. It was found that the antioxidant enzymes, especially superoxide dismutase (SOD), ascorbate peroxidase (APX), POX, catalase (CAT), and glutathione peroxidase (GPX) were greatly increased in the plants exposed to drought stress. The highest increase in antioxidant enzymes was observed in GPX, followed by APX and SOD, in the stressed ADU genotype, followed by the genotypes ADU-WW > AST-DS > AST-WW (Fig. 2g–k). In the stressed ADU genotype, a 2.38-fold increase in SOD (Fig. 2g), a 2.1-fold increase in APX (Fig. 2h), a 1.2-fold increase in catalase (Fig. 2i), a 4.1-fold increase in GPX (Fig. 2j), and a 2.4-fold increase in POX (Fig. 2k) were observed compared to the ADU-WW > AST-DS > ADU-WW genotypes. The increased activity of antioxidant enzymes may have helped in the detoxification of H2O2 produced during photosynthesis and photorespiration.
Sequencing and mapping statistics using reference-guided assembly
Root samples for RNA-seq were obtained from the highly tolerant ADU and moderately susceptible AST genotypes in three biological replicates for profiling the transcriptome, resulting in 308 million raw reads. High quality reads (trimmed and filtered) were mapped against the reference genome. Approximately 81.2%, 6%, and 12.8% of the reads were mapped onto the exonic, intronic, and intergenic regions, respectively, in the reference genome of ADU, whereas for the genotype of AST, 78.01 of the high-quality reads were mapped to exonic segments, and 6.28% and 13.08% were mapped to the intronic and intergenic regions, respectively (Table S1. Figure 3a, b). AST-DS, AST-WW, ADU-DS, and ADU-WW were the four samples tested for expression. The quantity of gene expression in the four samples was estimated using FPKM values. Overall 28,546 genes were seen to be expressed on the reference genome.
Mapping statistics of sequenced data: (a and b) An overview of mapped reads on referenced genomes of Arachis duranensis (ADU) and A. stenosperma (AST), respectively: the distribution of fragments represented as the percentage of reads mapped to exons, introns, and intergenic regions on reference peanut genomes., (c) The number of down- and up-regulated DEGs in different reference pairs, (d) Venn diagram showing the common and specific DEGs in different combinations of reference pairs.
Differentially expressed genes during drought stress
To filter out DEGs, a strict absolute log2 FC 2 and FDR 0.05 index was applied. The number of DEGs was 238 (downregulated 102/upregulated 136), 371 (downregulated 100/upregulated 271), 289 (downregulated 124/upregulated 165), and 567 (downregulated 262/upregulated 305), in AST-S compared with AST-WW, AST-WW compared with ADU-WW, ADU-DS compared with ADU-WW, and AST-DS compared with ADUS, respectively (Fig. 3c), and these genes were common DEGs in the three comparison groups (Table S2, Fig. 3d). Drought stress-induced expression of heat shock protein (HSP) coding genes including heat shock factor (Hsf)30, Dnaj/HSP40, Class and II HSPs, abscisic acid (ABA) insensitive 5, Aquaporin PIP-2, glutathione S-transferase, expansin 2, ethylene-responsive transcription factor (ERF), thioredoxin reductase and trehalose phosphate phosphatase, L-ascorbate oxidase, and peroxidase drought-responsive peroxidase genes (DRGs) were detected in ADU genotype, (Table S2). DEGs, induced in AST, comprise the genes linked to the cell wall and membrane-bound genes, such as expansin A10, ureide permease 2, xyloglucan endotransglucosylase hydrolase, and expansin-A8 when comparing stressed and control plants. Upregulated genes include those encoding oryzasin-1, asparticase, cysteine endopeptidase, and ubiquitin-E3ligase, as well as other drought-responsive DEGs such as abscisic acid (ABA) insensitive 5, aquaporin PIP -2, expansin 2, ethylene-responsive transcription factor, peroxidase, L-ascorbate oxidase, thioredoxin reductase heat shock proteins (HSPs) encoding genes such as HSPs of class II, Dnaj/HSP40, and heat shock factor (Hsf)30, as well as genes related to secondary metabolism, such as isoflavonoid and flavonoid production, were increased under stress in the wild genotype compared with the cultivar AST (Table S2).
A comparison of plants of both genotypes under untreated conditions revealed an intriguing result, namely the expression of genes that respond to drought stress, such as osmoprotective genes like abscisic acid 8-hydroxylase. Genes encoding 8-hydroxylase, peroxidase and proline imino peptidase, genes related to photosynthesis, accumulation of photosystem one1 (APO1), genes encoding photosystem II family proteins (D2 protein), and other genes encoding ethylene-responsive transcription factor (ERF) and lipid transfer protein genes were more highly expressed in the ADU genotype, indicating greater tolerance (Table S2).
Functional classification of DEGs
To explore the possible pathways involving different DEGs in their battle over drought stress, GO and KEGG pathway analyses were performed for DEGs found in different pairwise comparisons. A total of 143 GO annotation terms were found, of which 106 were involved in biological processes, 27 in molecular processes, and 10 in cellular components. When comparing the drought-tolerant genotype with the well-watered condition, among the DEGs, molecular function was the most enriched category, in which most DEGs were devoted to catalytic activity (GO:0,003,824), structural molecule activity (GO:0,005,198), and hydrolase activity (GO:0,016,787), while carbohydrate metabolic process (GO:0,005,975), ion transport (GO:0,006,811), and cell wall organization or biogenesis (GO:0,071,554) from the biological category and macromolecular complex (GO:0,032,991), membrane part (GO:0,044,425), and extracellular region (GO:0,005,576) from the cellular component were the other significantly enriched GO terms (Table S3. Supplementary Fig. 1). Annotation of GO in drought-sensitive genotypes compared with genotypes under good water conditions showed that most DEGs were related to metabolic processes (BP, GO:0,008,152), catalytic activity (MF, GO:0,003,824), and chloroplasts (CC, GO:0,009,507) (Table S4. Supplementary Fig. 1). When the drought-tolerant genotype was compared with the sensitive genotype under stress conditions, many DEGs were enriched molecular function, with most DEGs devoted to calcium ion binding (GO:0,005,509) and ATP binding (GO:0,043,531), while biological processes were the second enriched category, in which most DEGs were involved in DNA methylation or demethylation (GO:0,044,728), antioxidant activity (GO:0,016,209), response to stimuli (GO:0,050,896), response to stress (GO:0,006,950), RNA metabolic process (GO:0,016,070), and root system development (GO:0,022,622). Plasma membrane (CC, GO:0,005,886) and nucleus (CC, GO:0,005,634) are the other enriched GO annotation categories in which many DEGs were involved (Table S5. Supplementary Fig. 1). In the sensitive genotypes compared with the tolerant genotype pair under well-watered conditions, the three most enriched GO annotation categories were heterocyclic compound binding (MF, GO:1,901,363), membrane (CC, GO:0,016,020), and oxidation–reduction processes (BP, GO:0,055,114) (Table S6. Supplementary Fig. 1).
Most genes involved in root system development (184) and ethylene response (168) were found to be more abundant in ADU-DS, whereas genes involved in cell wall and membrane biogenesis were found to be more abundant in AST-DS (Fig. 4a and b). The ADU genotype had more genes involved in transcription and related processes such as tRNA modification, DNA methylation or demethylation, and transcription, whereas the AST genotype had more genes involved in photosynthetic membrane, metabolic processes, and secondary cell wall biogenesis of the plant type (Fig. 4b). The response to hormones was another pathway that was greatly enriched during drought stress (137). Genes encoding auxin (IAA) metabolism, jasmonic acid (JA), and abscisic acid (ABA) were the most abundant genes in this pathway. During drought, two AUX1 genes and four AUX /IAA genes were downregulated in the IAA pathway, whereas only one SAUR gene was upregulated (Fig. 4c). Six upregulated genes were detected in the ABA pathway, including four PP2C and two SnPK2 genes (Fig. 4c). The most frequent genes in this pathway were those encoding auxin (IAA) metabolism, jasmonic acid (JA), and abscisic acid (ABA). During drought, the IAA pathway’s two AUX1 genes and four AUX/IAA genes were downregulated, whereas only one SAUR gene was upregulated. Six genes were found to be upregulated in the ABA pathway, including four PP2C and two SnPK2 genes (Fig. 4c).
Fig. 4. a: Significant GO -terms (P-value < 0.01) enriched by 1002 responsive unigenes extracted from PLS-DA -analysis with a VIP -score greater than 1.0. The heat map was drawn based on the sum of VIP score values of the unigenes belonging to each enriched category (BP, MF and CC) shown separately for up- and down-regulated genes of ADU and AST genotypes in response to drought stress. The color bar shows the gradient of the sum of VIP score values. b: The heat map shows the relative abundance of the corresponding unigenes belonging to "Cell Wall" BP. The gray color indicates the genes that did not change significantly under stress compared to control conditions. c: Heatmap shows the relative abundance of the corresponding unigenes belonging to BPs "Responses to Hormones". The gray color indicates the proteins that did not change significantly under stress compared to well water conditions
Pathway analysis was performed with all identified DEGs from all four comparison pairs. The results showed 56 unique metabolic pathways, and the most enriched pathways were involved in secondary metabolite biosynthesis (413), metabolic pathways (316), genetic information processing (248), environmental information processing (229), signal transduction (218) involved, MAPK signal transduction (189), mismatch repair (185), arginine and proline metabolism (184), glycine, serine, and threonine metabolism (153), metabolism of cofactors and vitamins (153), biotin metabolism (149), and genetic information processing (137), etc. (Table S7, Fig. 5). During drought stress, all DEGs in the MAPK pathway were upregulated, including four protein phosphatase 2C family proteins, two protein kinase superfamily, and one chitinase family protein. There was downregulation of three genes involved in starch and sucrose metabolism, while three were upregulated (Fig. 5b). Three genes were downregulated, and six genes involved in the production of flavonoids and phenylpropanoids were upregulated (Table S2). Drought stress led to downregulation and upregulation of three genes and 20 upregulated genes related to glutathione metabolism, most of which were glutathione S-transferase family proteins. Moreover, several genes involved in hormone synthesis were shown to be differentially expressed during drought stress.
Genes and transcription factors responsive to drought stress
Transcription factors are crucial units in the transmission of stress signals (Wu et al. 2021). A total of 64 TF genes belonging to 10 families were differentially expressed in all three comparison pairs (Table S2). Of these, the top three TF families were bHLH, NAC, and WRKY. When compared under stress conditions in the AST-S vs. AST-WW, ADU-DS vs. ADU-WW, and ADU-DS vs. AST-DS, it was found that many transcription factors belonging to TF families, such as the AP2/ERF family (subfamily ERF or RAV), the WRKY group II b and III families, the HD-ZIP homeobox family or BZIP family, the ABC-2-type transporter, and the LBD (lateral organ boundary domain) TF were upregulated under stress conditions. The ABC transporter, other TF genes such as the NAC, bHLH family, cytochrome P-450, NF-Y, and WRKY were induced during drought stress. Over and above the identified TFs, some drought-responsive genes were also identified. DRGs such as Ardu.6ZX2I for proline and arginine metabolism and Ardu.EW2ZU for galactose metabolism were upregulated in response to stress in both AST-DS and AST-WW and ADU DS and ADU WW comparison groups. Similarly, DRGs such as AT3G27850 for propanoate metabolism, Aradu.XCD6I for stilbenoid, diarylhetanoid, gingerol biosynthesis, and pinene degradation, limonene Aradu.67ZY4 for oxidative phosphorylation, and Aradu.798H5 for Beta-Alanine metabolism and fatty acid degradation, etc., were upregulated in the ADU genotype in both the ADU-DS vs. ADU-WW and ADU-DS vs. AST-DS comparison groups. The upregulation of these genes in response to drought stress may provide understanding about the mechanisms essential for plant response to drought stress.
RNA-seq DEG approval by qRT-PCR
The RNA-seq data were further endorsed by qRT-PCR. In this work, an aggregate of 12 selected DEGs and 8 DE TFs genes were used to verify the RNA-seq results (Table S8). Despite slight differences in expression levels, almost all of the 15 selected genes had the same expression pattern, as shown by the RNA-seq data. The high correlation between the qRT-PCR results and the RNA-seq data (r = 0.96, p 2.2e-16, Fig. 6) confirmed the RNA-seq results.
SNP marker identification and validation
A total of 366 SNPs were identified between ADU and AST occurring in DEGs aligned for ADU (264) and AST (102) genotypes. The identified SNPs were located on a total of 134 DEGs, with 98 and 36 genes on the ADU and the AST genome, respectively. SNPs were found in four distinct combinations and classified as missense, splice region, 5′UTR, 3′UTR, stop gain, start lost, and stop loss variants (Table 1). Multiple missense variants on the genomes of ADU and AST in four combinations were 126 and 70, respectively. Overall 20 sets of allele-specific primers for 20 genes were designed to validate the polymorphism in ADU and AST (Table S9) by qRT-PCR. Differences in cycle threshold (Ct) values were used to distinguish homozygous and heterozygous alleles. If the Ct value for an allelic primer set of a gene is 30.0 in one genotype but not in the other, this indicates the existence of two distinct alleles, and vice versa. There was no polymorphism exhibited by 18 out of the 20 sets of allelic primers designed (Table 2), while there was some polymorphism in three SNPs representing two genes. The first SNP (Ct > 30 in ADU and Ct 30 in AST) is from the NFAY transcription factor (Aradu.YE2F8) gene, while the next SNP (Ct value of 20 in ADU and Ct > 25.0 in AST) is from the expansin alpha (Aradu.78HGD) gene, and the last SNP (Ct value of 20 in ADU and Ct > 25.0 in AST) is from cytokine (Aradu.U999X). These validated SNPs can be used to screen the first-generation breeding material obtained by crossing parent plants for high performance under drought stress. The distribution of SNP variants and different DEGs on the chromosome is shown in Fig. 7.
Discussion
Drought stress is often deliberated as one of the highly damaging environmental stresses limiting agricultural productivity worldwide (Fathi and Tari 2016). Peanut is drought-tolerant to a certain extent. Peanut varieties react to water stress differently depending on the growth stage of the crop, the farming system, and the duration of the water stress (Thangella et al. 2018). Nevertheless, water deficiency stress during the plant establishment stage would have a significant impact on peanut yield and productivity (Prasad et al. 2010). Thus, strengthening peanut drought tolerance is critical, and further study is necessary to investigate and comprehend drought stress. In this study, we conducted a physio-biochemical and transcriptome investigation of wild drought-tolerant peanut genotype at the seedling stage, in drought, well-watered, and re-watered conditions. Plants have evolved ways for adapting to and surviving drought stress. Increased ROS generation in the various cellular partitions, specifically the mitochondria, chloroplasts, and peroxisomes is an unavoidable effect of drought stress (Verma et al. 2019). ROS signaling is connected to sugar sensing and Ca2+ fluxes, and it is probable that during drought stress, ROS signaling is implicated upstream and downstream of the signaling pathways dependent on ABA. However, if drought stress continues to a certain level, ROS generation will exceed the anti-oxidant system’s scavenging function, leading to irreparable cellular damage (Qi et al. 2018). MDA is a marker for lipid peroxidation detection within plants and is often applied to assess plant tolerance to biological or non-biological stimuli (de Dios Alché 2019). Drought-resistant plants may show a very dual trend of increasing ROS and antioxidant enzyme levels in a stressed environment, while levels of lipid oxidation markers such as malondialdehyde (MDA) usually decrease in response to drought stress. In this experiment, the levels of ROS and MDA were higher in leaf tissues of AST-DS than in ADU-DS during drought (Fig. 2b), possibly due to the greater lipid membrane peroxidation caused by the excess of ROS (de Dios Alché 2019).
Plants have evolved efficient systems to protect themselves from such toxic ROS. These include two groups of enzymatic and non-enzymatic antioxidants: glutathione reductase (GR), catalase (CAT), ascorbate peroxidase (APX), peroxidase (POX), and superoxide dismutase (SOD) are the best known from the first class; and for the second group, tocopherols, glutathione, ascorbic acid, and carotenoids can be mentioned (Caverzan et al. 2016). In this experiment, the level of enzymatic antioxidants increased significantly in the tolerant genotype (ADU) after drought treatment. The concentration of some of these antioxidants varied two- to fourfold in ADU-DS compared with AST-DS (Fig. 2g–k). Considering the higher level of antioxidant enzymes and the lower level of ROS and MDA in ADU-DS, we might wrap up that the ADU genotype has great potential to reduce the deleterious effects of ROS under water deficit.
Proline accumulation under different abiotic stresses can be considered as a stress tolerance indicator (Shafi et al. 2019), since a surge in proline content allows plants to restore osmotic homeostasis by increasing water potential and protecting enzymes, cell organelles, and proteins from damage caused by abiotic stress (Bashir et al. 2020). In this study, ADU-DS exhibited higher proline content than the control and the post-irrigated variant, providing further evidence of how this genotype copes with osmotic stress caused by water deficit (Fig. 2a). Interestingly, the expression profile of genes encoding enzymes involved in proline metabolism showed upregulation in ADU-DS compared to the WW situation. Some key genes aldehyde dehydrogenase (such as Aradu.288JJ, Aradu.8BQ4V, Aradu.I79F7, Aradu.R63R7, and Aradu.ZIF6R), glutamate 5-semialdehyde dehydrogenase activity (Aradu.TN0QL and Aradu.QYZ18), and glutamate synthase (Aradu.02IKD), which are involved in the reduction of glutamate to Δ1-pyrroline-5-carboxylate, are particularly activated at ADU-DS (Table S2). Proline is produced from glutamate by the activity of three enzyme-encoding genes: Pyrroline-5-carboxylate reductase (P5CR), Pyrroline-5-carboxylate synthase (P5CS), and Pyrroline-5-carboxylate synthase-2 (P5CS2) (Frimpong et al. 2021). Again, the genes encoding pyrroline-5-carboxylate reductase activity (P5CR; Aradu.031BD), proline biosynthesis process (Aradu.K3KYD), and ornithine metabolic process (Aradu.MA8XX) were the most overexpressed genes in ADU genotype under drought stress.
When plants are exposed to stressful conditions such as high or low temperatures, salinity, drought, heavy metal pathogen attack, or even mechanical damage, gene expression begins to change as a direct result. However, it is well acknowledged that drought tolerance is a complicated occurrence involving the action and interaction of multiple genes (Mahmood et al. 2020; Yuan et al. 2019). We used RNA-seq to interpret the DEGs between the drought-tolerant ADU genotype and the drought-sensitive genotype AST to further investigate the mechanism of drought tolerance in peanut response to drought stress at the molecular level. Apparently, there was a remarkable difference in gene expression in response to drought stress. RNA-seq analysis revealed 1427 DEGS between the two genotypes. These transcripts exhibited different functions and belonged to different families such as TFs, membrane composition and signaling molecules, osmoprotectors, protection from oxidative damage, cellular transport systems, amino acid metabolism, hormone biosynthesis, and protein kinase.
Transcription factors
Transcription factors play crucial role in regulating plant transcription in response to biotic and abiotic stresses (Desai et al. 2021). In this study, the bHLH, NAC, GATA, and WRKY transcription factor families were the most overexpressed TFs. Therefore, these TFs most likely played an important role in plants facing water deficit stress. There are many reports on various crops indicating the importance of TFs in plant response to drought stress. MdCIB1, a bHLH gene in apples, acts as a positive regulator in drought stress by controlling stomata closure and recovery ROS (Ren et al. 2021). Here, 22 bHLHs genes were strongly induced when peanut was exposed to water deficit. Moreover, RNA-seq and quantitative real-time PCR studies in peanut showed that several NAC genes were activated upon salt and drought stress (Beillouin et al. 2021) (Yuan et al. 2020). In this study, 25 NAC genes were induced in response to drought stress, with NACs 65, 90, 87, 18, 100, 102, and 25 showing the highest increase in expression in response to stress. Among these NAC genes, NAC 18, AhNAC 87, AhNAC 102, and AhNAC 65 were reported by Yuan, Cuiling, et al. (2020) to have crucial roles in drought stress tolerance (Yuan, et al. 2020). It is well understood that changes in the expression patterns or activity of WRKY genes result in activation of numerous signaling and reprogramming pathways (Wani et al. 2021). Transgenic soybean overexpressing GmWRKY54 showed higher resistance to drought stress. This WRKY enhances stress tolerance by triggering the ABA/Ca2C signaling pathways involved in closure of stomata and by inducing the expression of various DRGs and stress-related TFs (Wei et al. 2019). Here, various WRKYs TFs were induced during drought stress, and most WRKY genes were upregulated in response to drought stress. Among the different WRKYs families, WRKYs 75 was highly unregulated in response to water deficit, which was previously reported to be involved in the development of root under stress (Khan et al. 2018). In addition to ABA-dependent regulation of DRGs, many DRGs’ expression is regulated by other TFs such as drought-responsive element (DRE) and C-repeat (CRT) cis-acting elements in collaboration with DRE-binding protein (DREB) or C-repeat-binding factor (CBF) transcription elements (Liu et al. 2018). In the present experiment, DREB2A-regulated genes LEA and DREB2A were upregulated in response to drought treatment (Fig. 8).
Overview of the drought stress signaling pathway. The Ca2 + channel acts as an osmosensor and increases cytosolic free Ca2 + in response to drought stress, which eventually activates CIPKs and CDPKs. Under osmotic stress, SnRK2 are activated by Ca2 + , which is a prerequisite for the accumulation of ABA, and control osmotic adjustment under stress conditions. In the presence of ABA, the ABA receptors PYR /PIL bind to ABA and inhibit PP2C activity, leading to autoactivation of SnRK2s, which phosphorylate their downstream targets such as transcription factors and modulate their expression. The MAP kinase cascade pathway carries the signal for drought stress tolerance response the products of these genes confer drought stress tolerance. A group of TFs such as bZIP, NAC, MYB, DREB, WRKY, and HDZF are also involved in ABA -inducible gene expression and drought responses
Hormone biosynthesis
Numerous genes related to the biosynthesis of hormones, including those involved in ethylene, ABA, auxins, and gibberellins, have been shown to be expressed differentially during drought. With the exception of gibberellin, genes related to biosynthesis of hormones were generally upregulated under drought stress treatments. In our study, genes involved in ABA biosynthesis were also constitutively induced in the tolerant genotype (ADU). The plant hormone ABA plays an important part in many signal transductions (Yang et al. 2019), and is triggered by various stresses such as drought and salinity (Ku et al. 2018). In this study, a gene encoding LEA-like proteins (Aradu.0124 J) was highly upregulated in response to stress conditions. Overexpression of LEA-like proteins in rice increases stress tolerance in stressed plants (Xiao et al. 2007). In this study, upregulation of 9-cis-epoxycarotenoid dioxygenase (NCED), including Aradu.Y48CE and Aradu.Y6MST, which are key enzymes in the biosynthesis of ABA, was also observed, whereas genes involved in gibberellin production were typically downregulated in tolerant plants under treatment. In Arabidopsis thaliana, At-NCED3 was strongly upregulated under salinity and dehydration stress, and overexpression of At-NCED3 improved tolerance to dehydration stress in transgenic plants (Truong et al. 2021). Lv et al. (2021) indicated that in wheat subjected to drought stress, the accumulation of ABA in grains increased sharply, while the GA content dropped significantly (Lv, et al. 2021). Thus, the inhibition of GA biosynthesis in tolerant lines could be regulated by promoting ABA production in the root tissue.
The accumulation of flavonoids in response to various abiotic stresses such as UV-B (Schenke et al. 2019), cold (Zhao et al. 2019), photooxidative stress (Csepregi and Hideg 2018), and drought (Gharibi et al. 2019) has been previously identified as an important non-enzymatic antioxidant. In this study, DEGs exhibited significantly increased transcript levels in anthocyanin and flavonoid production when the tolerant genotype (ADU) was exposed to drought stress. All these abiotic stresses lead to the accumulation of ROS in the cell; the accumulation of flavonoids may serve as a scavenger of ROS produced by such stresses (Czarnocka and Karpiński 2018). Simply put, these results show that flavonoids and anthocyanins accumulate more in response to water deficiency, which may explain the higher tolerance to drought-induced oxidative damage in this genotype. In addition, the expression of genes involved in auxin signal transduction, such as ARF and the Aux/IAA families, was higher in ADU-DS than in AST-DS (Fig. 8). It is well known that the Aux/IAA families and auxin response factor (ARF) play a crucial role in auxin-mediated responses in water deficit (Luo et al. 2018; Song et al. 2019).
Cellular transport systems
Tonoplast intrinsic proteins (TIP) are among the five aquaporin subfamilies of higher plants. The expression of TIPs can be affected by various abiotic stresses such as cold, drought, and salinity (Kurowska 2020). In Nicotiana glauca, two homologous TIP, NgMIP3 and NgMIP2 genes were downregulated in response to long-term water deficiency (Smart et al. 2001); and in Arabidopsis, the AtTIP2;2, AtTIP2;1, AtTIP1;2, and AtTIP1;1 genes were downregulated more than fourfold after a 12-day drought treatment (Alexandersson et al. 2005). On the contrary, when the expression levels of AtTIP2;1 and AtTIP1;1 were examined after 48 h of drought treatment, all of these genes were found to be upregulated in tolerant genotypes (Feng et al. 2018). Among the various DEGs identified, 14 ion transporter genes encoding oxidoreductases, glutamate receptors, and nucleobase ascorbate transporters were explicitly upregulated in the ADU genotype under drought stress. In addition, several genes encoding ion transporters for Ca2+, Na+, and K+ were activated in both genotypes (but with higher expression levels in the ADU genotype) under drought stress, indicating the possible role of these genes in induced drought stress signal transduction.
Signaling and other abiotic stress regulated genes
Genes involved in signal transduction, such as calmodulin (CML), calcium-dependent protein kinases (CDPKs), and calmodulin-related calcium sensing proteins, ABA responsive receptor-like cytoplasmic kinases (RLCKs), and calcineurin B-like protein-interacting protein kinases (CIPKs) showed bipolar expression patterns in response to drought stress treatments. A large number of ABA responsive genes were induced under both conditions (well-watered and drought-treated), but with a higher expression level during drought. The results suggest that a protein serine/threonine kinase activity (Aradu.4AW6R) was specifically activated in drought-treated plants. Lim et al. showed in a study of pepper roots under water-deficit stress that serine/threonine kinase activity responded strongly to ABA (Lim et al. 2020).
The calcineurin B-like protein interacting protein kinases (CIPKs) and receptor-like cytoplasmic kinase (RLCK) play an important role in plant under drought stress (Lu et al. 2020; Mahmood, et al. 2020). We observed that a large number of members of the CIPK and RLCK families were highly expressed in drought-stressed plants compared to well-watered plants. Transgenic Arabidopsis plants overexpressing mammalian inositol polyphosphate 5-phosphatase showed greater tolerance to drought and lost less water than non-transgenic control plants (Rabara et al. 2015). Interestingly, in this work, the expression of inositol polyphosphate 5-phosphatase-like genes was strongly upregulated in drought-treated plants compared to well-watered and re-watered plants.
The transcription levels of some stress-responsive genes, namely dehydrins (DHN1) and heat shock protein (HSP70), signaling genes such as Map kinase (MAPK2/4) and genes involved in synthesis of sugars such as isoflavanoid reductase (IFR) and myoinositol phosphate synthase (MIPS), and eight TFs were analyzed by qRT-PCR under all three conditions (Fig. 7). Notwithstanding a slight variation in log2C of some genes such as DHN1, HSP70, GNAC, and MAPK2/4, the expression pattern was like that observed by RNA-seq. Earlier studies have revealed that several stress-responsive genes, especially HSP70, MIPS, IFR, and LEA, were strongly induced under drought stress (Kumar et al. 2020; Xiong et al. 2017). These results suggest that DHN1, HSP70, LEA-5, IFR, DHN1, and MAPK2/4 genes, when upregulated, contribute to drought tolerance of wild type when exposed to drought treatment. Transcription factors (TFs) have a vital role in conferring drought tolerance by controlling gene expression (Fig. 8). NAC, one of the most important plant-specific transcription factors, was found to be strongly upregulated during drought, indicating its influence on genes involved in drought tolerance (Puranik et al. 2013). Furthermore, the study on SNP validation has given more information about genetic markers that, once validated in a larger number of genotypes, can be utilized in genome-assisted breeding to improve tolerance to drought stress.
Conclusions
Here, we sought to elucidate the mechanism of drought resistance in peanut by performing phenotypic, physiological, and genome-wide transcriptome assessments of two wild peanut genotypes. The results propose that the higher buildup of LTR, CRPK, CHS, LEA, and also HSP in the tolerant genotype provides better biochemical, physiological, and molecular responses and therefore improved tolerance to drought stress. RNA-seq analysis identified 1465 DEGs mainly associated with secondary metabolite biosynthesis, phytohormone signal transduction, flavonoid metabolism, carbohydrate metabolism, and transcriptional regulator activity. The results show that hormone transduction and signaling can enhance the expression of drought tolerance-related DEGs involved in encoding auxin-response factors, flavonoid biosynthesis, ABA signal transduction, higher osmolyte accumulation, and also producing additional soluble sugars for enhanced metabolism of carbon, leading to increased root biomass in the tolerant genotypes under drought stress conditions. In addition, discovered SNPs associated with lipid transfer and F-box proteins showed variations between ADU and AST genotypes, which can be used as linked markers in genome-assisted breeding programs to perform first-generation selection. The finding of this research may pave the way for creating drought resistance in peanut and other related crops through techniques such as molecular breeding or genetic engineering. The DEGs identified through this study can be considered as good candidates for further functional analysis, to clearly interpret their roles in providing drought tolerance.
Abbreviations
- ABA:
-
Abscisic Acid
- ABFs:
-
ABA response element-binding factors
- APX:
-
Ascorbate Peroxidase
- CAT:
-
Catalase
- cDNA:
-
Complementary DNA
- DEGs:
-
Differentially expressed genes
- DGRs:
-
Drought Response Genes
- DHAR:
-
Dehydroascorbate Reductase
- EL:
-
Electrolytic Leakage
- FDR:
-
False Discovery Rate
- FPKM:
-
Fragments per kb per million fragments
- GLCAKG:
-
Glucosyluronic Acid Kinase
- GO:
-
Gene Ontology
- GPX:
-
Glutathione Peroxidase
- GSH:
-
Reduced Glutathione
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- MDA:
-
Malondialdehyde
- NCBI:
-
National Center for Biotechnology Information
- OP:
-
Osmotic Potential
- PAL:
-
Phenylalanine Ammonia Lyase
- POX:
-
Peroxidase
- PP2C:
-
Protein Phosphatase 2C
- qRT-PCR:
-
Quantitative real-time PCR
- RNA-seq:
-
RNA sequence
- ROS:
-
Reactive Oxygen Species
- RWC:
-
Relative Water Content
- SA:
-
Salicylic Acid
- SNRK2:
-
SNF1-related kinase
- SOD:
-
Superoxide Dismutase
References
Abdelrahman M, Jogaiah S, Burritt DJ, Tran LSP (2018) Legume genetic resources and transcriptome dynamics under abiotic stress conditions. Plant Cell Environ 41:1972–1983
Alexandersson E, Fraysse L, Sjövall-Larsen S, Gustavsson S, Fellert M, Karlsson M, Johanson U, Kjellbom P (2005) Whole gene family expression and drought stress regulation of aquaporins. Plant Mol Biol 59:469–484
Balyan S, Joseph SV, Jain R, Mutum RD, Raghuvanshi S (2020) Investigation into the miRNA/5′isomiRNAs function and drought-mediated miRNA processing in rice. Funct Integr Genomics:1–14
Barrs H, Weatherley P (1962) A re-examination of the relative turgidity technique for estimating water deficits in leaves. Aust J Biol Sci 15:413–428
Bashir MA, Silvestri C, Ahmad T, Hafiz IA, Abbasi NA, Manzoor A, Cristofori V, Rugini E (2020) Osmotin: a cationic protein leads to improve biotic and abiotic stress tolerance in plants. Plants 9:992
Bates LS, Waldren RP, Teare I (1973) Rapid determination of free proline for water-stress studies. Plant Soil 39:205–207
Beillouin D, Ben‐Ari T, Malezieux E, Seufert V, Makowski D (2021) Positive but variable effects of crop diversification on biodiversity and ecosystem services. Global Change Biology. https://doi.org/10.1111/gcb.15747
Bhauso TD, Thankappan R, Kumar A, Mishra GP, Dobaria JR, Rajam MV (2014) Over-expression of bacterial mtlD gene confers enhanced tolerance to salt-stress and water-deficit stress in transgenic peanut (Arachis hypogaea) through accumulation of mannitol. Aust J Crop Sci 8:413
Borden KA, Thomas SC, Isaac ME (2020) Variation in fine root traits reveals nutrient-specific acquisition strategies in agroforestry systems. Plant Soil 453:139–151
Brasileiro AC, Morgante CV, Araujo AC, Leal-Bertioli SC, Silva AK, Martins AC, Vinson CC, Santos CM, Bonfim O, Togawa RC (2015) Transcriptome profiling of wild Arachis from water-limited environments uncovers drought tolerance candidate genes. Plant Mol Biol Report 33:1876–1892
Brown J, Pirrung M, McCue LA (2017) FQC Dashboard: integrates FastQC results into a web-based, interactive, and extensible FASTQ quality control tool. Bioinformatics 33:3137–3139
Caverzan A, Casassola A, Brammer SP (2016) Reactive oxygen species and antioxidant enzymes involved in plant tolerance to stress. SHANKER AK & SHANKER C Abiotic and biotic stress in plants-Recent advances and future perspectives Publisher InTech:463–480
Che Omar S, Bentley MA, Morieri G, Preston GM, Gurr SJ (2016) Validation of reference genes for robust qRT-PCR gene expression analysis in the rice blast fungus Magnaporthe oryzae. PloS one 11:e0160637
Csepregi K, Hideg É (2018) Phenolic compound diversity explored in the context of photo-oxidative stress protection. Phytochem Anal 29:129–136
Czarnocka W, Karpiński S (2018) Friend or foe? Reactive oxygen species production, scavenging and signaling in plant response to environmental stresses. Free Radical Biol Med 122:4–20
Dang PM, Chen CY, Holbrook CC (2012) Identification of drought-induced transcription factors in peanut (Arachis hypogaea L.). J Mol Biochem 1
de Dios Alché J (2019) A concise appraisal of lipid oxidation and lipoxidation in higher plants. Redox Biol 23:101136
Desai H, Hamid R, Ghorbanzadeh Z, Bhut N, Padhiyar SM, Kheni J, Tomar RS (2021) Genic microsatellite marker characterization and development in little millet (Panicum sumatrense) using transcriptome sequencing. Sci Rep 11:1–14
Dobin A, Gingeras TR (2015) Mapping RNA‐seq reads with STAR. Curr Protoc Bioinformatics 51:11.14. 11–11.14. 19
Eybishtz A, Peretz Y, Sade D, Akad F, Czosnek H (2009) Silencing of a single gene in tomato plants resistant to tomato yellow leaf curl virus renders them susceptible to the virus. Plant Mol Biol 71:157–171
Fathi A, Tari DB (2016) Effect of drought stress and its mechanism in plants. Int J Life Sci 10:1–6
Feng Z-J, Xu S-C, Liu N, Zhang G-W, Hu Q-Z, Xu Z-S, Gong Y-M (2018) Identification of the AQP members involved in abiotic stress responses from Arabidopsis. Gene 646:64–73
Frimpong F, Windt CW, van Dusschoten D, Naz AA, Frei M, Fiorani F (2021) A wild allele of pyrroline-5-carboxylate synthase1 leads to proline accumulation in spikes and leaves of barley contributing to improved performance under reduced water availability. Front Plant Sci 12:180
Garg R, Shankar R, Thakkar B, Kudapa H, Krishnamurthy L, Mantri N, Varshney RK, Bhatia S, Jain M (2016) Transcriptome analyses reveal genotype-and developmental stage-specific molecular responses to drought and salinity stresses in chickpea. Sci Rep 6:1–15
Gharibi S, Tabatabaei BES, Saeidi G, Talebi M, Matkowski A (2019) The effect of drought stress on polyphenolic compounds and expression of flavonoid biosynthesis related genes in Achillea pachycephala Rech. f. Phytochemistry 162:90–98
Ghosh S, Chan C-KK (2016) Analysis of RNA-seq data using TopHat and Cufflinks plant Bioinformatics. Springer, pp. 339–361.
Gui Y-W, Sheteiwy MS, Zhu S-G, Batool A, Xiong Y-C (2021) Differentiate effects of non-hydraulic and hydraulic root signaling on yield and water use efficiency in diploid and tetraploid wheat under drought stress. Environ Exp Bot 181:104287
Hamid R, Jacob F, Marashi H, Rathod V, Tomar RS (2020) Uncloaking lncRNA-meditated gene expression as a potential regulator of CMS in cotton (Gossypium hirsutum L.). Genomics 112:3354–3364
Hamid R, Marashi H, Tomar RS, Malekzadeh Shafaroudi S, Sabara PH (2019) Transcriptome analysis identified aberrant gene expression in pollen developmental pathways leading to CGMS in cotton (Gossypium hirsutum L.). PloS one 14:e0218381
Hamid R, Tomar RS, Marashi H, Shafaroudi SM, Golakiya BA, Mohsenpour M (2018) Transcriptome profiling and cataloging differential gene expression in floral buds of fertile and sterile lines of cotton (Gossypium hirsutum L.). Gene 660:80–91
Hiscox J, Israelstam G (1979) A method for the extraction of chlorophyll from leaf tissue without maceration. Can J Bot 57:1332–1334
Jiang C, Li X, Zou J, Ren J, Jin C, Zhang H, Yu H, Jin H (2020) Comparative transcriptome analysis of genes involved in two peanut varieties under drought stress.
Jiang C, Li X, Zou J, Ren J, Jin C, Zhang H, Yu H, Jin H (2021) Comparative transcriptome analysis of genes involved in the drought stress response of two peanut (Arachis hypogaea L.) varieties. BMC Plant Biol 21:1–14
Joshi R, Wani SH, Singh B, Bohra A, Dar ZA, Lone AA, Pareek A, Singla-Pareek SL (2016) Transcription factors and plants response to drought stress: current understanding and future directions. Front Plant Sci 7:1029
Khan A, Pan X, Najeeb U, Tan DKY, Fahad S, Zahoor R, Luo H (2018) Coping with drought: stress and adaptive mechanisms, and management through cultural and molecular alternatives in cotton as vital constituents for plant stress resilience and fitness. Biol Res 51
Ku Y-S, Sintaha M, Cheung M-Y, Lam H-M (2018) Plant hormone signaling crosstalks between biotic and abiotic stress responses. Int J Mol Sci 19:3206
Kumar J, Sen Gupta D, Djalovic I, Kumar S, Siddique KH (2021) Root-omics for drought tolerance in cool-season grain legumes. Physiol Plant 172:629–644
Kumar S, Ayachit G, Sahoo L (2020) Screening of mungbean for drought tolerance and transcriptome profiling between drought-tolerant and susceptible genotype in response to drought stress. Plant Physiol Biochem 157:229–238
Kurowska MM (2020) TIP aquaporins in plants: role in abiotic stress toleranceabiotic stress in plants. IntechOpen.
Lamaoui M, Jemo M, Datla R, Bekkaoui F (2018) Heat and drought stresses in crops and approaches for their mitigation. Front Chem 6:26
Launio CC, Luis JS, Angeles YB (2018) Factors influencing adoption of selected peanut protection and production technologies in Northern Luzon, Philippines. Technol Soc 55:56–62
Lim J, Lim CW, Lee SC (2020) Pepper novel serine-threonine kinase CaDIK1 regulates drought tolerance via modulating ABA sensitivity. Front Plant Sci 11:1133
Liu S, Lv Z, Liu Y, Li L, Zhang L (2018) Network analysis of ABA-dependent and ABA-independent drought responsive genes in Arabidopsis thaliana. Genet Mol Biol 41:624–637
Lorrain C, Marchal C, Hacquard S, Delaruelle C, Pétrowski J, Petre B, Hecker A, Frey P, Duplessis S (2018) The rust fungus Melampsora larici-populina expresses a conserved genetic program and distinct sets of secreted protein genes during infection of its two host plants, larch and poplar. Mol Plant Microbe Interact 31:695–706
Lu L, Chen X, Zhu L, Li M, Zhang J, Yang X, Wang P, Lu Y, Cheng T, Shi J (2020) NtCIPK9: a calcineurin B-like protein-interacting protein kinase from the halophyte Nitraria tangutorum, enhances arabidopsis salt tolerance. Front Plant Sci 11:1112
Luo J, Zhou J-J, Zhang J-Z (2018) Aux/IAA gene family in plants: molecular structure, regulation, and function. Int J Mol Sci 19:259
Lv X, Ding Y, Long M, Liang W, Gu X, Liu Y, Wen X (2021) Effect of foliar application of various nitrogen forms on starch accumulation and grain filling of wheat (Triticum aestivum L.) under drought stress. Frontiers Plant Sci 12:463
Maga TJ (2021) Evaluation of faba bean (Vicia faba L.) genotypes for drought tolerance under the subtropical region of New South Wales.
Mahmood T, Khalid S, Abdullah M, Ahmed Z, Shah MKN, Ghafoor A, Du X (2020) Insights into drought stress signaling in plants and the molecular genetic basis of cotton drought tolerance. Cells 9:105
Marshall AH, Collins RP, Humphreys MW, Scullion J (2016) A new emphasis on root traits for perennial grass and legume varieties with environmental and ecological benefits. Food Energy Security 5:26–39
Martignago D, Rico-Medina A, Blasco-Escámez D, Fontanet-Manzaneque JB, Caño-Delgado AI (2020) Drought resistance by engineering plant tissue-specific responses. Front Plant Sci 10:1676
Oliveros JC (2007) VENNY. An interactive tool for comparing lists with Venn diagrams. http://bioinfogp.cnb.csic.es/tools/venny/index.html
Oshunsanya SO, Nwosu NJ, Li Y (2019) Abiotic stress in agricultural crops under climatic conditions Sustainable agriculture, forest and environmental management. Springer, pp. 71–100.
Polania J, Poschenrieder C, Rao I, Beebe S (2017) Root traits and their potential links to plant ideotypes to improve drought resistance in common bean. Theor Exp Plant Physiol 29:143–154
Prasad PV, Kakani VG, Upadhyaya HD (2010) Growth and production of groundnut. UNESCO Encyclopedia:1–26
Puranik S, Sahu PP, Mandal SN, Parida SK, Prasad M (2013) Comprehensive genome-wide survey, genomic constitution and expression profiling of the NAC transcription factor family in foxtail millet (Setaria italica L.). PloS one 8:e64594
Qi J, Song CP, Wang B, Zhou J, Kangasjärvi J, Zhu JK, Gong Z (2018) Reactive oxygen species signaling and stomatal movement in plant responses to drought stress and pathogen attack. J Integr Plant Biol 60:805–826
Rabara RC, Tripathi P, Reese RN, Rushton DL, Alexander D, Timko MP, Shen QJ, Rushton PJ (2015) Tobacco drought stress responses reveal new targets for Solanaceae crop improvement. BMC Genomics 16:1–23
Rathod V, Hamid R, Tomar RS, Padhiyar S, Kheni J, Thirumalaisamy P, Munshi NS (2020a) Peanut (Arachis hypogaea) transcriptome revealed the molecular interactions of the defense mechanism in response to early leaf spot fungi (Cercospora arachidicola). Plant Gene 23:100243
Rathod V, Hamid R, Tomar RS, Patel R, Padhiyar S, Kheni J, Thirumalaisamy P, Munshi NS (2020b) Comparative RNA-seq profiling of a resistant and susceptible peanut (Arachis hypogaea) genotypes in response to leaf rust infection caused by Puccinia arachidis. 3 Biotech 10:1–15
Reddy YN, Reddy YP, Ramya V, Suma L, Reddy AN, Krishna SS (2021) Drought adaptation: approaches for crop improvement Millets and Pseudo Cereals. Elsevier, pp. 143–158.
Ren Y-R, Yang Y-Y, Zhao Q, Zhang T-E, Wang C-K, Hao Y-J, You C-X (2021) MdCIB1, an apple bHLH transcription factor, plays a positive regulator in response to drought stress. Environ Exp Bot 104523
Saxena RK, Cui X, Thakur V, Walter B, Close TJ, Varshney RK (2011) Single feature polymorphisms (SFPs) for drought tolerance in pigeonpea (Cajanus spp.). Funct Integr Genomics 11:651–657
Schenke D, Utami HP, Zhou Z, Gallegos M-T, Cai D (2019) Suppression of UV-B stress induced flavonoids by biotic stress: is there reciprocal crosstalk? Plant Physiol Biochem 134:53–63
Schmieder R, Edwards R (2011) Quality control and preprocessing of metagenomic datasets. Bioinformatics 27:863–864
Shafi A, Zahoor I, Mushtaq U (2019) Proline accumulation and oxidative stress: diverse roles and mechanism of tolerance and adaptation under salinity stress Salt stress, microbes, and plant interactions: mechanisms and molecular approaches. Springer, pp. 269–300.
Shen Y, Zhiguo E, Zhang X, Liu Y, Chen Z (2015) Screening and transcriptome analysis of water deficiency tolerant germplasms in peanut (Arachis hypogaea). Acta Physiol Plant 37:103
Smart LB, Moskal WA, Cameron KD, Bennett AB (2001) MIP genes are down-regulated under drought stress in Nicotiana glauca. Plant Cell Physiol 42:686–693
Song S, Hao L, Zhao P, Xu Y, Zhong N, Zhang H, Liu N (2019) Genome-wide identification, expression profiling and evolutionary analysis of auxin response factor gene family in potato (Solanum tuberosum group phureja). Sci Rep 9:1–13
Stansell J, Shepherd J, Pallas J, Bruce R, Minton N, Bell D, Morgan L (1976) Peanut responses to soil water variables in the Southeast. Peanut Sci 3:44–48
Tamir HD Root system architecture and its reaction in cereals under water stressed environment. Adv Crop Sci Tech 8:2
Thakur O, Randhawa GS (2018) Identification and characterization of SSR, SNP and InDel molecular markers from RNA-seq data of guar (Cyamopsis tetragonoloba, L. Taub.) roots. BMC Genomics 19:1–14
Thangella PA, Pasumarti SN, Pullakhandam R, Geereddy BR, Daggu MR (2018) Differential expression of leaf proteins in four cultivars of peanut (Arachis hypogaea L.) under water stress. 3 Biotech 8:1–21
Toomer OT (2018) Nutritional chemistry of the peanut (Arachis hypogaea). Crit Rev Food Sci Nutr 58:3042–3053
Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25:1105–1111
Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, Van Baren MJ, Salzberg SL, Wold BJ, Pachter L (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28:511
Truong HA, Lee S, Trịnh CS, Lee WJ, Chung E-H, Hong S-W, Lee H (2021) Overexpression of the HDA15 gene confers resistance to salt stress by the induction of NCED3, an ABA biosynthesis enzyme. Front Plant Sci 12:631
Tulsani NJ, Hamid R, Jacob F, Umretiya NG, Nandha AK, Tomar RS, Golakiya BA (2020) Transcriptome landscaping for gene mining and SSR marker development in Coriander (Coriandrum sativum L.). Genomics 112:1545–1553
Verma G, Srivastava D, Tiwari P, Chakrabarty D (2019) ROS modulation in crop plants under drought stress. Reactive oxygen, nitrogen and sulfur species in plants: production, metabolism, signaling and defense mechanisms:311–336
Wang X, Zenda T, Liu S, Liu G, Jin H, Dai L, Dong A, Yang Y, Duan H (2019) Comparative proteomics and physiological analyses reveal important maize filling-kernel drought-responsive genes and metabolic pathways. Int J Mol Sci 20:3743
Wang Z, Zhang Y, Huang Z, Huang L (2008) Antioxidative response of metal-accumulator and non-accumulator plants under cadmium stress. Plant Soil 310:137–149
Wani SH, Anand S, Singh B, Bohra A, Joshi R (2021) WRKY transcription factors and plant defense responses: latest discoveries and future prospects. Plant Cell Rep:1–15
Wei W, Liang DW, Bian XH, Shen M, Xiao JH, Zhang WK, Ma B, Lin Q, Lv J, Chen X (2019) GmWRKY54 improves drought tolerance through activating genes in abscisic acid and Ca2+ signaling pathways in transgenic soybean. Plant J 100:384–398
Wissuwa M, Kretzschmar T, Rose TJ (2016) From promise to application: root traits for enhanced nutrient capture in rice breeding. J Exp Bot 67:3605–3615
Wu M, Zhang K, Xu Y, Wang L, Liu H, Qin Z, Xiang Y (2021) The moso bamboo WRKY transcription factor, PheWRKY86, regulates drought tolerance in transgenic plants. Plant Physiol Biochem
Xiao B, Huang Y, Tang N, Xiong L (2007) Over-expression of a LEA gene in rice improves drought resistance under the field conditions. Theor Appl Genet 115:35–46
Xiong J, Sun Y, Yang Q, Tian H, Zhang H, Liu Y, Chen M (2017) Proteomic analysis of early salt stress responsive proteins in alfalfa roots and shoots. Proteome Sci 15:1–19
Xiong R, Liu S, Considine MJ, Siddique KH, Lam HM, Chen Y (2021) Root system architecture, physiological and transcriptional traits of soybean (Glycine max L.) in response to water deficit: a review. Physiol Plant 172:405–418
Yan C, Song S, Wang W, Wang C, Li H, Wang F, Li S, Sun X (2020) Screening diverse soybean genotypes for drought tolerance by membership function value based on multiple traits and drought-tolerant coefficient of yield. BMC Plant Biol 20:1–15
Yang J, Duan G, Li C, Liu L, Han G, Zhang Y, Wang C (2019) The crosstalks between jasmonic acid and other plant hormone signaling highlight the involvement of jasmonic acid as a core component in plant response to biotic and abiotic stresses. Front Plant Sci 10:1349
Yuan C, Li C, Lu X, Zhao X, Yan C, Wang J, Sun Q, Shan S (2020) Comprehensive genomic characterization of NAC transcription factor family and their response to salt and drought stress in peanut. BMC Plant Biol 20:1–21
Yuan X, Wang H, Cai J, Bi Y, Li D, Song F (2019) Rice NAC transcription factor ONAC066 functions as a positive regulator of drought and oxidative stress response. BMC Plant Biol 19:1–19
Zhao M, Jin J, Gao T, Zhang N, Jing T, Wang J, Ban Q, Schwab W, Song C (2019) Glucosyltransferase CsUGT78A14 regulates flavonols accumulation and reactive oxygen species scavenging in response to cold stress in Camellia sinensis. Front Plant Sci 10:1675
Zhao X, Li C, Wan S, Zhang T, Yan C, Shan S (2018) Transcriptomic analysis and discovery of genes in the response of Arachis hypogaea to drought stress. Mol Biol Rep 45:119–131
Acknowledgements
We acknowledge the ICAR Directorate of Groundnut Research, Junagadh, Gujarat, India, for providing wild-type peanut seeds and also the Department of Biotechnology, Junagadh Agricultural University, Junagadh (Gujarat) for providing the resources, laboratory facilities and infrastructure required for the experiments.
Funding
This study is funded by the Department of Agricultural Biotechnology, Junagadh Agricultural University, Junagadh, Gujarat.
Author information
Authors and Affiliations
Contributions
FJ: sample collection, executed laboratory procedures of the project, and prepared the initial draft of the manuscript; RH: project administration, performed data analysis, data visualization, diagram preparation, as well as writing, review, and editing the manuscript; ZGH: assisted in improving the manuscript and data analysis as well as diagram preparation; AV: guided throughout the experiment: MJ: executed laboratory procedures of the project.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Highlights
• Transcriptome analysis revealed an aggregate of 1,465 DEGs and among the identified DEGs there were 366 single nucleotide polymorphisms (SNPs).
• These DEGs were mostly involved in DNA methylation or demethylation, phytohormone signal transduction, and flavonoid production.
• The bHLH, NAC, and WRKY transcription factor families were the most overexpressed TFs respectively, indicating that, these TFs most likely played an important role in dealing with water deficiency stress in plants.
• Three SNPs of the gene encoding transcription factor NFAY (Aradu.YE2F8), expansin alpha (Aradu.78HGD) and cytokinin dehydrogenase 1-like (Aradu.U999X) exhibited polymorphism in the different selected genotypes.
• Twenty unigenes involved in responses to drought stress were validated using qRT-PCR.
Supplementary Information
Below is the link to the electronic supplementary material.

Rights and permissions
About this article
Cite this article
Thoppurathu, F.J., Ghorbanzadeh, Z., Vala, A.K. et al. Unravelling the treasure trove of drought-responsive genes in wild-type peanut through transcriptomics and physiological analyses of root. Funct Integr Genomics 22, 215–233 (2022). https://doi.org/10.1007/s10142-022-00833-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10142-022-00833-z