Abstract
Bacillus subtilis is a remarkably diverse bacterial species that displays many ecological functions. Given its genomic diversity, the strain Bacillus subtilis EA-CB0575, isolated from the rhizosphere of a banana plant, was sequenced and assembled to determine the genomic potential associated with its plant growth promotion potential. The genome was sequenced by Illumina technology and assembled using Velvet 1.2.10, resulting in a whole genome of 4.09 Mb with 4332 genes. Genes involved in the production of indoles, siderophores, lipopeptides, volatile compounds, phytase, bacilibactin, and nitrogenase were predicted by gene annotation or by metabolic pathway prediction by RAST. These potential traits were determined using in vitro biochemical tests, finding that B. subtilis EA-CB0575 produces two families of lipopeptides (surfactin and fengycin), solubilizes phosphate, fixes nitrogen, and produces indole and siderophores compounds. Finally, strain EA-CB0575 increased 34.60% the total dry weight (TDW) of tomato plants with respect to non-inoculated plants at greenhouse level. These results suggest that the identification of strain-specific genes and predicted metabolic pathways might explain the strain potential to promote plant growth by several mechanisms of action, accelerating the development of plant biostimulants for sustainable agricultural.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plant growth-promoting rhizobacteria (PGPR) have been used as biostimulants and biopesticides in different crops of economic relevance. They promote plant growth through diverse mechanisms like phytohormone production, nitrogen fixation, siderophore, antibiotic production, induced systemic resistance (ISR), and competition between others (Backer et al. 2018). From the commercially available PGPRs, Bacillus subtilis outstand for its resistance to stress conditions due to spore formation, broad metabolite spectrum, fast growth in liquid media, and the ability to colonize plant surfaces (Kumar et al. 2011). Therefore, different efforts have been made to elucidate its potential and mechanisms of action from different omics perspectives.
From the genome perspective, the first sequenced strain was B. subtilis Marbug 168, which has also become the most studied Gram-positive bacterium, revealing important information about sporulation, germination, nutrition, and development for B. subtilis group. Its genome is 4.2 Mb composed of 4100 genes that encode proteins from which 4% is devoted to antibiotic production and another 4% for sporulation and germination processes (Kunst et al. 1997; Moszer 1998). Since then, the whole-genome sequence of around 200 strains of B. subtilis group have been sequenced (https://www.ncbi.nlm.nih.gov/assembly/?term=Bacillus%20subtilis), generating information on the genomic diversity but also on the systematics of Bacillus genus and the adaptations of this taxon in their natural habitat (Li et al. 2016; Smith et al. 2014; Zeigler 2011). Access to whole-genome sequences and their comparative analyses with different Bacillus subtilis strains and close relatives is important to understand gene flow, speciation, diversity, and genomic dynamics in bacilli for physiological, ecological, and evolutive studies (Earl et al. 2012; Shaligram et al. 2016). Currently, there are studies where several genomes of Bacillus strains are elucidated in order to establish the safety and the efficiency for industrial microbiology (Sulthana et al. 2019), to investigate the genetics of fermenting strains and its relationship with the productivity of metabolites (Deng et al. 2019; Kamada et al. 2015), to clarify the phylogeny of Bacillus groups (Stevens et al. 2019), or to determine plasmid-independent species-species markers (Ivanova et al. 2003). The study of the genome of microorganisms used for biofertilizers and biostimulants production is important to bioinoculants technology because it helps identify genes that contribute to the beneficial activity and increasing knowledge of the molecular mechanisms related to plant growth potential. Also, this provides information for inoculants biosafety and the compliance with existing and developing regulations for this kind of products whose supply and consumption is growing (EPA 2019). The use of next generation sequencing (NGS) has allowed whole-genome sequencing of PGPRs (plant growth-promoting rhizobacteria) isolated from different crops such as wheat, pepper, cotton, and coconut (Gupta et al. 2014; Kai-Jium et al. 2018); and it is an opportunity to obtain information about strains used and to design strategies for development and use of such PGPRs to support sustainable agriculture.
In this research, we present the sequencing, assembling, and characterization of B. subtilis EA-CB0575 whole genome, isolated from a commercial crop of Musa AAA in Colombia; this microorganism has shown successful results for plant growth promotion of banana crops, maize, and tomato (Posada et al. 2018). We hypothesize that the genome sequence of B. subtilis EA-CB0575 will reveal genes involved in plant growth promotion mechanisms, which will be expressed in vitro and potentially promote growth of tomato plants. Therefore, the presence of homologous genes associated with mechanisms of plant growth promotion such as indole and siderophores production, phosphate solubilization, nitrogen fixation, LPs production, and production of volatile compounds was determined for the assembled genome. Biochemical traits, related to these characteristics, were evaluated at in vitro level, and the strain’s potential for growth promotion of tomato crop was determined at greenhouse level as evidence of the relationship between the genomic potential and the capacity of growth promotion in a plant.
Methodology
Microorganisms and culture conditions
B. subtilis EA-CB0575 was isolated from roots of a banana plant cv. Valery from Urabá – Colombia (Northeastern 07° 51′ 58.6″ N, − 76° 37′ 39″ W) in 2009 (Posada et al. 2016). The microorganism was identified by analysis of 16s rRNA gene sequencing (Accession number KC170988) and characterized as a Gram-positive rod (1.85 ± 0.31 μm), forming central spores (0.72 ± 0.15 μm), and producing irregular and matt white colonies with crateriform elevation in TSA X 0.5 (105458, Merck). The strain was stored in TSB medium (TSB, 105459, Merck) with glycerol (20% v/v) at − 80 °C and registered on the bacterial collection RNC 191 (Instituto Alexander Von Humboldt). B. subtilis 168 (Zeigler et al. 2008) and B. velezensis FZB42 (Fan et al. 2018), previously B. amyloliquefaciens FZB42 (Fan et al. 2012), were donated by Dr. Camilo Ramírez (Universidad de Antioquia, Colombia) and Dr. Rainer Borriss (ABiTEP GmbH ABI, Germany) respectively.
Genomic DNA preparation
B. subtilis EA-CB0575 genomic DNA was obtained from cells harvested in the exponential phase of a submerged culture in TSB. Briefly, the strain was cultured 14 h at 150 rpm and 30 °C, centrifugated, and obtained pellet used for DNA extraction using Ultraclean Microbial DNA isolation (Mobio). DNA quality was determined using NanoDrop and agarose 1% gel electrophoresis.
Sequencing and annotation of B. subtilis EA-CB0575 genome
B. subtilis EA-CB0575 genome was sequenced using Illumina HiSeq 2000 paired-end method and 100 bp reads with 265 bp average insert size were obtained. The quality of the FASTQ files was verified with FastQC (Andrews 2010) and reads were trimmed to ensure high quality (Phred score > 30) using Trimmomatic version 0.35 (Bolger et al. 2014). The genome was assembled with Velvet 1.2.10 (Zerbino and Birney 2008) using k-mer = 95 and derived statistics of the assembly process were determined (genome total length, N50, N90, time for running (min), cluster numbers, cluster average size (pb)). Subsequently, OriFind and Rast software (Aziz et al. 2008) were used for ORI and ORF prediction, respectively, and the order of alignments with ProgressiveMauve aligner and Murasaki software (Popendorf et al. 2010) with B. subtilis 168 as the reference strain. Once the genome was assembled, gene prediction and annotation were performed using RAST (http://rast.nmpdr.org/), and circular genome representation was done with CGView webserver (http://stothard.afns.ualberta.ca/cgview_server/). RAST metabolic predictions were used to determine indole, siderophores, and volatile compounds production. Phosphate solubilization and nitrogen fixation capacity were determined to evaluate the presence of phy and nif genes inside the genome. This search was done using the reference strains B. velezensis FZB42 (GenBank Accession number: CP000560.1) for indole, specifically IAA (indole acetic acid), volatiles, and siderophores production; B. subtilis 168 for phytase production; and Klebsiella pneumoniae (Genbank accession number X13303.1) for nitrogen fixation. Local BLAST between references and B. subtilis EA-CB0575 was performed and subsequently, a combined procedure between the RAST metabolic annotation (KEGG tool) and the search for genes of interest was used to find related genes to the PGPR traits in the assembled genome of the B. subtilis strain EA-CB0575. The nif and phy cluster genes search was performed comparing the homology of these genes with sections of the evaluated strains.
Identification and characterization of NRPS genomic clusters
The presence of non-ribosomal peptide synthetases (NRPS) coding regions in B. subtilis EA-CB0575 genome was evaluated using homologous sequences to coding regions of B. velezensis FZB42 strain, which produce three families of LPs (Koumoutsi et al. 2004). BLASTn algorithm was used to determine differences in operon sequences throughout the genome, and Murasaki software 1.68.6 (Popendorf et al. 2010) was used to determine ORFs. RAST and FgenesB operon predictor (Softberry, Inc., NY, USA) was used to evaluate coding sequences. NCBI Conserved Domain Database (CDD) (Marchler-Bauer et al. 2011) was used to contrast reported information with B. subtilis EA-CB0575 genome information and to predict structural domains related to each ORF previously found.
Comparative genomics of B. subtilis EA-CB0575 and phylogenetic study
B. velezensis FZB42, B. subtilis 168, and B. subtilis EA-CB0575 genomes were compared to locate gene clusters involved in the secondary metabolites production using Murasaki 1.68.6 software (Popendorf et al. 2010) and GMV tool. A phylogenetic reconstruction analysis was performed to infer the evolutionary relationships of B. subtilis EA-CB0575 and others Bacillus spp. with whole genomes reported in GenBank NCBI. This analysis was done using five housekeeping genes (rpoB, gyrA, purH, polC, and groEL) for thirty-one Bacillus spp. strains and using Peptoclostridium difficile as the outgroup (Table 1). Bayesian inference (BI) and maximum likelihood (ML) were used for the phylogenetic reconstruction. Sampled genes were aligned independently in an amino acid–based fashion using RevTrans. First, nucleotide coding sequences were translated to amino acids using VirtualRibosome from RevTrans 1.4 package and using genetic bacterial code table # 11 (https://www.ncbi.nlm.nih.gov/Taxonomy/Utils/wprintgc.cgi). Amino acid sequences of each gene were aligned independently using MAFFT 7.213 in automatic mode. Then, alignments were reverse-translated from amino acids to nucleotides with RevTrans 1.4 software (Wernersson and Pedersen 2003). Aligned nucleotide sequences for each gene were concatenated into a single FASTA file and converted to NEXUS format by ReadSeq 2.1.19 (Gilbert 2003), and to Phylip using Biopython utilities. Partitionfinder 1.1 software (Lanfear et al. 2014) was used to find the best partitioning scheme for the data and the appropriate nucleotide substitution models for each partition. The BI analysis was done using the software MrBayes 3.2.2 (Ronquist et al. 2012). Four independent MCMC (Markov Chain Monte Carlo) analyses were carried out with 10 million generations and eight chains (seven hot and one cold) for each one and a relative burn-in fraction of 0.35. Consensus tree for BI was obtained using the remaining trees. ML analysis was performed with Garli HPC-MPI 2.07 (Bazinet et al. 2014). A search was carried out with eight replicates and the one with the highest probability was selected (likelihood), followed by an analysis with 1000 bootstrap replicates. SumTrees 3.3.1 (Sukumaran and Holder 2010) was used to map bootstrap replicates on the best tree from the first analysis. The visualization of the obtained trees was done with FigTree 1.4.1 (Rambaut 2014). The roots of both trees were fixed based on the external group used (Peptoclostridium difficile).
Phenotypic characterization of B. subtilis EA-CB0575 and reference strains
Production of total indoles was determined by the colorimetric technique using Salkowski’s reagent as described by Patten and Glick (2002). Siderophore production was determined by the colorimetric CAS method (Schwyn and Neilands 1987); phosphate solubilization by growing on NBRIP medium (NaCl 1 g; CaCl2*2H2O 0.2 g; MgSO4*7H2O 0.4 g; NH4NO3 1 g; glucose 10 g, benomyl 300 ppm, bactoagar 7 g, and phosphoric rock 3.5 g, using phosphoric rock instead tricalcium phosphate) (Bashan et al. 2013; Kim et al. 1997; Murphy and Riley 1962) and nitrogen fixation were determined by growth on NFb medium (Eckert et al. 2001). Antagonistic potential against banana pathogens as Fusarium oxysporum (strain EAHP-0015, donated by Banana Growers Association from Colombia, AUGURA) and Ralstonia solanacearum (EA-EP009, isolated of banana plant at Universidad EAFIT, Medellin) was evaluated by using the dual-culture plate method previously described by Lemessa and Zeller (2007). Antagonistic potential against Pseudocercospora fijiensis (isolated of banana leaves plant using necrotic tissue by ascospore discharge method) was evaluated using the varnish technique modified by Talavera et al. (1998) and percent inhibition of germ tube was determined by Gutierrez-Monsalve et al. (2015). Finally, production and purification of lipopeptides was carried out by a methodology previously described (Villegas-Escobar et al. 2013). Briefly, 20 mL of an overnight culture of B. subtilis EA-CB0575 (12 h, 30 °C) was transferred to 500-mL Erlenmeyer flask containing 180 mL of MOLP medium (Jacques et al. 1999) and incubated for 3 days at 30 °C and 150 rpm. After 12 h of incubation, 4% (w/v) of amberlite resin XAD-16 (Alfa Aesar®) was added to the culture. After incubation, the resin was recovered, washed with 400 mL of sterile distilled water, and adsorbed metabolites were eluted with 200 mL of methanol. The methanolic extract was evaporated under reduced pressure in a rotary evaporator (50 °C, − 50 psig) obtaining 99 mg of solid residue from 200 mL total culture. The solid residue was suspended in distilled water (37 mg/mL) and applied to a solid-phase extraction (SPE C18) cartridge (Baker®, 10 g). The cartridge was then rinsed successively with 80 mL of distilled water, 80 mL of 50% methanol, and 160 mL of 100% methanol. The methanolic fraction was evaporated and the solid residue dissolved in methanol (50 mg/mL) for RP-HPLC analysis. The methanolic fraction was purified by RP-HPLC using an Eclipse XDB C18 column (250 × 4.6 mm, 5 μm) connected to an Agilent G1311A quaternary pump and with a solvent system consisting of 0.1% in water (solvent A) and 0.1% TFA in acetonitrile (solvent B). Forty microliters of the sample was injected into the column and the compounds were eluted by a gradient program (30/100/100% B in 0/25/35 min) at a flow rate of 1 mL/min and UV detection at 214 nm. Peaks with different retention times were collected, vacuum evaporated (Eppendorf Concentrator Plus™, Harburg, Germany), and stored at 4 °C. Purified HPLC fractions and surfactin standard (Sigma Aldrich, S3523) were analyzed by high-resolution mass spectrometry (Iowa State University’s Protein Facility, Ames, IO, USA) using an HPLC (Agilent 1260) coupled to Thermo Scientific Hybrid Q-Exactive ™ quadrupole-orbitrap tandem mass spectrometer equipped with a NanoSpray Flex ionization source and cyano cinnamic acid as matrix. Solvent A (water + 0.1% formic acid) and B (acetonitrile with 0.1% formic acid) with a gradient of 20–100% were used for 13 min. Data were acquired in a range between 200 and 2000 m/z in positive ionization mode. MS/MS analysis was performed using an HCD ion collision cell and the software Xcalibur (Thermo Scientific, version 2.2). Proteowizard 3.0.10.200 (Chambers et al. 2012) was used for the analysis of the results.
Plant growth promotion of B. subtilis EA-CB0575 in tomato
Plant growth promotion effect of B. subtilis EA-CB0575 and two reference strains (B. velezensis FZB42 and B. subtilis 168) were evaluated in tomato variety Chonto-Santa Cruz at greenhouse level using a completely randomized design (CRD). Tomato seeds were disinfected successively with 70% ethanol for 15 min, 5% sodium hypochlorite for 5 min, and three washes with sterile distilled water. After scarification at 4 °C for 24 h, seeds were inoculated by immersing the roots in a bacterial suspension (1 × 108 CFU/mL) or in sterile distilled water (control) for 1 h. The seeds were sowed in pots with 500 g of commercial soil (1 sand: 0.5 black soil: 0.5 rice husk) and kept at greenhouse conditions (22–30 °C, 12 h light/12 h dark, 60% of maximum water-holding capacity) for 1 month. Plants were fertilized after 15 days of inoculation with 15-15-15 NPK. The growth-promoting effect of the different strains was performed by measuring shoot length (SL) and dry weights (shoot dry weight (SDW), root dry weight (RDW), and total dry weight (TDW)). This experiment was conducted twice with 8 replicates per treatment and two plants per pot.
Statistical analysis
Analysis of variance (ANOVA) was used to analyze each experiment in RStudio. The assumptions of normality (Shapiro-Wilks test), homoscedasticity (Levenne’s test and Barlett’s test), and independence (graphic residues vs. run order and Durbin-Watson statistics) were determined. In the case of significant P value (P < 0.05), means were compared by using LSD or Dunnett multiple comparison test.
Results
Genome analysis of B. subtilis subsp. subtilis EA-CB0575 reveals the potential for plant growth promotion
16S rRNA gene sequencing was performed to define the strain taxonomy, finding that it belongs to Bacillus subtilis species with a query cover of 100%, an identity of 99%, and an E-value of 0.0. The complete genome of B. subtilis EA-CB0575 was assembled in 16 contigs, reaching an average size of 255,665 bp. For this assembly, an average G-C content of 43.7% and a total genome size of 4.09 Mb was determined (Fig 1). The average coverage of the cluster was 110× and half of the genome bases were found in 2 contigs (N50), 90% in 6 (N90), and 95% in 7 contigs (N95). The maximum coverage of the genome was 2000×. Table 2 shows the number of genes annotated and involved in B. subtilis EA-CB0575 metabolic and cellular processes. There is a high similarity among the number of genes in each category between the strain EA-CB0575 and B. subtilis 168 type strain; but the former has more genes in several metabolism-related functions (fatty acids, lipids and isoprenoids, sulfur, carbohydrates, aromatic compounds, and phosphorus) and in cellular processes (division and cellular cycle, cell wall formation and capsule formation, motility and chemostasis, regulation and cellular signaling, and genes related to phages, prophages, transposable elements, plasmids). The genome contains 4332 genes, 4237 genes encoding proteins, 95 RNA genes (including 10 copies of the 16S rRNA), 2917 genes encoding proteins with predicted function, and 1320 genes encoding hypothetical proteins. The presence of related genes to PGPR mechanisms or the metabolic pathway prediction of RAST was found from the gene annotation. The production of enzymes involved in the metabolism of indole via the tryptophan pathway was predicted, suggesting that B. subtilis EA-CB0575 has the potential to produce these compounds. Indole, indole acetate, and indole-acetamide production was predicted (Fig. S1). The potential for production of siderophores as bacillibactin type, enterochelins (enterobacins), vibriobacins, and related ones synthesized by NPRSs was found. Some enzymes related to the production of compounds such as salicylate, isocorismate, and dihydroxybenzoate would likely be produced by B. subtilis EA-CB0575 (Fig. S2). The gene cluster that encodes to produce bacilibactin (dhbACEBF) was found in the strain genome when compared with the reference genomes (Table 3). Volatile compounds as 2,3-butanediol and acetoin might be produced by this strain (Fig. S3) given that it has the potential to produce the enzymes α−acetolactate synthetase, α−acetolactate decarboxylase, acetoin-reductase, acetylacetoin-synthase, and others reported (Qi et al. 2014). The phyC genes, coding for enzymes that sequentially remove phosphates of phytate, were found in the genome of the strain B. subtilis EA-CB0575, suggesting the capacity of this strain for organic phosphate solubilization. Also, some genes related to enzyme activity to fully perform citric acid pathway (TCA cycle) were found in the strain 575 genome, indicating a possible inorganic phosphate solubilization by organic acid production (Vyas and Gulati 2009).
Finally, the environmental nitrogen fixation capacity of strain EA-CB0575 was evaluated. The genes nifU, nifS, nifV, and nifF were present in the strain genome which are involved in nitrogenase enzymatic activity responsible for the biological fixation of nitrogen. However, the presence of all the genes of the nitrogenase cluster which are essential for the nitrogenase activity was not found. The assembled and annotated genome was saved on Universidad EAFIT server, operating under Ubuntu 14.04 and accessed through the GBrowse2 genome browser (Stein et al. 2002). An analysis of the cluster ends was performed, finding that 50% of the genes in this section correspond to rRNA, 26.67% to non-ribosomal peptide synthases, followed by 6.67% for tRNAs, and the rest corresponds to phages and repeated GXT proteins.
The genome of B. subtilis EA-CB0575 harbors an array of gene clusters involved in synthesis of secondary metabolites
The presence of NRPS coding sequences involved in the synthesis of surfactins, fengycins, and iturins in B. subtilis EA-CB0575 genome was evaluated using B. velezensis FZB42 as the genome reference. Coding regions for surfactins and fengycins production were found; however, no coding regions were found for iturins production (Fig. 2). The coding regions for NRPSs that produce surfactins were found in clusters 5, 6, and 7 and for fengycins production in clusters 9, 10, and 11. B. subtilis EA-CB0575 loci for NRPS was compared respect to the same loci for references B. velezensis FZB42 and B. subtilis 168 and 4 ORFs were reconstructed for the surfactin operon, with two gaps of a few nucleotides between assembly clusters 5 and 6, and 6 and 7 in the srfAA and srfAC genes, respectively. Seven domains of adenylation and a thioesterase/acyltransferase were obtained from the amino acid sequences of the ORFs. Three of 5 ORFs of the operon for fengycin production were obtained, corresponding to fenA, fenC, and fenE genes. Two domains of adenylation were obtained for the first, two for the second, and one for the adenylation and one thioesterase for the third. For the reference strains, the coding regions for the synthases related to the production of surfactins and fengycins were found; however, the region of iturin was determined for FZB42 strain and not for 168 strain. Comparative genomics of B. subtilis EA-CB0575 was performed with the two reference strains B. subtilis 168 and B. velezensis FZB42 to produce some secondary metabolites (Table 3). Similarities were observed in genome sizes and genes. There is a greater similarity between the percentages of the genome dedicated to the production of secondary metabolites in B. subtilis strains than B. velezensis, this latter with the highest percentage of the genome dedicated to the production of these compounds (8.02%). Some important clusters of genes, such as those related to the production of antibiotics, siderophore bacilibactin, and some LPs, are shared among the evaluated species.
Determination of the loci to produce the NRPSs of surfactins, fengycins, and iturins in B. velezensis FZB42, B. subtilis EA-CB0575, and B. subtilis 168 genomes. a Diagram of location for responsible loci to the synthesis of NRPS related to surfactin in B. subtilis EA-CB0575 strain in contrast to the reference strains B. subtilis 168 and B. velezensis FZB42 b Diagram of location for responsible loci for genes involved in the production of the iturin bacillomycin and fengycin B in B. subtilis EA-CB0575 strain in contrast to the reference strains B. subtilis 168 and B. velezensis FZB42
B. subtilis EA-CB0575 belongs to the clade of B. subtilis subsp. subtilis
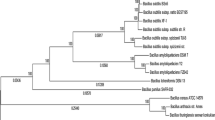
The phylogenetic relationship of B. subtilis EA-CB0575 with other Bacillus sp. strains was performed by comparing a set of housekeeping genes (Fig. 3). The best partitioning scheme of the alignment for the concatenated genes was composed of 6 subsets, with the 15 preliminary partitions given, one for each position of codons (1, 2, 3) in each of the 5 genes analyzed. Two major clades were observed: the first containing B. thuringiensis, B. anthracis, B. cereus, B. mycoides, and B. weihenstephanensis, which are Bacillus spp. pathogens of various hosts. The second clade was composed of B. pumilus, B. licheniformis, B. subtilis, B. amyloliquefaciens, and B. velezensis.
Phylogeny of Bacillus spp. from housekeeping genes (rpoB, gyrA, purH, polC, and groEL) obtained by Bayesian inference and maximum likelihood methods. The support values are shown for the internal nodes: posterior probability (PP) for BI and percentage of bootstrap (BP) for ML. The study strains are highlighted in red
PGPR traits found in B. subtilis EA-CB0575 genome are expressed in vitro
To determine if genes related to PGPR traits were functional, different in vitro evaluations were performed (Table 4). Among evaluated strains, B. subtilis EA-CB0575 produced IAA (15.0 ± 2.0 μg/mL) and siderophores (15.4 ± 1.1 μM), solubilized phosphate, and had the ability to fix environmental nitrogen while B. subtilis 168 produced 2.0-fold lower concentrations of IAA (6.9 ± 1.4 μg/mL) and siderophores (7.6 ± 0.9 μM), and no capacity to solubilized phosphate or fix nitrogen. On the other hand, B. velezensis FZB42 produced IAA and siderophores, solubilized phosphate, but has no potential to fix nitrogen in vitro. B. subtilis EA-CB0575 inhibited the growth of the phytopathogens R. solanacearum (halo 1.2 ± 0.3 cm on BGA medium), F. oxysporum (reduction of growth of 21 ± 0.1% on PDA medium), and P. fijiensis (reduction of 33.1 ± 4.3% on germinative tubes of ascospores) when Bacillus was evaluated in coculture with the fungi tested in vitro (Table 4). Finally, to determine if gene clusters related to NRPS were expressed in B. subtilis EA-CB0575, different purified fractions obtained by HPLC were analyzed by mass spectrometry analysis. Two families of lipopeptides were identified: surfactins and fengycins (Table 4; Fig. 4 and supplementary material Tables S1 and S2 and, Fig. S5, S6, S7, S8). In particular, the precursor ions observed at m/z [M + H]+ 1008.66, 1022.67, 1036.69 and at m/z [M + Na]+ 1030.64, 1044.66, 1058.67, and 1072.69 [M + Na]+ (Fig. S5) were used for further MS/MS analysis, and results showed product ions characteristic of surfactins with different lengths of fatty acid chains (C12, C13, C14, C15, and C16) and therefore different molecular weights. For the other purified fraction, the precursor ions at m/z 1408, 1435.77, 1463.81, 1477.82, 1491.83, and 1505.85 corresponding to protonated molecules [M + H]+, and m/z 1444, 1460 to sodiated [M + Na]+, and potassium [M + K]+ adducts respectively (Fig. S7), were also used for further MS/MS analysis. In general, product ions obtained were characteristic of fengycins A (m/z 1080.50 and 966.45) and B (m/z 1108.58 and 994.49). These results all together suggest that genes found in the genome of B. subtilis EA-CB0575 are functional in vitro and may confer potential for promoting plant growth.
Lipopeptides analysis production by HPLC chromatogram for SPE-100% methanol extract from B. subtilis EA-CB0575 culture in MOLP medium. HPLC operation conditions: 214 nm, injection of 40 μL (concentration 10 mg/mL), flow 1 mL/min, 30 °C, column eclipse XDB C18 250 × 4.6 mm 5 um. A: water HPLC + 0, 1% TFA. B: CH3CN + 0.1% TFA. Gradient 30/100/100% B in 0/25/35 min
B. subtilis is a potential PGPR for tomato crops
Tomato seeds variety Chonto-Santa Cruz were inoculated with suspensions of B. subtilis EA-CB0575 as spores or vegetative cells and the reference strains B. subtilis 168 and B. velezensis FZB42 as vegetative cells. We determined an average increase of 29% for the variable TDW of the plants with application of B. subtilis strains respect to the control. B. subtilis EA-CB0575 was the strain with a higher increase of this variable (34.60%). SDW and RDW shown differences between B. subtilis EA-CB0575 and reference strains and the control, respectively; with percentage increase related to control of 48.4% for SDW, when vegetative cells of B. subtilis EA-CB0575 were applied; and 54.2% for RDW when spores were inoculated. B. velezensis FZB42 did not show increase with respect to control samples without application of the microorganism (Table 5). These results are consistent with assays previously reported, indicating a possible relation between B. subtilis EA-CB0575 colonization root potential and plant growth potential for tomato (Posada et al. 2018).
Discussion
The results of the genomic assembly of B. subtilis EA-CB0575 are similar to those reported for other genomes of B. subtilis strains (Guo et al. 2015; Liu et al. 2018; Rahimi et al. 2018). The genome presented some breaks in the corresponding sections to genes encoding rRNA, tRNAs, non-ribosomal peptide synthases, inserts of phages, and transposases; it could be due to the difficulty for the assembler to work with repeating sequences (Nederbragt et al. 2010), because the algorithm of graphs does not solve copies separately (Compeau et al. 2011). In addition, regions with modular structure preserved at the sequence level, such as NRPSs, are another cut-off point of the assembly, because of the ambiguity at the ends that is not computationally solved with the available information. Fragmentation of the genome obtained (16 clusters, with an average length of 255,665 bp) is good for genomic-derived analyses. When the obtained number of contigs is compared with the number for other similar projects, in our case, there is a smaller division than for other Bacillus (205 partitions for the type strain, and 390 for B. natto BEST195, among others) (Kunst et al. 1997; Ulyanova et al. 2016).
When some genes related to PGPR mechanisms were annotated and metabolic predictions were performed, we determined that B. subtilis EA-CB0575 genome has genes or enzymes related to the conversion pathway of tryptophan to indole, which is consistent with the determined indole production (15 μg/mL), concentration reported as not deleterious to the plant (Barazani and Friedman 1999). Metabolic prediction was evaluated for siderophores production, finding its metabolic machinery inside the B. subtilis EA-CB0575 genome, which was biochemically corroborated with an in vitro production of 15.4 μM. These compounds give to the plant and microorganisms possibilities to improve iron assimilation and are elicitors of systemic resistance in the plant (Aznar and Dellagi 2015). The presence of nifU, nifS, nifV, and nifF, all essential for encoding the components of the enzymatic modules of nitrogenase (Li et al. 2016), was determined. It suggests the possibility of the strain to fix environmental nitrogen, which was determined experimentally by B. subtilis EA-CB0575 growth on NFb medium. The phyC genes, coding for enzymes that sequentially remove phosphates of phytate, were found in the genome of the strain B. subtilis EA-CB0575, indicating a possibility for organic phosphate solubilization. It has been reported, about their presence in B. subtilis 168 (Idriss et al. 2002). Several genes with potential enzyme activity to fully perform citric acid pathway (TCA cycle) were found in the strain 575 genome, such intermediate acids (e.g., oxalic acid, malic acid, citric acid) could be potentially involved in inorganic phosphate solubilization exhibited by B. subtilis EA-CB0575. This was corroborated in biochemical evaluations for strain EA-CB0575, which could grow in medium NBRIP with phosphoric rock and showed positive results using molybdate blue test (Murphy and Riley 1962). The type strain 168 did not grow in the medium and did not show positive results for the test, indicating that phosphate solubilization for this strain could be related to organic solubilization processes, using phytates (Ahmad et al. 2017). In addition, it was found that B. subtilis EA-CB0575 possesses the machinery to produce 2,3-butanediol and acetoin, but this was not corroborated at in vitro level.
The characterization of regions for coding to NPRSs of the LPs was carried out. B. velezensis FZB42 and B. subtilis 168 strains were used as the reference. It was determined that B. subtilis EA-CB0575 genome has homologous regions to regions that codify for surfactins and fengycins NRPSs. No coding regions were found for iturin production, consistent with experimental results (Villegas-Escobar et al. 2013). LPs production in MOLP medium was evaluated using HPLC and mass spectrometry. We determined the production of surfactins C12 to C16, as well as fengycins A (C14, C15, C16, C17) and B (C16 and C17). These compounds could be responsible for antagonistic activity against evaluated phytopathogens, as reported by several authors (Fan et al. 2018; Villegas-Escobar et al. 2013). We determined the presence of loci for surfactins and fengycins codification for the reference strains, but their in vitro evaluations were not done. Different reports indicate lack of detection for these that compounds at in vitro cultures of the B. subtilis 168 as being due to a mutation in the sfp gene, which affects the production of surfactins (Chen et al. 2007). Loci to encode NRPS production of iturins was found for FBZ42 strain; and experimental results for reported researches shown in vitro production of the three LP families announced (Koumoutsi et al. 2004).
B. subtilis EA-CB0575 has more elements inserted in the genome than the reference strains, possibly because of their wild character and the domestication of the reference strain 168 (Veening et al. 2006). In addition, 3.9% of its genome is associated with secondary metabolism. However, its potential is related not only to the number of genes involved but also to the levels being expressed by these genes. We suggest that transcriptomic analyses should be performed in order to know this. It was determined that many of the gene clusters that are present in the reference strain B. amyloliquefaciens FZB42, active principle of Biomex® and Rhizovital®42, are present in B. subtilis EA-CB0575; this makes this strain promissory for bioproducts development. The topologies of the phylogenetic trees obtained were coincident and have high values of support in most branches (BP, > 75; PP, > 0.95). It was determined that the offered resolution by trees based on the 16S gene is not enough to differentiate the strains in the B. amyloliquefaciens, B. subtilis, and B. licheniformis groups, probably because of the high similarity in 16s rDNA sequences (Rooney et al. 2009). Two clades were found: one for B. thuringiensis, B. anthracis, B. cereus, B. mycoides, and B. weihenstephanensis, considered pathogens of several hosts; and a second clade formed by B. pumilus, B. licheniformis, B. subtilis, B. velezensis, and B. amyloliquefaciens, all present in the soil. B. amyloliquefaciens, B. velezensis, and B. subtilis differ in well-resolved siblings clades and independent of B. licheniformis clade, which did not happen in phylogeny inferred with information from the 16S ribosomal gene (Fig. S4).
We could determine the PGPR potential of B. subtilis EA-CB0575 and B. subtilis 168 strains in tomato plants. We evaluated this potential for B. subtilis EA-CB075 in others crops as banana (Posada et al. 2016), and we established a potential colonization capacity on tomato and banana roots using FISH and CARD-FISH methods (Posada et al. 2018). These results are related with bacterial genomic and biochemical potential evaluated in this research. Better performance of B. subtilis microorganisms than B. velezensis to promote plant TDW was found with percentages from 25.40 to 34.60%. B. velezensis did not show differences with control; however, this strain has been reported as a growth promoter through indirect promotion mechanisms such as biocontrol (Chowdhury et al. 2015).
Conclusion
B. subtilis EA-CB0575 is a potential PGPR due to its genomic and phenotypic capacity related to the mechanisms of plant growth promotion. Its genome was assembled in this project in 16 contigs, and genes related to plant growth promotion mechanisms were annotated and analyzed. It was found genes related to IAA, siderophores, acetoin, 2,3-butanediol and LPs production, nitrogen fixation, phosphate solubilization and antagonistic activity against phytopathogens. The biochemical test allowed to determine the possibility of the microorganism for the production of LPs of surfactin families (surfacins C12, C13, C14, C15, and C16), fengycins A (C14, C15, C16, C17), and fengycins B (C16 and C17). Also, we corroborate its possibility for IAA and siderophores production, to solubilize phosphates and fix environmental nitrogen. It was determined that the microorganism has 3.9% of its genome dedicated to the production of secondary metabolites and belongs to B. subtilis subtilis clade. In addition, B. subtilis EA-CB0575 and its type strains promote TDW of the tomato plant.
References
Ahmad Z, Wu J, Chen L, Dong W (2017) Isolated Bacillus subtilis strain 330-2 and its antagonistic genes identified by the removing PCR. Sci Rep 7:1777. https://doi.org/10.1038/s41598-017-01940-9
Andrews S (2010) FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc. Accessed 29 Mar 2018
Aziz RK, Bartels D, Best AA, De Jongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O (2008) The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75
Aznar A, Dellagi A (2015) New insights into the role of siderophores as triggers of plant immunity: what can we learn from animals? J Exp Bot 66(11):3001–3010
Backer R, Rokem JS, Ilangumaran G, Lamont J, Praslickova D, Ricci E, Subramanian S, Smith DL (2018) Plant growth-promoting rhizobacteria: context, mechanisms of action, and roadmap to commercialization of biostimulants for sustainable agriculture. Front Plant Sci 9:1473. https://doi.org/10.3389/fpls.2018.01473
Barazani O, Friedman J (1999) Is IAA the major root growth factor secreted from plant-growth-mediating bacteria? J Chem Ecol 25(10):2397–2406
Bashan Y, Kamnew A, De-Bashan LE (2013) Tricalcium phosphate is inappropriate as a universal selection factor for isolating and testing phosphate-solubilizing bacteria that enhance plant growth: a proposal for an alternative procedure. Biol Fertil Soils 49:465–479
Bazinet AL, Zwickl DJ, Cummings MP (2014) A gateway for phylogenetic analysis powered by grid computing featuring GARLI 2.0. Syst Biol 63(5):812–818
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120
Cai J, Liu F, Liao X, Zhang R (2014) Complete genome sequence of Bacillus amyloliquefaciens LFB112 isolated from chinese herbs, a strain of a broad inhibitory spectrum against domestic animal pathogens. J Biotechnol 10(175):63–64
Chambers MC, Maclean B, Burke R, Amodei D, Ruderman DL, Neumann S, Gatto L, Fischer B, Pratt B, Egertson J, Hoff K, Kessner D, Tasman N, Shulman N, Frewen B, Baker TA, Brusniak MY, Paulse C, Creasy D, Flashner L, Kani K, Moulding C, Seymour SL, Nuwaysir LM, Lefebvre B, Kuhlmann F, Roark J, Rainer P, Detlev S, Hemenway T, Huhmer A, Langridge J, Connolly B, Chadick T, Holly K, Eckels J, Deutsch EW, Moritz RL, Katz JE, Agus DB, MacCoss M, Tabb DL, Mallick P (2012) A cross-platform toolkit for mass spectrometry and proteomics. Nat Biotech 30(10):918–920
Chen X, Koumoutsi A, Scholz R, Eisenreich A, Schneider K, Heinemeyer I, Morgenstern B, Voss B, Hess W, Reva O, Junge H, Voigt B, Jungblut P, Vater J, Sussmuth R, Liesegang H, Strittmatter A, Gottschalk G, Borriss R (2007) Comparative analysis of the complete genome sequence of the plant growth-promoting bacterium Bacillus amyloliquefaciens FZB42. Nat Biotechnol 25(9):1007–1014
Chowdhury SP, Hartmann A, Gao X, Borriss R (2015) Biocontrol mechanism by root-associated Bacillus amyloliquefaciens FZB42 – a review. Fron Microbiol 6(780):1–11
Compeau PEC, Pevzner PA, Tesler G (2011) How to apply de Bruijn graphs to genome assembly. Nat Biotech 29(11):987–991
Costa F, Sérgio J, Schmitt K, de Oliveira R, Farias C, da Silva P, Dellagostin O, Galdino F, de Menezes R, de Sousa O, Maggioni R, Marins L (2018) Complete genome sequence of native Bacillus cereus strains isolated from intestinal tract of the crab Ucides sp. Data Brief 16:381–385
Deng Y, Zhu Y, Wang P, Zhu L, Zheng J, Li R, Ruan L, Peng D, Sun M (2011) Complete genome sequence of Bacillus subtilis BSn5, an endophytic bacterium of Amorphophallus konjac with antimicrobial activity for the plant pathogen Erwinia carotovora subsp. carotovora. J Bacteriol 193(8):2070–2071. https://doi.org/10.1128/JB.00129-11
Deng Q, Wang R, Sun D et al (2019) Biochem Genet. https://doi.org/10.1007/s10528-019-09927-z
Earl AM, Eppinger M, Fricke WF, Rosovotzz MJ, Rasko DA, Daugherty S, Losick R, Kolter R, Ravel J (2012) Whole-genome sequences of Bacillus subtilis and close relatives. J Bacteriol 194(9):2378–2379
Eckert B, Weber OB, Kirchhof G, Halbritter A, Stoffels M, Hartmann A (2001) Azospirillum doebereinerae sp. nov., a nitrogen-fixing bacterium associated with the C4-grass Miscanthus. Int J Syst Evol Microbiol 51(Pt 1):17–26
EPA (Environmental Protection Agency) (2019) Draft guidance for pesticide registrants on plant regulator label claims, including plant biostimulants; notice of availability. Government Publishing Office. https://www.epa.gov/pesticides/epa-releases-public-comment-draft-guidance-plant-regulators-including-plant-biostimulants. Accessed 20 july 2019
Fan B, Borriss R, Bleiss W, Wu X (2012) Gram-positive rhizobacterium Bacillus amyloliquefaciens FZB42 colonizes three types of plants in different patterns. J Microbiol 50(1):38–44
Fan B, Wang C, Song X, Ding X, Wu L, Wu H, Gao X, Borriss R (2018) Bacillus velezensis FZB42 in 2018: the gram-positive model strain for plant growth promotion and biocontrol. Fron Microbiol 9:2491. https://doi.org/10.3389/fmicb.2018.0249
Gilbert D (2003) Sequence file format conversion with command-line readseq. Curr Protoc bioinformatics appendix 1, appendix 1E
Guo S, Li X, He P, Ho H, Wu Y, He Y (2015) Whole-genome sequencing of Bacillus subtilis XF-1 reveals mechanisms for biological control and multiple beneficial properties in plants. J Ind Microbiol Biotechnol 42(6):925–937
Gupta A, Gopal M, Thomas GV, Manikandan V, Gajewski J, Thomas G, Seshagiri S, Schuster SC, Rajesh P, Gupta R (2014) Whole genome sequencing and analysis of plant growth promoting bacteria isolated from the rhizosphere of plantation crops coconut, cocoa and arecanut. PLoS One 9(8):e104259. https://doi.org/10.1371/journal.pone.0104259
Gutierrez-Monsalve J, Mosquera S, González-Jaramillo L, Mira J, Villegas-Escobar V (2015) Effective control of black Sigatoka disease using a microbial fungicide based on Bacillus subtilis EA-CB0015 culture. Biol Control 87:39–46
Idriss EE, Makarewicz OM, Farouk A, Rosner K, Greiner R, Bochow H, Richter T, Borriss R (2002) Extracellular phytase activity of Bacillus amyloliquefaciens FZB45 contributes to its plant-growth-promoting effect. Microbiology 148(Pt 7):2097–2109
Ivanova N, Sorokin A, Anderson I, Galleron N, Candelon B, Kapatral V, Bhattacharyya A, Reznik G, Mikhailova N, Lapidus A, Chu L, Mazur M, Goltsman E, Larsen N, D'Souza M, Walunas T, Grechkin Y, Pusch G, Haselkorn R, Fonstein M, Dusko Ehrlich S, Overbeek R, Kyrpides N (2003) Genome sequence of Bacillus cereus and comparative analysis with Bacillus anthracis. Nature 423(6935):87–91
Jacques P, Hbid C, Destain J, Razafindralambo H, Paquot M, De Pawn E, Thonart P (1999) Optimization of biosurfactant lipopeptide production from Bacillus subtilis S499 by Plackett-Burman Design. Appl Biochem Biotechnol 77(1–3):223–233
Kai-Jium L, Shin-Shun L, Chia-Wei L, Chih-Horng K, Chi-Te L (2018) Whole-genome sequencing and comparative analysis of two plant-associated strains of Rhodopseudomonas palustris (PS· and YCS3). Sci Rep 8:12769
Kamada M, Hase S, Fujii K, Miyake M, Sato K, Kimura K, Sakakibara Y (2015) Whole-genome sequencing and comparative genome analysis of Bacillus subtilis strains isolated from non-salted fermented soybean foods. PLoS One 10(10):e0141369. https://doi.org/10.1371/journal.pone.0141369
Khatri I, Sharma S, Ramya TNC, Subramanian S (2016) Complete genomes of Bacillus coagulans S-lac and Bacillus subtilis TO-A JPC, two phylogenetically distinct probiotics. PLoS One 11(6):e0156745
Kim KY, McDonald GA, Jordan D (1997) Solubilization of hydroxyapatite by Enterobacter agglomerans and cloned Escherichia coli in culture medium. Biol Fertil Soils 2(4):347–352
Kim BY, Lee SY, Ahn JH, Song J, Kim WG, Weon HY (2015) Complete genome sequence of Bacillus amyloliquefaciens subsp. plantarum CC178, a phyllosphere bacterium antagonistic to plant pathogenic fungi. Genome Announc 3(1):e01368–e01314
Koumoutsi A, Chen XH, Henne A, Liesegang H, Hitzeroth G, Franke P, Vater J, Borriss R (2004) Structural and functional characterization of gene clusters directing nonribosomal synthesis of bioactive cyclic lipopeptides in Bacillus amyloliquefaciens strain FZB42. J Bacteriol 186(4):1084–1096
Kumar A, Prakash A, Johri BN (2011) Bacillus as PGPR in crop ecosystem. In: Maheshwari D (ed) Bacteria in agrobiology: crop ecosystems. Springer, Berlin
Kunst F, Ogasawara N, Moszer I, Albertini AM, Alloni G, Azevedo V, Bertero MG, Bessieres P, Bolotin A, Borchert S, Borriss R, Boursier L, Brans A, Braun M, Brignell SC, Bron S, Brouillet S, Bruschi CV, Caldwell B, CapuanoV CNM, Choi SK, Codani JJ, Connerton IF, Cummings NJ, Daniel RA, Denizot F, Devine KM, Dusterhoft A, Ehrlich SD, Emmerson PT, Entian KD, Errington J, Fabret C, Ferrari E, Foulger D, Fritz C, Fujita M, Fujita Y, Fuma S, Galizzi A, Galleron N, Ghim SY, Glaser P, Goffeau A, Golightly EJ, Grandi G, Guiseppi G, Guy BJ, Haga K, Haiech J, Harwood CR, Henaut A, Hilbert H, Holsappel S, Hosono S, Hullo MF, Itaya M, Jones L, Joris B, Karamata D, Kasahara Y, Klaerr-Blanchard M, Klein C, Kobayashi Y, Koetter P, Koningstein G, Krogh S, Kumano M, Kurita K, Lapidus A, Lardinois S, Lauber J, Lazarevic V, Lee SM, Levine A, Liu H, Masuda S, Mauel C, Medigue C, Medina N, Mellado RP, Mizuno M, Moestl D, Nakai S, Noback M, Noone D, O’Reilly M, Ogawa K, Ogiwara A, Oudega B, Park SH, Parro V, Pohl TM, Portetelle D, Porwollik S, Prescott AM, Presecan E, Pujic P, Purnelle B et al (1997) The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390(6657):249–256
Lanfear R, Calcott B, Kainer D, Mayer C, Stamatakis A (2014) Selecting optimal partitioning schemes for phylogenomic datasets. BMC Evol Biol 14(1):1–14
Lemessa F, Zeller W (2007) Screening rhizobacteria for biological control of Ralstonia solanacearum in Ethiopia. Biol Control 42:336–344
Li Q, Xu LZ, Zou T, Ai P, Huang GH, Li P, Zheng AP (2015) Complete genome sequence of Bacillus thuringiensis strain HD521. Stand in Genomic Sci 10:62. https://doi.org/10.1186/s40793-015-0058-1
Li XX, Liu Q, Liu XM, Shi HW, Chen SF (2016) Using synthetic biology to increase nitrogenase activity. Microb Cell Factories 20(1):15–43
Li P, Tian W, Jiang Z, Liang Z, Wu X, Du B (2018) Genomic characterization and probiotic potency of Bacillus sp. DU-106, a highly effective producer of L-lactic acid isolated from fermented yogurt. Fron Microbiol 9:2216. https://doi.org/10.3389/fmicb.2018.02216
Liu H, Yin S, An L, Zhang G, Cheng H, Xi Y, Cui G, Zhang F, Zhang L (2017) Complete genome sequence of Bacillus subtilis BSD-2, a microbial germicide isolated from cultivated cotton. J Biotechnol 230:26–27
Liu Y, Lai Q, Shao Z (2018) Genome analysis-based reclassification of Bacillus weihenstephanensis as a later heterotypic synonym of Bacillus mycoides. Int J Syst Evol Microbiol 68(1):106–112
Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Lu F, Marchler GH, Mullokandov M, Omelchenko MV, Robertson CL, Song JS, Thanki N, Yamashita RA, Zhang D, Zhang N, Zheng C, Bryant SH (2011) CDD: a conserved domain database for the functional annotation of proteins. Nucleic Acids Res 39(Database issue):D225–D229
Md. Anisur R, Md. Sanaullah N, Md. Anayet H, Md. Rakib U, Md. Hafijur R, Md. Amzad H, Yeasmeen A, Md. Saiful I (2014) Identification of potential drug targets by subtractive genome analysis of Bacillus anthracis A0248: an in-silico approach. Comput Biol Chem 52:66–72
Moszer I (1998) The complete genome of Bacillus subtilis: from sequence annotation to data management and analysis. FEBS Lett 430(1–2):28–36
Murphy J, Riley JP (1962) A modified single solution method for the determination of phosphate in natural waters. Anal Chim Acta 27:31–36
Nederbragt AJ, Rounge TB, Kausrud KL, Jakobsen KS (2010) Identification and quantification of genomic repeats and sample contamination in assemblies of 454 pyrosequencing reads. Sequencing 2010, Article ID 782465, 12 pages
Nye TM, Schroeder JW, Kearns DB, Simmons LA (2017) Complete genome sequence of undomesticated Bacillus subtilis strain NCIB 3610. Genome Announc 5(20):e00364–e00317
Patten CL, Glick BR (2002) Regulation of indoleacetic acid production in Pseudomonas putida GR12-2 by tryptophan and the stationary-phase sigma factor RpoS. Can J Microbiol 48(7):635–642
Pengfei G, Guoqiang Y, Weichen B, Jing L, Heping Z, Wenyi Z (2015) Complete genome sequence of Bacillus licheniformis BL-09, a probiotic strain isolated from naturally fermented congee. J Biotechnol 206:58–59
Pereira FL, Júnior CAO, Silva RO, Dorella FA, Carvalho AF, Almeida GM, Lobato F, Figueiredo HC (2016). Complete genome sequence of Peptoclostridium difficile strain Z31. Gut pathogens 8(1):11
Popendorf K, Tsuyoshi H, Osana Y, Sakakibara Y (2010) Murasaki: a fast, parallelizable algorithm to find anchors from multiple genomes. PLoS One 5(9):e12651
Posada LF, Ramírez M, Ochoa-Gómez N, Cuellar-Gaviria TZ, Argel-Roldan LE, Ramírez CA, Villegas-Escobar V (2016) Bioprospecting of aerobic endospore-forming bacteria with biotechnological potential for growth promotion of banana plants. Sci Hortic 212:81–90
Posada LF, Álvarez JC, Romero-Tabarez M, de-Bashan L, Villegas-Escobar V (2018) Enhanced molecular visualization of root colonization and growth promotion by Bacillus subtilis EA-CB0575 in different growth systems. Microbiol Res 217:69–80
Qi G, Kang Y, Li L, Xiao A, Zhang S, Wen Z, Xu D, Chen S (2014) Deletion of meso 2,3-butanediol dehydrogenase gene budC for enhanced D-2,3-butanediol production in Bacillus licheniformis. Biotechnol Biofuels 7(1):16
Rahimi T, Niazi A, Deihimi T, Taghavi S, Ayatollahi S, Ebrahimie E (2018) Genome annotation and comparative genomic analysis of Bacillus subtilis MJ01, a new bio-degradation strain isolated from oil-contaminated soil. Funct Integr Genomics 18:533–543
Rambaut A (2014) FigTree 1.4.1. http://tree.bio.ed.ac.uk/software/figtree/. Accessed 1 June 2018
Ronquist F, Teslenko M, Van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61(3):539–542
Rooney AP, Price NP, Ehrhardt C, Swezey JL, Bannan JD (2009) Phylogeny and molecular taxonomy of the Bacillus subtilis species complex and description of Bacillus subtilis subsp. inaquosorum subsp. nov. Int J Syst Evol Microbiol 59(Pt 10):2429–2436
Rückert C, Blom J, Chen X, Reva O, Borriss R (2011) Genome sequence of B. amyloliquefaciens type strain DSM7(T) reveals differences to plant-associated B. amyloliquefaciens FZB42. J Biotechnol 20(155):78–85
Schroeder JW, Simmons LA (2013) Complete genome sequence of Bacillus subtilis strain PY79. Genome Announc 1(6):e01085–e01013
Schwyn B, Neilands JB (1987) Universal chemical assay for the detection and determination of siderophores. Anal Biochem 160(1):47–56
Shaligram S, Kumbhare SV, Dhotre DP, Muddeshwar M, Kapley A, Joseph N, Purohit H, Shouche Y, Pawar S (2016) Genomic and functional features of the biosurfactant producing Bacillus sp. AM13. Funct Integr Genomics 16:557–566
Smith JL, Goldberg JM, Grossman AD (2014) Complete genome sequences of Bacillus subtilis subsp. subtilis laboratory strains JH642 (AG174) and AG1839. Genome Announc 2(4). https://doi.org/10.1128/genomeA.00663-14
Stein LD, Mungall C, Shu S et al (2002) The generic genome browser: a building block for a model organism system database. Genome Res 12(10):1599–1610. https://doi.org/10.1101/gr.403602
Stevens MJ, Tasara T, Klumpp J, Stephan R, Ehling-Schulz M, Johler S (2019). Whole-genome-based phylogeny of Bacillus cytotoxicus reveals different clades within the species and provides clues on ecology and evolution. Sci Rep 9(1):1–14
Sukumaran J, Holder MT (2010) DendroPy: a Python library for phylogenetic computing. Bioinformatics 26(12):1569–1571
Sulthana A, Lakshmi SG, Madempudi RS (2019) Genome sequencing and annotation of Bacillus subtilis UBBS-14 to ensure probiotic safety. J Genomics 29(7):14–17
Talavera S, Bustamante E, Gonzales R, Sánchez V (1998) Selección y evaluación en laboratorio y campo de microorganismos glucanolíticos antagonistas a Mycosphaerella fijiensis. Manejo Integrado Plagas 47:24–30
Tirumalai MR, Rastogi R, Zamani N, O’Bryant Williams E, Allen S et al (2013) Candidate genes that may be responsible for the unusual resistances exhibited by Bacillus pumilus SAFR-032 spores. PLoS One 8(6):e66012
Ulyanova V, Shah Mahmud R, Malanin S, Vershinina V, Ilinskaya O (2016) Improving Bacillus altitudinis B-388 genome scaffolding using mate-pair next-generation sequencing. BioNanoScience 7(1):85–87
Veening JW, Kuipers OP, Brul S, Hellingwerf KJ, Kort R (2006) Effects of phosphorelay perturbations on architecture, sporulation, and spore resistance in biofilms of Bacillus subtilis. J Bacteriol 188(8):3099–3109
Veith B, Herzberg C, Steckel S, Feesche J, Maurer KH, Ehrenreich P, Bäumer S, Henne A, Liesegang H, Merkl R, Ehrenreich A, Gottschalk G (2004) The complete genome sequence of Bacillus licheniformis DSM13, an organism with great industrial potential. J Mol Microbiol Biotechnol 7:204–211
Villegas-Escobar V, Ceballos I, Mira JJ, Argel LE, Orduz Peralta S, Romero-Tabarez M (2013) Fengycin C produced by Bacillus subtilis EA-CB0015. J Nat Prod 76:503–509
Vyas P, Gulati A (2009) Organic acid production in vitro and plant growth promotion in maize under controlled environment by phosphate-solubilizing fluorescent Pseudomonas. BMC Microbiol 9:174. https://doi.org/10.1186/1471-2180-9-174
Wernersson R, Pedersen AG (2003) RevTrans: multiple alignment of coding DNA from aligned amino acid sequences. Nucleic Acids Res 31(13):3537–3539
Zeigler DR (2011) The genome sequence of Bacillus subtilis subsp. spizizenii W23: insights into speciation within the B. subtilis complex and into the history of B. subtilis genetics. Microbiology 157(Pt 7):2033–2041
Zeigler DR, Prágai Z, Rodriguez S, Chevreux B, Muffler A, Albert T, Perkins JB (2008) The origins of 168, W23, and other Bacillus subtilis legacy strains. J Bacteriol 190(21):6983–6995
Zerbino DR, Birney E (2008) Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18(5):821–829
Zhaohui X, Yan J, Danhua Q, Huaibao L, Fan Y, Jian Y, Lihong C, Lilian S, Xingye X, Ying X, Yafang Z, Qi J (2009). Complete genome sequence of the extremophilic Bacillus cereus strain Q1 with industrial applications. J Bacterio 191(3):1120-1121
Zheng-Bing G, Yu-Jie C, Yan-Zhou Z, Hong Z, Xiang-Ru L (2015) Complete genome sequence of Bacillus pumilus W3: a strain exhibiting high laccase activity. J Biotechnol 207:8–9
Acknowledgments
The authors acknowledge supercomputing resources made available by the Centro de Computación Científica Apolo at Universidad EAFIT (http://www.eafit.edu.co/apolo). This research was made possible by the Subscribe Contract Numbers 166 and 139 from 2017 with the Ministerio de Medio Ambiente y Desarrollo Territorial in the category “Contrato de Acceso a Recursos Genéticos y Productos Derivados para la Investigación Científica” and “Contrato de Acceso a Recursos Genéticos y Productos Derivados con Fines Comerciales,” respectively.
Funding
This study was financially supported by the Universidad EAFIT, the Association of Banana Growers of Colombia (AUGURA) and Colciencias (contract number 0836-2012).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 799 kb)
Rights and permissions
About this article
Cite this article
Franco-Sierra, N.D., Posada, L.F., Santa-María, G. et al. Bacillus subtilis EA-CB0575 genome reveals clues for plant growth promotion and potential for sustainable agriculture. Funct Integr Genomics 20, 575–589 (2020). https://doi.org/10.1007/s10142-020-00736-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10142-020-00736-x