Abstract
In this study, whey proteins were fermented with 34 lactic acid bacteria for 48 h at 37 °C and their ability to inhibit angiotensin 1-converting enzyme (ACE) activity were compared. All the lactic acid bacteria displayed varying proteolytic abilities in whey. Their fermentates also displayed varying abilities to inhibit ACE in vitro. Seven fermentates showed strong ACE inhibitory abilities between 84.70 ± 0.67 and 52.40 ± 2.1% with IC50 values between 19.78 ± 1.73 and 2.13 ± 0.7 mg/ml. Pediococcus acidilactici SDL1414 showed the strongest ACE inhibitory activity of 84.7 ± 0.67% (IC50 = 19.78 ± 1.73 μg/ml). Mass spectrometry revealed that more than half (57.7%) of the low molecular weight peptides (< 7 kDa) in the P. acidilactici SDL1414 fermented samples were ACE inhibitory peptides. Our results show that P. acidilactici SDL1414 could be used as a starter culture in the dairy industry to develop antihypertensive functional foods for hypertension management.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Food proteins usually contain several biologically active peptides. However, these peptides remain inactive as long as they remain bonded to other amino acids in the primary structure (Daliri et al., 2017a). Once released through enzymatic proteolysis and/or microbial fermentation, the free forms of the peptides demonstrate health effects in the gut or after systemic absorption into blood circulation. Several bioactive peptides have been reported to lower blood sugar, serum cholesterol, high blood pressure and inhibit microbial and cancer growth (Sánchez and Vázquez, 2017). Existing synthetic antihypertensive drugs have numerous side effects and this has increased scientific interest in searching for antihypertensive peptides as alternative therapeutics to control systemic blood pressure and to prevent cardiovascular diseases (Daliri et al., 2017b). One important enzyme involved in blood pressure regulation is angiotensin I-converting enzyme (ACE). The enzyme is a transmembrane metallopeptidase found in biological fluids and in many tissues such as the lung, thoracic aorta, heart, kidney, and liver (Nakamura et al., 2013). The enzyme hydrolyzes angiotensin I (a decapeptide) to yield angiotensin II (an octapeptide) which binds to AT1 receptors on vascular smooth muscles and endothelial cells leading to vasoconstriction. Angiotensin II also inactivates the endothelium-dependent vasodilator bradykinin (a nanopeptide) and this leads to high blood pressure (Manzanares et al., 2015). Therefore, inhibition of ACE activity is a good target for antihypertensive agents. Whey proteins such as beta-lactoglobulin, alpha-lactalbumin, bovine serum albumin, and immunoglobulin exhibit diverse physiological functions and their hydrolysates have been shown to have ACE inhibitory activities (Ibrahim et al., 2017). For instance the fragment 208–216 sequence of serum albumin, fragments 102–105 and f146–149 sequences of β-lactoglobulin and also fragments 50–53 sequences of α-lactalbumin have ACE inhibitory properties (Ahn et al., 2009; Tavares et al., 2011). Many lactic acid bacteria (LAB) have been shown to degrade milk proteins to release bioactive peptides during diary fermentation. Such bacteria contain well developed proteolytic systems that break down proteins in the growth media into short amino acid chains (Daliri et al., 2017c). Given the proteolytic nature of LAB such as Lactobacillus acidophilus, L. brevis, Lactobacillus animalis and Lactococcus lactis, their use as microbial catalysts for producing ACE inhibitory peptides have been well documented (Brzozowski and Lewandowska, 2014; Hayes et al., 2007). Lactobacillus helveticus CP790 and Saccharomyces cerevisiae have been used as mixed cultures to produce potent ACE inhibitors such as Val-Pro-Pro and Ile-Pro-Pro from casein (Nakamura et al., 1995). Consuming milk containing the peptides effectively reduced high blood pressure in antihypertensive patients (Jauhiainen et al., 2010).
In this study we investigated the in vitro ACE inhibitory activity of 34 lactic acid bacteria during whey fermentation. Whey fermentates with the strongest ACE inhibition was analyzed by liquid chromatography-electrospray ionization-quantitative time-of-flight tandem mass spectrometry to identify the biopeptides that might have been involved in the inhibition.
Materials and methods
Chemicals and cultures
Unless specified, all chemical reagents were of analytical grade and were obtained from Sigma-Aldrich, South Korea. N-Hippuryl-His-Leu hydrate powder, ≥ 98% (HHL), ACE from rabbit lung, Phe-Gly, O-phthaldialdehyde (OPA), sodium tetraborate, sodium dodecyl sulfate, and whey from bovine milk (spray-dried) were purchased. Thirty-four lactic acid bacteria strains namely Pediococcus pentosaceus SDL1416, Pediococcus acidilactici SDL1405, Enterococcus lactis SCL1421, Lactobacillus plantarum JDFM44, Weissella confusa SCSB2320, Lactobacillus rhamnosus JDFM6, Pediococcus acidilactici SDL1414, Enterococcus faecalis MAD13, Streptococcus thermophilus SCML300, Pediococcus pentosaceus SDL1415, Lactobacillus rhamnosus JDFM33, Streptococcus thermophilus SCML337, Pediococcus pentosaceus SDL1409, Pediococcus pentosaceus SDL1401, Pediococcus acidilactici SDL1406, Leuconostoc mesenteroides JBNU10, Leuconostoc citreum SC53, Pediococcus acidilactici SKL1418, Lactobacillus pentosus SC48, Enterococcus faecium CK5, Pediococcus pentosaceus MAC11, Leuconostoc paramesenteroides SC46, Weissella koreensis JBNU2, Lactobacillus curvatus JBNU38, Weissella confusa SCKB2318, Enterococcus faecium SC54, Lactobacillus brevis SDL1411, Lactobacillus brevis SDL1408, Pediococcus acidilactici SCL1420, Lactobacillus plantarum SDL1413, Weissella cibaria SCCB2306, Lactobacillus arizonensis SC25, Pediococcus acidilactici DN9 and Pediococcus acidilactici SDL1402 were obtained from Soonchang Jang Ryu Saupso Company-Korea. Stock cultures were maintained at − 80 °C in de Man, Rogosa and Sharpe broth (MRS, MBCell-Korea), containing 20% (v/v) glycerol. Cultures were streaked on MRS agar and cultured at 37 °C for 24 h. Single colonies were then transferred into MRS broth at 37 °C and harvested at the exponential phase of growth. The viable bacterial count was determined by plate count on MRS agar. The basal growth medium for the lactic acid bacteria consisted of 2% (w/v) reconstituted whey powder in distilled water, supplemented with 1% glucose and 0.5% yeast extract (Becton–Dickinson Co., Cockeysville, MD). Whey powder was composed of 11% protein. The whey-based growth media were autoclaved at 121 °C for 15 min prior to inoculation with lactic acid bacteria.
Cultivation of lactic acid bacteria in whey media
To grow the lactic acid bacteria in whey-based growth media, 2 × 108 cfu/ml of each strain in the 24 h MRS broth at 37 °C were inoculated into 500 mL Erlenmeyer flask containing 200 ml of whey medium (pH 6). Cultivation was carried out at 37 °C with 150 rpm of agitation. After growth for 48 h, the media was centrifuged (10,000×g; 10 min) to remove precipitates. The supernatant (hydrolysate) was freeze-dried using TFD5505 table top freeze dryer (ilshinBioBase Co. Ltd, South Korea) and the freeze-dried samples were stored at − 80 °C for further analysis.
Determination of degree of protein hydrolysis
The degree of whey protein hydrolysis was carried out as reported earlier (Church et al., 1983). In brief, O-phthaldialdehyde (OPA) solution containing 40 mg of OPA dissolved in 1 ml of methanol, 100 ml of β-mercaptoethanol solution, 25 ml of 100 mM sodium tetraborate and 2.5 ml of 20% (w/w) sodium dodecyl sulfate (Sigma), was diluted to a final volume of 50 ml with water. The freeze-dried samples (5 mg/ml) were incubated with 1 ml OPA for 2 min at room temperature (~ 28 °C) and absorbance at 340 nm was measured. The amount of peptide liberated during the fermentation process was calculated using Phe-Gly as a standard.
Recovery of low molecular weight peptides (< 7000 Da)
The hydrolysate (700 mg) was dissolved in 2 ml distilled water and dialyzed against water using a 7000 Da molecular weight cut-off dialysis bag (Viskase Corporation, Chicago, Illinois) for 48 h to recover low molecular weight peptides. We chose this molecular cut-off because most reported ACE inhibitory peptides are less than 10 kDa (Abdel-Hamid et al., 2017; Georgalaki et al., 2017). After discarding the retentates, the dialysates (permeates) were freeze-dried. The protein concentrations were determined using Bradford reagent according to the manufacturer’s instructions.
In-vitro assay for ACE inhibitory activity
ACE inhibitory activity was measured by the procedures summarized in Table 1 (Cushman and Cheung, 1971). Briefly, 20 μl of the ACE inhibitor (LMW peptides) solution was mixed with 50 μl of 5 mM HHL in 100 mM sodium borate buffer (pH 8.3) containing 0.3 M NaCl and incubated at 37 °C for 5 min. The reaction was initiated by adding 10 μl of 0.1 U/ml ACE solution and the mixture was incubated at 37 °C for 30 min. The reaction was stopped by adding 100 μl of 1 M HCl and the reaction mixture was mixed with 1.0 ml of ethyl acetate. The mixture was vortexed for 60 s and centrifuged at 2000 × g for 5 min. An aliquot (800 µl) of ethyl acetate layer was transferred to a clean tube and evaporated on a water bath. Distilled water (800 µl) was then added to dissolve the hippuric acid (HA) remaining in the tube and the amount of HA formed was measured at 228 nm using a biospectrometer (Eppendorf Biospectrometer® fluorescence). The amount of HA liberated from HHL under this reaction conditions without an inhibitor was used as control. The extent of inhibition was calculated as 100% × [(B − A/B)] where A is the optical density (OD) in the presence of ACE and ACE inhibitory component, B is the OD without ACE inhibitor. For the determination of IC50, series of dilutions containing 5000, 500, 50, 5, 0.5, and 0.05 μg/ml of the peptides were prepared. The amount of peptides required to suppress 50% ACE activity was calculated from the regression curves observed for each fraction.
Identification of peptides by mass spectrometry
The peptide profile of the samples fermented with P. acidilactici SDL 1414 was determined using Liquid chromatography-electrospray ionization-quantitative time-of-flight tandem mass spectrometry experiments (LC-ESI-TOF–MS/MS). This fermentate was chosen for further analysis because it showed the lowest IC50 (highest potency). LC-ESI-TOF–MS/MS was performed at the National Instrumentation Center for Environmental Management of Seoul National University-Korea, using a method already described (Chang et al., 2014). Mass spectrometry was carried out using an integrated system comprising an autosampler (TempoTM nano LC system; MDS SCIEX, Canada), an auto-switching nano pumpand a hybrid quadrupole-time-of-flight (TOF) mass spectrometer (QStar Elite; Applied Biosystems, USA) with a fused silica emitter tip (New Objective, USA). Nano-electrospray ionization (ESI) was applied for sample ionization. The sample (2 μl) was injected into the LC-nano ESI–MS/MS system. The sample was initially trapped on a ZORBAX 300SB-C18 trap column (300-μm i.d × 5 mm, 5-μm particle size, 100 pore size, Agilent Technologies, part number 5065-9913) and washed for 6 min with gradient with 98% solvent A and 2% solvent B at a flow rate of 5 μL/min. Solvents A and B consisted of [water/acetonitrile (98:2, v/v), 0.1% formic acid] and [Water/acetonitrile (2:98, v/v), 0.1% formic acid] respectively. Separation was carried out using a ZORBAX 300SB-C18 capillary column (75-μm i.d × 150 mm, 3.5 μm particle size, 100 pore size, part number 5065-9911) at a flow rate of 300 nl/min with gradient at 2 to 35% solvent B over 30 min, then from 35 to 90% over 10 min, followed by 90% solvent B for 5 min, and finally 5% solvent B for 15 min. Electrospray was performed through a coated silica tip (FS360-20-10-N20-C12, PicoTip emitter, New Objective) at an ion spray voltage of 2000 eV. Using Analyst QS 2.0 software (Applied Biosystems, USA), the peptides were analyzed automatically. The range of m/z values was 200-2000.
Statistical analysis
All experiments were carried out in triplicates and the results were expressed as the mean ± standard deviation. The statistical analysis of data was performed using GraphPad Prism 5.0 (2007) statistical software system (GraphPad Software Inc. CA 92037 USA). P < 0.05 was considered significant.
Results and discussion
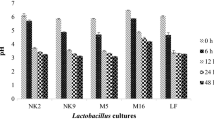
Degree of whey hydrolysis by lactic acid bacteria
The degree of whey protein hydrolysis by each of the 34 LAB was tested after fermentation for 48 h. LAB require exogenous amino acids for growth and they derive these amino acids from the protein substrates in which they are cultured. They do this by hydrolyzing proteins into oligopeptides using their cell-enveloped proteinase (Liu et al., 2010). Specific peptide transport systems then transport the oligopeptides into the cells where they are hydrolyzed into amino acids by various intracellular peptidases (Daliri et al., 2017c). All the LAB in this study showed varying abilities to hydrolyze whey (Fig. 1) and this agrees with earlier reports that showed that LAB protein digestion is strain dependent (Bounouala et al. 2017). We observed that L. plantarum JDFM44 showed the highest ability to hydrolyze whey proteins followed by L. plantarum SDL1413 (Fig. 1). This could be due to the release of large amounts of exopeptidases which hydrolyzed N-terminal amino acids from the proteins during the fermentation process. Ent. faecium CK-5, P. pentosaceus SDL1401, L. pentosus SC48 and P. acidilactici SDL1414 also showed > 50% degree of hydrolysis while the remaining strains showed low degrees of hydrolysis during the fermentation process.
The degree of whey hydrolysis by 34 lactic acid bacteria after 48 h fermentation at 37 °C. Degree of hydrolysis is mean ± SEM of three independent experiments. Bars with different alphabets are significantly different (p < 0.05). Legend: 1—L. rhamnosus JDFM6; 2—P. Pentosaceus SDL1415; 3—P. Pentosaceus SDL1416; 4—L. rhamnosus JDFM33; 5—E. lactis SCL1421; 6—P. acidilacti SDL1405; 7—W. confusa SCSB2320; 8—P. acidilactici SDL1414; 9—S. thermophillus SCML300; 10—Ent. Faecalis MAD13; 11—L. planterum JDFM44; 12—Ent. fecium SC54; 13—P. acidilactici SCL1420; 14—W. koreensis JBNU2; 15—L. pentosus SC48; 16—Leu. paramesenteroides SC46; 17—P. acidilactici SKL1418; 18—L. brevis SDL1408; 19—L. brevis SDL1411; 20—W. confusa SCKB2318; 21—L. curvatus JBNU38; 22—Ent. fecium CK5; 23—S. thermophilus SCML337; 24—L. plantarum SDL1413; 25—L. arizonensis SC27; 26—L. pentosaceus SDL1409; 27—L. pentosus SDL1401; 28—P. acidilactici SDL1406; 29—Leu. mesenteroides JBNU10; 30—W. cibaria SCCB2306; 31—P. pentosaceus SDL1416; 32—P. acidilactici DM9; 33—P. pentosaceus MAC11; 34—L. citrium SC53
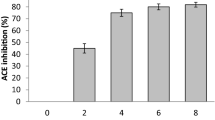
Determination of in vitro ACE inhibitory (ACEI) activity in low molecular weight (LMW) peptides
All the fermentates showed varying ACEI ability (data not shown) however, the fermentates that had ≥ 50% is shown in Table 2 and their IC50 were determined. When LMW peptides (< 7 kDa) were recovered from these samples, 17 fermentates had ACEI ability (data not shown). LMW peptides from P. acidilactici SDL1414 fermentation showed the strongest in vitro ACEI activity of 84.7 ± 0.67 with an IC50 value of 19.78 ± 1.73 μg/ml (Table 2). Captopril (a synthetic ACEI compound) showed an inhibition of 90.82 ± 10.3 and an IC50 value of 5 ± 0.1 μg/ml. Interestingly, we did not find any correlation between the degree of hydrolysis and ACE inhibitory potency. Since LMW peptides from P. acidilactici SDL 1414 fermentation showed the highest ACE inhibition, they were chosen for further studies using LC–ESI–MS/MS to identify the antihypertensive peptides generated during the fermentation process.
Peptides generated from whey during fermentation by Pediococcus acidilactici SDL1414
LC-ESI-TOF–MS/MS analysis of the LMW peptide profile showed that the peptides were generated from casein, beta lactoglobulin, lactophorin, polymeric immunoglobulin receptor and uncharacterized protein GP2 (Table 3). This indicates that P. acidilactici SDL1414 has the ability to hydrolyze different substrates present in the media relative to other lactic acid bacteria such as Bifidobacterium longum which preferentially hydrolyze only casein (Ha et al., 2015). During the fermentation process, a large number of peptides were generated from the C-terminus of β-CN but the N-terminal was resistant to hydrolysis probably due to the presence of several phosphoserine residues at that region (Kaspari et al., 1996). The resistance of the N-terminal region of β-CN to LAB hydrolysis has also been reported in earlier studies (Ha et al., 2015). Hydrolysis of the C-terminal region on the other hand could be due to its hydrophobic nature which makes it more accessible for hydrolysis (Chang et al., 2014). Other LAB including Lactobacillus helveticus (Griffiths and Tellez, 2013), Lactobacillus delbrueckii subsp. lactis lactis (Tsakalidou et al., 1999), S. thermophilus (Miclo et al., 2012), and Lactobacillus lactis (Juillard et al., 1995) also readily hydrolyze the C-terminal region of β-CN to generate large numbers of peptides during milk fermentation.
The N terminus of αS1-CN was also found to be resistant to P. acidilactici SDL1414 hydrolysis and this could also be due to the presence of phosphoserine residues in that region (Ha et al., 2015). The bacterium however readily hydrolyzed the C-terminus of the protein during the fermentation process.
The caseinomacropeptide or glycomacropeptide (f 106-169) segment of κ-CN is composed of glycan chains and has been reported to be resistant to hydrolysis in many studies (Svanborg et al., 2016). The resistance has been attributed to the presence of hydrophilic amino acids and their negative charges which cause increased electrostatic repulsion (Zahraa et al., 2010). However, in the present study, 17 different peptides were obtained from the region (Table 3), 9 of which have bear sequences already reported to have ACE inhibition (Table 4). P. acidilactici SDL1414 fermentation might have induced structural changes in the glycomacropeptide region to allow hydrolysis.
Many ACEI peptides have been reported to be present in whey. LC-ESI-TOF–MS/MS analysis of our low molecular weight peptides showed the presence of 52 different peptides (Table 3). Among these peptides, we identified 1 peptide, αS1-CN (f14-23) EVLNENLLRF, which has already been reported to be an ACE inhibitor (Torres-Llanez et al., 2011). Also, 28 peptides generated in this study had their C-terminal amino acid sequences homologous with previously identified ACEI peptides (Table 4), yet, the complete sequences observed in this study have not been reported before (Kumar et al., 2014; Minkiewicz et al., 2008).
The abundance of antihypertensive segments in these peptides may at least, in part, account for the strong ACE inhibitory activity of the fermentate and the low IC50 (19.78 ± 1.73 μg/ml) observed in this study. Though our ACEI peptides are less potent than captopril (IC50 = 5 μg/ml), natural antihypertensive peptides have high tissue affinities and hence may be more slowly eliminated from tissues compared to synthetic drugs (Koyama et al., 2014). The slow elimination could enhance a more sustained antihypertensive effect in consumers. There is therefore a demand for such bioactive functional ingredients and that warrants this type of research.
The current study shows that P. acidilactici SDL1414 could be used as a starter culture in the dairy industry to develop antihypertensive functional foods. The peptides in the fermentates could also be purified and used as nutraceuticals or supplements to reduce high blood pressure. However, in vivo studies regarding the stability of these peptides against gastrointestinal enzymes as well as their blood pressure lowering ability is warranted.
References
Abdel-Hamid M, Otte J, De Gobba C, Osman A, Hamad E. Angiotensin I-converting enzyme inhibitory activity and antioxidant capacity of bioactive peptides derived from enzymatic hydrolysis of buffalo milk proteins. Int Dairy J. 66:91–98 (2017)
Ahn J, Park S, Atwal A, Gibbs B, Lee B. Angiotensin I‐converting enzyme (ACE) inhibitory peptides from whey fermented by Lactobacillus species. J Food Biochem. 33:587–602 (2009)
Bounouala FZ, Roudj S, Karam N-E, Recio I, Miralles B. Casein hydrolysates by Lactobacillus brevis and Lactococcus lactis proteases. Peptide profile discriminates strain-dependent enzyme specificity. J. Agric. Food Chem. 65: 9324–9332 (2017)
Brzozowski B, Lewandowska M. Prolyl endopeptidase-Optimization of medium and culture conditions for enhanced production by Lactobacillus acidophilus. Electron J Biotechnol.17:204–210 (2014)
Chang OK, Roux É, Awussi AA, Miclo L, Jardin J, Jameh N, Dary A, Humbert G, Perrin C. Use of a free form of the Streptococcus thermophilus cell envelope protease PrtS as a tool to produce bioactive peptides. Int Dairy J. 38:104–115 (2014)
Church FC, Swaisgood HE, Porter DH, Catignani GL. Spectrophotometric assay using O-phthaldialdehyde for determination of proteolysis in milk and isolated milk proteins. J Dairy Sci. 66:1219–1227 (1983)
Cushman D, Cheung H. Spectrophotometric assay and properties of the angiotensin-converting enzyme of rabbit lung. Biochem. Pharmacol. 20:1637–1648 (1971)
Daliri EB-M, Oh DH, Lee BH. Bioactive peptides. Foods 6:32–42 (2017a)
Daliri EB-M, Lee BH, Oh DH. Current trends and perspectives of bioactive peptides. Crit. Rev. Food. Sci. Nutr. (2017b). https://doi.org/10.1080/10408398.2017.1319795
Daliri EB-M, Lee BH, Oh DH. Current perspectives on antihypertensive probiotics. Probiotics Antimicro Prot. 9:91–101 (2017c)
Georgalaki M, Zoumpopoulou G, Mavrogonatou E, Van Driessche G, Alexandraki V, Anastasiou R, Papadelli M, Kazou M, Manolopoulou E, Kletsas D, Devreese B. Evaluation of the antihypertensive angiotensin-converting enzyme inhibitory (ACE-I) activity and other probiotic properties of lactic acid bacteria isolated from traditional Greek dairy products. Int Dairy J. 75:10–21 (2017)
Griffiths MW, Tellez AM. Lactobacillus helveticus: the proteolytic system. Front Microbiol. 4:30 (2013). https://doi.org/10.3389/fmicb.2013.00030
Ha GE, Chang OK, Jo S-M, Han G-S, Park B-Y, Ham J-S, Jeong S-G. Identification of antihypertensive peptides derived from low molecular weight casein hydrolysates generated during fermentation by Bifidobacterium longum KACC 91563. Korean J Food Sci Anim Resour. 35:738 (2015)
Hayes M, Stanton C, Slattery H, O’Sullivan O, Hill C, Fitzgerald G, Ross R. Casein fermentate of Lactobacillus animalis DPC6134 contains a range of novel propeptide angiotensin-converting enzyme inhibitors. Appl. Environ. Microbiol. 73:4658–4667 (2007)
Ibrahim HR, Ahmed AS, Miyata T. Novel angiotensin-converting enzyme inhibitory peptides from caseins and whey proteins of goat milk. J. Adv. Res. 8:63–71 (2017)
Jauhiainen T, Rönnback M, Vapaatalo H, Wuolle K, Kautiainen H, Groop P, Korpela R. Long-term intervention with Lactobacillus helveticus fermented milk reduces augmentation index in hypertensive subjects. Eur J Clin Nutr. 64: 424–431 (2010)
Jin Y, Yu Y, Qi Y, Wang F, Yan J, Zou H. Peptide profiling and the bioactivity character of yogurt in the simulated gastrointestinal digestion. J. Proteom. 141:24–46 (2016)
Juillard V, Laan H, Kunji E, Jeronimus-Stratingh CM, Bruins AP, Konings WN. The extracellular PI-type proteinase of Lactococcus lactis hydrolyzes beta-casein into more than one hundred different oligopeptides. J. Bacteriol. 177:3472–3478 (1995)
Kaspari A, Diefenthal T, Grosche G, Schierhorn A, Demuth H-U. Substrates containing phosphorylated residues adjacent to proline decrease the cleavage by proline-specific peptidases. Biochim Biophys Acta Protein Struct Molec Enzym. 1293:147–153 (1996)
Koyama M, Hattori S, Amano Y, Watanabe M, Nakamura K. Blood pressure-lowering peptides from neo-fermented buckwheat sprouts: a new approach to estimating ACE-inhibitory activity. PloS one. 9:e105802 (2014)
Kumar R, Chaudhary K, Sharma M, Nagpal G, Chauhan JS, Singh S, Gautam A, Raghava GP. AHTPDB: a comprehensive platform for analysis and presentation of antihypertensive peptides. Nucleic acids research. 43:D956-D62 (2014)
Liu M, Bayjanov JR, Renckens B, Nauta A, Siezen RJ. The proteolytic system of lactic acid bacteria revisited: a genomic comparison. BMC genomics. 11:36 (2010)
Manzanares P, Salom JB, García-Tejedor A, Fernández-Musoles R, Ruiz-Giménez P, Gimeno-Alcañíz JV. Unraveling the mechanisms of action of lactoferrin-derived antihypertensive peptides: ACE inhibition and beyond. Food & function. 6:2440–52 (2015)
Miclo L, Roux Em, Genay M, Brusseaux Em, Poirson C, Jameh N, Perrin C, Dary A. Variability of hydrolysis of β-, αs1-, and αs2-caseins by 10 strains of Streptococcus thermophilus and resulting bioactive peptides. J. Agric. Food Chem. 60:554–565 (2012)
Minkiewicz P, Dziuba J, Iwaniak A, Dziuba M, Darewicz M. BIOPEP database and other programs for processing bioactive peptide sequences. J. AOAC Int. 91:965–980 (2008)
Nagpal R, Behare P, Rana R, Kumar A, Kumar M, Arora S, Morotta F, Jain S, Yadav H. Bioactive peptides derived from milk proteins and their health beneficial potentials: an update. Food & function. 2:18–27 (2011)
Nakamura K, Naramoto K, Koyama M. Blood-pressure-lowering effect of fermented buckwheat sprouts in spontaneously hypertensive rats. J Funct Foods.5:406–415 (2013)
Nakamura Y, Yamamoto N, Sakai K, Takano T. Antihypertensive effect of sour milk and peptides isolated from it that are inhibitors to angiotensin I-converting enzyme. J Dairy Sci. 78:1253–1257 (1995)
Oh NS, Lee JY, Oh S, Joung JY, Kim SG, Shin YK, Lee K-W, Kim SH, Kim Y. Improved functionality of fermented milk is mediated by the synbiotic interaction between Cudrania tricuspidata leaf extract and Lactobacillus gasseri strains. Appl Microbiol Biotechnol. 100:5919-5932 (2016)
Sánchez A, Vázquez A. Bioactive peptides: A review. Food Quality Safety.1:29–46 (2017)
Svanborg S, Johansen A-G, Abrahamsen RK, Schüller RB, Skeie SB. Caseinomacropeptide influences the functional properties of a whey protein concentrate. Int Dairy J. 60:14–23 (2016)
Tavares T, del Mar Contreras M, Amorim M, Pintado M, Recio I, Malcata FX. Novel whey-derived peptides with inhibitory effect against angiotensin-converting enzyme: in vitro effect and stability to gastrointestinal enzymes. Peptides. 32:1013–1019 (2011)
Torres-Llanez M, González-Córdova A, Hernandez-Mendoza A, Garcia H, Vallejo-Cordoba B. Angiotensin-converting enzyme inhibitory activity in Mexican Fresco cheese. J Dairy Sci. 94:3794–800 (2011)
Tsakalidou E, Anastasiou R, Vandenberghe I, Van Beeumen J, Kalantzopoulos G. Cell-wall-bound proteinase of Lactobacillus delbrueckii subsp. lactis ACA-DC 178: characterization and specificity for β-casein. Appl. Environ. Microbiol. 65:2035–2040 (1999)
Zahraa N. Le peptide κ-CN (f106-109) du lait: propriétés nutritionnelles, biologiques et techno-fonctionnelles. Mémoire de M2 UHP Nancy 1, France (2010)
Acknowledgements
This work was funded by the Ministry of Small and Medium scale Enterprises and Startups. Grant Number C0502529. We also thank the central laboratory of Kangwon National University for their assistance in LC-MS analysis.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Daliri, E.BM., Lee, B.H., Park, BJ. et al. Antihypertensive peptides from whey proteins fermented by lactic acid bacteria. Food Sci Biotechnol 27, 1781–1789 (2018). https://doi.org/10.1007/s10068-018-0423-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10068-018-0423-0