Abstract
In this study, Lactobacillus bulgaricus NCDC (09) and Lactobacillus fermentum TDS030603 (LBF) were evaluated for their ACE-inhibitory activity and peptides production under optimized conditions from fermented camel milk (Camelus dromedarius). Lactic cultures were evaluated for their pepX activity, proteolytic activity and ACE-inhibitory activity. 09 culture exhibited higher PepX and ACE-inhibitory activity than LBF. 2% rate of inoculation and 12 h of incubation were optimized on the basis of pepX and proteolytic activity. Purified peptides from fermented camel milk were characterized by amino acids profiling through the search in BlastP, Protein information resource (PIR) databases. ACE-inhibitory activity of different peptides from fermented camel milk were also confirmed by the database of antihypertensive peptides (AHTPDB). Fermented camel milk produced by Lactobacillus cultures could be a novel source of ACE-inhibitory peptides.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Milk proteins are known for many biological activities that play significant role in improving human health and nutrition (Park 2009). Bioactive peptides can be produced from milk proteins by digestive enzymes (proteases), microbial or plant enzymes or by fermenting with proteolytic dairy starter cultures (Hayes et al. 2007). Bioactive peptides are well-known with health benefits like cholesterol-lowering (Hartmann and Meisel 2007), antimicrobial (López-Expósito and Recio 2006), immunomodulatory, opioid, antioxidative, antihypertensive (Jäkälä and Vapaatalo 2010) and mineral-binding (Vegarud et al. 2000). The main bioactive peptides studied across the world are those with antihypertensive effect (Korhonen and Pihlanto 2006), which are known for their angiotensin-converting enzyme (ACE) inhibiting function. ACE-inhibitions plays key role in blood pressure regulation (De Leo et al. 2009). It has been reported that, milk derived ACE-inhibitory peptides exhibit the inhibition of conversion of angiotensin I to angiotensin II; thus these peptides have a blood pressure lowering effect (Vermeirssen et al. 2004). ACE-inhibitory peptides have been isolated from variety of fermented dairy products including yoghurt (Donkor 2007), cheese (Hartmann and Meisel 2007) and fermented bovine milk (Qian et al. 2011). Use of proteolytic lactic acid bacteria for the milk fermentation is one of the economical and practical methods for the production of fermented dairy products enriched with bioactive peptides (Hayes et al. 2007). The proteolytic system of LAB can contribute to the liberation of health enhancing bioactive peptides from milk. The latter may improve absorption in the intestinal tract, stimulate the immune system, exert antihypertensive or antithrombotic effects and antioxidative activity or function as carriers for minerals especially calcium (Padghan et al. 2016). As per the previous reports, bovine milk fermented with different proteolytic lactic acid bacterial strains such as Lactobacillus delbrueckii subsp. bulgaricus (Papadimitriou et al. 2007), Lactobacillus helveticus JCM1004, Lactobacillus helveticus MTCC5463 (Pan et al. 2005; Hati et al. 2016), Lactobacillus acidophilus DPC6026 and Lactococcus lactis YT2027 contain health-beneficial bioactive peptides. Fermented milk products are produced from different species like bovine, goat, sheep, buffalo, etc (Chandan 2004). Camel milk is well suitable for fermented milk production. It contains well-balanced nutrients and biological components. Camel milk is high in vitamin C and niacin, as well as richer in Cu and Fe than bovine milk (El-Agamy 2006). Moreover, camel milk lacks beta-lactoglobulin and contains alpha-lactalbumin, a similar situation to that in human milk. Fermented camel milk is now available in different countries and traditionally produced by the communities. In Ethiopia, dromedary camel milk was used as a potential treatment for a number of diseases like Malaria, Jaundice, to clear the stomach, flatulence, to detoxify snake venom, constipation and post pregnancy care of women (Seifu 2007; Mehari et al. 2007). Also used in treating other health problems such as dropsy, tuberculosis, asthma and leishmaniasis or kala-azar in different parts of the world including India, Russia and Sudan (Abdelgadir et al. 1998). Fermented camel milk (FCM) is also considered as probiotic with its unique antibodies and medicinal properties. FCM may provide beneficial effects like anti-inflammation and anti-obesity (Badkook 2013). Fermented camel milk is also a source of different bioactive peptides. Bioactive peptides like ACE-inhibitory peptides and antioxidative peptides were also derived from fermented camel milk (Moslehishad et al. 2013). Furthermore, some reports are available on peptides with a wide range of functionalities in fermented bovine milk with proteolytic strains of LAB (FitzGerald and Murray 2006; Hayes et al. 2007; Papadimitriou et al. 2007), but there is a lack of information in the literature about biological activity of camel milk-derived peptides by lactic acid fermentation. Therefore, it would be interesting to investigate the functionalities of bioactive peptides produced during the fermentation of camel milk. In this study, L. bulgaricus NCDC (09) and L. fermentum TDS030603 (LBF) were considered because of their high proteolytic activity and ACE-inhibitory activity in camel milk medium. The objective of this study was to purify and characterize the ACE-inhibitory peptides derived from fermented camel milk (Camelus dromedarius).
Materials and Methods
Collection of Lactobacillus Cultures and their Maintenance
The LAB cultures used in the present study i.e. L. bulgaricus NCDC (09) and L. fermentum TDS030603 (LBF) were obtained from the Culture Collection of Dairy Microbiology Department, SMC College of Dairy Science, Anand, Gujarat, India. The lactic cultures were propagated in sterilized reconstituted skim milk (10% TS) and stored at 5 ± 2 °C. The transfer was given every week during the course of the study. Purity and activity was tested every time before sub-culturing.
Camel Milk Procurement
Fresh camel milk (Camelus dromedarius) sample was procured from a farm at Ankalav village of Anand District, Gujarat, India.
Assessment of X-prolyl-dipeptidyl Aminopeptidase Activity (PepX)
Pure lactic cultures were activated in MRS broth at the rate of 2% of inoculation and incubated for 24 h at 37 °C. Freshly grown cultures were inoculated at the rate of 2% in 100 mL MRS broth and incubated for 0, 3, 6, 9 and 12 h. Pellets were collected after centrifugation at 3500 rpm for 15 min at 4 °C. (Hermle centrifuge, Germany). Then, pellets were washed with 25 mL of sodium phosphate buffer and finally pellets were dissolved in 10 mL of sodium phosphate buffer. Then cell pellets were sonicated by probe sonicator (Labman, India). After sonication, the supernatants were collected by centrifugation at 13,000 rpm for 13 min at 4 °C. Then, crude supernatants collected were used as samples for the enzymatic assay.
Enzymatic Reaction of pepX Activity
X-prolyl-dipeptidyl aminopeptidase (pepX) activity of the cultures was assayed with glycyl-prolyl p-nitroanilide (Sigma, USA) (Gly-Pro-pNA) as the substrate with some modifications (Donkor et al. 2007). The incubation mixture contained 50 μL of 6.4 mmol/L of substrate glycyl-prolyl p-nitroanilide, 2.85 mL of 50 mmol L−1 of Tris–HCl buffer (pH 7.0) and 100 μL of sample (supernatant). The mixture was incubated at 37 °C for 30 min and the reaction was stopped by adding 500 μL of 30% acetic acid. The extent of hydrolysis was measured with the spectrophotometer (Systronics Spectrophotometer 2202, India) at 410 nm. The activity of each sample was evaluated in triplicate.
Preparation of Fermented Camel Milk
Fresh camel milk was collected and then filtered through muslin cloth and heated at 90 °C for 10 min and then stored at 5 ± 1 °C. 2% of previously grown active cultures from reconstituted milk were inoculated in camel milk and incubated for 24 h at 37 °C for activation in camel milk medium.
Preparation of the Supernatant (Water Soluble Extract) from Fermented Camel Milk
Fermented camel milk samples were centrifuged at 13,000 rpm for 20 min at 4 °C (Hermle centrifuge, Germany). The supernatant (Water soluble extract) was collected and it was filtered through 0.45 μm membrane filter (Millex® - HV, MERK, Ireland). This supernatant was further assayed for ACE-Inhibitory activity.
Determination of ACE-inhibitory Activity
The ACE-inhibitory activity percentage (ACEi %) was determined according to the method described by Hati et al. (2015) with some modifications as described below. 50 μL of 5 mM HHL (Sigma, USA) (10.74 mg HHL in 5 mL sodium borate buffer, pH 8.3) solution was mixed with 500 μL deionized water and 200 μL of sample (Water soluble extract). The residue containing hippuric acid was dissolved in 2 mL of deionized water and it was filtered through 0.45 μm membrane filter (Millex® - HV, MERK Ireland). The absorbance of the solution was measured spectrophotometrically (Systronics Spectrophotometer 2202, India) at 228 nm. The activity of each sample was tested in triplicate. The formula used for the determination of ACE-inhibitory peptides is such as; ACEi% = {(Absorbance of HA control – Absorbance of HA sample/Absorbance of HA control) × 100}. Where, HA control was the absorbance of concentration of hippuric acid produced by the ACE in buffer without lactic cultures. HA sample was the absorbance of the concentration of hippuric acid produced by the ACE in the presence of lactic cultures. In the case of fractionated 3, 5 and 10 kDa, HA control was the absorbance of concentration of hippuric acid produced by the ACE in camel milk without lactic cultures. HA sample was the absorbance of the concentration of hippuric acid produced by the ACE in the presence of lactic cultures in camel milk medium.
Determination of Proteolytic Activity of Lactic Cultures in Camel Milk Medium
Growth conditions for the production peptides of the selected lactic acid bacteria was optimized by measuring the proteolytic activity through o-phthaldialdehyde (OPA) method (Hati et al. 2016).
Sample Preparation for the Proteolytic Activity
Both the lactic cultures were activated by growing in heat treated camel milk. Different rates of inoculation like 1.0, 1.5 and 2% were used with different incubation periods to optimize the growth conditions for the production of peptides. The culture tubes were incubated at 37 °C for different intervals i.e. 0, 3, 6, 9 and 12 h, the tubes were taken out for the evaluation of peptide content (Proteolytic activity) after each interval. The degree of proteolysis during fermentation of milk was determined by measuring the release of free NH3 groups following the o-phthaldialdehyde (OPA) method (Hati et al. 2016).
Enzymatic Reaction of Proteolytic Activity
An aliquot of 3 mL from each fermented camel milk sample was mixed with 5 mL of 0.75% tri-chloroacetic acid (TCA) and vortex for 1 min, and then the mixture was filtered using Whatman™ no. 42 filter papers (UK). The filtrate (200 μL) was added to 3 mL of OPA reagent and after the incubation at room temperature (20 °C) for 2 min, absorbance of the solution was measured by a spectrophotometer (Systronics PC based double beam Spectrophotometer 2202, India) at 340 nm. The proteolytic activity of these lactic cultures was expressed as the absorbance of free amino groups measured at 340 nm. A relative degree of proteolysis was determined as the difference between proteolytic activities in fermented camel milk to that of unfermented camel milk. All the analyses were carried out in triplicate.
Peptides Purification by RP-HPLC
Relative proteolytic activity was assessed by peptides mapping of the fermented camel milks, performed using a RP-HPLC (Shimadzu LC-20, Japan), for the separation of different peaks. A binary gradient RP-HPLC system was used, fitted with C18 column (Se Quant® ZIC®-cHILIC) white pore analytic column (3 μ, 250 × 4.6 mm). Sample was applied using microinjector (HAMILTON Bonaduz AG, Switzerland) with 20 μL loop. Eluent-A was 1% (v/v) of TFA in deionized water and Eluent-B was 1% (v/v) of TFA in mixture of 80:20 of acetonitrile and deionized water. Separation was conducted at room temperature at flow rate 0.25 mL/min with eluent-A for 0–10 min. and linear gradient, from 0 to 80% of eluent-B, for 0–10 min. The column finally eluted with 100% eluent-B for 11–20 min. Peaks were a function of absorbance observed by an ultra violet/visible wavelength detector (Shimadzu, SPD-20A) operating at 214 nm. Total peak area was obtained by the integration of all the peaks observed. All the analysis were carried out in triplicate. The change of peptide profile was expressed as relative proteolytic activity, Rpa %, calculated from the given equation as; Rpa %= {(TPE b – TPE e / TPE e ) × 100}. Where TPE b = Total peak area at the beginning (unfermented camel milk sample), TPE e = Total peak area at the end of hydrolysis (fermented camel milk sample).
Amino Acid Characterization of ACE-Inhibitory Peptides Through RP-LC/MS
Fractionations of 3 kDa permeate and 10 kDa permeate samples was carried out through RP-HPLC. Each of the sample was injected to collect different fractions eluted at different time intervals (retention times) and these fractions were used for the characterization of amino acid sequence of the peptides through RP-LC/MS. Ekspert ultra LC 100 (Eksigent, USA) was used in conjunction with ABSCIEX QTRAP 4500 Ion Trap Mass Spectrometer via Electron Spray Ionization (ESI) interface. In built MASCOT script was used for the characterization of the peptides following the protocols given by Jakubczyk and Baraniak (2014); Tagliazucchi et al. (2015) with some modifications (viz., search parameter such as peptide mass tolerance set was 1.2 and MS/MS tolerance was 0.6). RP-LC/MS was used for identification of peptides through Enhanced Mass Spectra (EMS) following three Enhanced Product Ion (EPI) scan using Information Dependent Analysis (IDA) survey scan.
Chromatography
The sequence analysis of peptides in the fractions of 3 and 10 kDa permeates were carried out using liquid chromatography (LC) connected to a mass spectrometer (MS). For liquid chromatography, Ekspert ultra LC 100 from Eksigent, USA was used in conjunction with ABSCIEX QTRAP 4500 ion trap mass spectrometer via Electron Spray Ionization (ESI) interface. The LC-100 was equipped with degasser, quaternary pump, column oven and auto-sampler. Gradient elution was optimized for getting good resolution and noise control during acquisition. For Liquid Chromatography ACQUITY UPLC-BEH C18 (2.1 × 50 mm, 1.7 µm) column (WATERS Company make, UK) was used. The column temperature and sample temperature were maintained at 65 and 4 °C respectively. The mobile phase was composed of solvent A (water) and solvent B (Acetonitrile) with 0.1% formic acid each. The gradient column elution was as follows: (i) 0 min, 95% A and 5% B; (ii) 0–2 min, 90% A and 10% B; (iii) 2–15 min, 50% A and 50% B; (iv) 15–22 min, 50% A and 50% B; (v) 22–24 min, 10% A and 90% B; (vi) 24–25 min, 10% A and 90% B; (vii) 25–35 min, 10% A and 90% B and (viii) 35–60 min, 95% A and 5% B. The flow rate of the mobile phase was set to 0.4 mL/min with an injection volume fixed at 5 µL.
Peptide Identification (Mass Spectrometry MS/MS)
Identification of peptides sequences from fractions exhibiting remarkable ACE-inhibitory activity was carried out by RP-LC/MS. In this study an Information Dependent Analysis (IDA) workflow was optimized for protein identification consisting EMS scan followed by 3 EPI scan. IDA was set by providing Rolling Collision Energy excluding former target ion for 20 s. Optimized IDA methods for identification of peptides was given in Table 1.
Data Processing
Acquired raw data were processed by Analyst software followed by Mascot Search (Matrix Science, London, UK, onsite license) against Swiss-Prot database. Search parameters for peptide and product ions mass tolerance were 1.2 and 0.6 Da, respectively. Enzyme specificity was no cleavage for HPLC purified undigested samples with missed cleavage sites allowed were 0, and fixed modification of cysteine was by carbamidomethylation and variable modification was set to carboxymethylation and methionine oxidation. Peptides with Mascot Score exceeding the threshold value corresponding to <5% false positive rate, calculated by Mascot procedure, and with the Mascot significant score were considered to be positively identified.
In Silico Analysis
Peptide sequences derived from MASCOT with significant score were matched in the databases with the Taxonomy ID-9838 for one humped camel milk protein (Camelus dromedarius) database search against (I) NCBI (blastp tool) (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastp&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome) (II) Protein Information Resource (PIR) to confirm the camel milk protein sequences (3–30 amino acids) (http://research.bioinformatics.udel.edu/peptidematch/index.jsp) (Barker et al. 2001). Also, the peptide sequences were matched with the Database of Antihypertensive peptides (AHTPDB) for confirming the ACE-inhibitory activity (http://crdd.osdd.net/raghava/ahtpdb/pep.php) (Kumar et al. 2015).
Statistical Analysis
All the parameters under the study were analyzed by statistical methods. All determinations were performed at least in triplicate, and the results were expressed as means ± standard deviations (SD). Data obtained were analyzed by statistical designs and softwares. Significant differences between treatments were tested by analysis of variance (ANOVA) with a level of significance of p < 0.05.
Results and Discussions
Assessment of X-prolyl-dipeptidyl Aminopeptidase Activity (PepX)
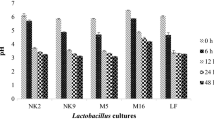
Both the lactic cultures were activated individually in sterilized reconstituted skim milk at 2% rate of inoculation and incubated for 24 h at 37 °C. The PepX activity was evaluated for growing all the active lactic cultures in MRS broth, by inoculating at the rate of 2% (v/v) and incubating for 0, 3, 6, 9 and 12 h at 37 °C. As the PepX is an intracellular enzyme, we had taken intracellular extracts which may be have higher PepX activity as compared to extracellular extract for the same cultures in a same medium. The PepX activity was analyzed spectrophotometrically as absorbance at 410 nm (optical density). X-prolyl-dipeptidyl aminopeptidase (PepX) activity of dairy lactic cultures is an important characteristic due to high proline content in milk protein. The capability of PepX is to cleave Xaa-Pro dipeptides from the (X-Pro ↓ Y…) N terminus of peptides (Pan et al. 2005). PepX releases Xaa-Pro dipeptides from peptides ranging from three to seven amino acid residues; the exhibition of PepX activity from lactic acid bacteria indicates its ability to produce bioactive peptides (Pan et al. 2005). Different studies had been reported about the PepX activity of LAB in milk and milk products. Extracellular and Intracellular extracts were used to determine the PepX activity. PepX activity of both the culture was presented in the Table 2. PepX activity was differing significantly (p < 0.05) with incubation periods. Also, there was a significant difference (p < 0.05) observed within the cultures. Also it was found that, the PepX activity of both the lactic cultures was increased significantly with the time of incubation.
Pastar et al. (2003) and Savijoki et al. (2006) also reported that, Lb. rhamnosus possesses a proteolytic system that includes a proline-specific peptidase like proline-specific aminopeptidase (PepR) and X-prolyl-dipeptidyl aminopeptidase (PepX) that subsequently may result in accumulation of bioactive ACE-inhibitors in fermented milk. Pan et al. (2005) similarly identified X-prolyl-dipeptidyl aminopeptidase activity from IE cell-free extract of L. helveticus JCM1004 in the hydrolysis of skimmed milk proteins and expressed as 23.6 ± 1.6 units per gram. Degraeve and Martial (2003) also reported the PepX acticity of Lactobacillus helveticus ITG LH1 and expressed as 35 ± 7 Ug.protein−1, a strain used for Swiss-type cheese and also purified X-prolyl dipeptidyl aminopeptidase. It was also reported that, the proline specific aminopeptidases such as prolyl iminopeptidases (E.C. 3.4.11.5) and X-prolyl dipeptidyl aminopeptidases (E.C. 3.4.14.5) were found in Streptococcus thermophilus, Lb. helveticus and Lb. delbrueckii subsp. lactis which were used as a starter bacteria in Swiss-type cheese production (Degraeve and Martial 2003). Kunji et al. (1996) also reported the presence of pepX in all species of LAB.
Determination of ACE-inhibitory Activity
In this study, the ACE-inhibitory activity was assessed by fermenting heat treated camel milk (90 °C for 10 min.) with two pure lactic cultures at the rate of 2% inoculation, incubated at 37 °C for different time intervals (0, 12, 24 and 48 h). Different intervals 0, 3, 6 and 9 h of incubation was also tested to check %ACE-inhibitory activity but it showed very less difference in % ACE-inhibitory activity, so, finally the periods of 0, 12, 24 and 48 h of incubation was tested. ACE-inhibitory activity was differing significantly (p < 0.05) with incubation periods. Also, there was a significant difference (p < 0.05) observed within the cultures. ACE-inhibitory inhibitory activity was increased from 0 to 48 h in both the lactic cultures. This might be suggested that, the ACE-inhibitory activity was increasing with the growth of lactic cultures during their growth. It was observed that, ACE-inhibitory activity was differing significantly (p < 0.05) with incubation periods (Table 3). Also, there was a significant difference (p < 0.05) observed within the cultures.
The measured ACE-inhibitory activity had been varied from 55.66 to 76.75% during the incubation period of 0–48 h (Table 3). It was also observed that, the ACE-inhibitory activity was increased from 0 to 48 h in all the lactic cultures. 09 exhibited highest ACE-inhibitory activity (76.75%) than LBF (73.93%) ACE-inhibitory activity up to 48 h of incubation period at 37 °C. This might be suggested that, the ACE-inhibitory activity was increasing with the growth of lactic cultures during their growth. Moslehishad et al. (2013) studied the ACE-inhibitory activity of camel milk and bovine milk fermented by Lb. rhamnosus PTCC 1637. They utilized water soluble extract as a sample, similar to our study and indicated that the IC50 values, especially in camel milk (2.223 ± 0.052–3.930 ± 0.118 mg mL−1), were considerably lower than that reported by Chen et al. (2010) for koumiss whey (52.47 ± 2.87 mg mL−1). They also suggested that, the more pronounced ACE-inhibitory activity of camel milk than bovine milk leads to the development of fermented camel milk by LAB as a novel food product containing ACE-inhibitory peptides. Hati et al. (2015) studied the ACE-inhibitory activity of Lactobacillus rhamnosus (NS4 and NS6), Lactobacillus helveticus MTCC 5463 (V3), Lactobacillus delbruckii (09), Enterococcus feacalis (ND3), Enterococcus feacalis (ND11), Lactobacillus rhamnosus (SH8) and Lactobacillus rhamnosus (I4) in skim milk. The results indicated that, the production of ACE-I inhibitors was not confined to single species or strain of bacteria but all the strains tested, produced peptide, which showed in-vitro ACE-I-inhibitory activity. L. rhamnosus (NS4) and L. bulgaricus (09) gave maximum ACE-I inhibitory activity 79.66 and 67.09% respectively compared to other isolates. Same isolate 09 gave exhibited highest ACE-inhibitory compared to skim milk after comparing our data with Hati et al. (2015). In our study, it may also be suggested that camel milk become a unique growth medium for the growth of lactic acid bacteria which exhibited higher ACE-inhibitory activity as compared to skim milk medium.
Determination of Proteolytic Activity
Selected proteolytic lactic cultures i.e. L. bulgaricus NCDC (09) and L. fermentum TDS030603 (LBF) were used to determine the growth conditions for the production of peptides through the measurement of proteolytic activity. Both the cultures were inoculated at the rate of 1, 1.5 and 2% (v/v) in heat treated camel milk and incubated at 37 °C for 0, 3, 6, 9 and 12 h. Then, the sample prepared was used to optimize the growth conditions through OPA method (Hati et al. 2015). The proteolytic activity of these lactic cultures was expressed as the amount of free amino groups measured as difference in absorbance values at 340 nm, after subtraction of values for the un inoculated control camel milk.
The OPA-based spectrophotometric assay detects released α-amino groups, which result from the proteolysis of milk proteins, thus giving a direct measurement of proteolytic activity. Individually each lactic culture 09 and LBF were statistically analyzed for the optimization of growth condition for the production of peptides through the best inoculation rate (1.0, 1.5 and 2.0%) and the optimum incubation period. Peptide production of individual lactic cultures 09 and LBF in heat treated camel milk was represented in the Tables 4 and 5 respectively.
It was observed that the proteolytic activity of 09 in camel milk was significantly (p < 0.05) increased with the incubation periods (0, 3, 6, 9 and 12 h) (Table 4). The proteolytic activity was significantly higher at the 12 h {(0.915) = periodic mean} of incubation for all the inoculation rates (1.0%, 1.5% or 2.0%) than 0 h (0.573), 3 h (0.629), 6 h (0.727) and 9 h (0.854). The proteolytic activity was found highest at 12 h (1.025) of incubation and 2% of inoculation than 1 and 1.5% inoculation. Overall proteolytic activity through 09 was ranged from 0.558 (1.5% inoculation in 0 h of incubation) to 1.025 (2% inoculation at 12 h of incubation).
It was observed that the peptide production of LBF in camel milk was significantly (p < 0.05) increased with the incubation periods (0, 3, 6, 9 and 12 h) and their interaction with different inoculation rates were found significant (Table 5).
The proteolytic activity was significantly higher at the 12 h {(0.928) = periodic mean} of incubation for all the inoculation rates (1, 1.5 and 2%) than 0 h (0.570), 3 h (0.603), 6 h (0.679) and 9 h (0.776). The proteolytic activity was found highest at 12 h (1.023) of incubation and 2% of inoculation as compared to 1 and 1.5% inoculation. Overall proteolytic activity of LBF was ranged from 0.542 (1.0% inoculation in oh of incubation) to 1.023 (2% inoculation in 12 h of incubation).
From the above Tables 4 and 5, it was found that both the cultures exhibited proteolytic activity differently. It differs with different inoculation rates and incubation periods. Increase in proteolytic activity with the different incubation periods was directly related to the amount amino acids required by the lactic cultures during their growth phases based upon which the release of free NH3 groups differs with the inoculation rates. It was reported that the extent of proteolysis varied among strains examined and showed to be the time and strain dependent (Donkor et al. 2007). This report had a similar observation from our study.
In another study, Hati et al. (2015) studied the proteolytic activity of Lactobacillus rhamnosus (NS4 and NS6), Lactobacillus helveticus MTCC 5463 (V3), Lactobacillus delbruckii (09), Enterococcus feacalis (ND3), Enterococcus feacalis (ND11), Lactobacillus rhamnosus (SH8) and Lactobacillus rhamnossus (I4) by growing in skim milk @ of 1% 37 °C for 24 h. They found that, NS4 liberated highest amount of amino acids after 24 h of fermentation at 37 °C, while 09 and ND3 also efficiently released amino acids during fermentation. However, V3, NS6, SH8, ND11 and I4 showed comparatively lower proteolytic activity. So, they also concluded that, NS4, 09 and ND3 having strong proteolytic system and maximum proteolytic enzymes producing ability in skim milk medium compared to the other isolates.
Similarly, Rahman et al. (2009) studied the changes in proteolytic activities of L. acidophilus, L. bulgaricus, L. lactis, St. thermophilus and mixed cultures of L. bulgaricus and St. thermophilus (1:1) during fermentation of camel milk. The proteolytic activities exhibited by different cultures were observed after 0, 1.5, 3, 4.5 and 6 h of incubation at 43 °C. They compared the proteolytic activity after 1.5 h of incubation with the 6 h of incubation and found significant increase in proteolysis as similar to our study. In general these results showed that Lactobacillus strains had higher proteolytic activity than the Lactococcus lactis strain (Rahman et al. 2009).
Donkor et al. (2007) studied the proteolyitc activity of L. acidophilus (L10), Bifidobacterium (B94), L. casei (L26), S. thermophilus (St 1342), L. delbrueckii ssp. bulgaricus (Lb 1466), L. acidophilus (La 4962), Bifidobacterium (Bl 536) and L. casei (Lc 279) by growing in RSM @ of 1% and providing incubation period of 0, 6, 12, and 24 h at 42 °C. They found that, the amount of liberated amino groups and peptides increased only slightly during fermentation from 0 to 12 h for some strains (L. acidophilus L10, L. acidophilus La 4962, B. lactis B94, B. longum Bl 536, L. casei L26, and L. casei Lc 279) but increased significantly (p < 0.05) for all strains from 12 to 24 h. They reported that, Lactobacillus casei L26 showed the highest proteolytic activity followed in order by L. delbrueckii ssp. bulgaricus Lb 1466, S. thermophilus St 1342 and L. acidophilus (La 4962 and L10), Bifidobacterium and L. casei (Lc 279) with the activity apparently strain specific (p < 0.05). Similar to above study, L. delbrueckii ssp. bulgaricus (09) strain from our study exhibited strong proteolytic activity after 12 h of incubation with the 2% inoculation rate.
Peptide Production (% Relative Proteolytic Activity) Through RP-HPLC
The Relative proteolytic activity was determined according to the method of Vasiljevic and Jelen (2002). Sample was prepared after inoculating in heat treated camel milk at the rate of 2% and incubated at 37 °C for 12 h. The supernatant from fermented sample was injected in RP-HPLC and the change of peptide profile was determined by integrating area under peaks obtained in unfermented camel milk and the fermented camel milk and expressed as Rpa %.
It was observed that 09 exhibited highest peptide production (47.50%) than LBF (41.81%) (Table 6). Figure 1 represent the RP-HPLC chromatogram of control (unfermented camel milk) and fermented camel milk (09) under optimized growth conditions. And Fig. 2 represent the RP-HPLC chromatogram of 3 and 10 kDa permeate of 09 lactic culture under optimized growth conditions. A comparison of lactic culture’s ability for hydrolyzing milk protein was shown by recording area counts from the peptides chromatographic profiles. Peptides produced by the 09 were maximum compared to control milk peptides (unfermented camel milk). Similarly, peptides produced by LBF was found maximum as compared to control milk. Small fractions of peptides were exhibited in control due to some native proteolytic enzymes (Hati et al. 2015). Relative proteolytic activity (%peptide production) of fermented camel milk under optimized growth conditions was presented in the Fig. 1.
Amino Acid Characterization of ACE-Inhibitory Peptides Through LC-MS Analysis
Fractionation of 3 and 10 kDa permeates of fermented camel milk was carried out through RP-HPLC. Each of the sample was injected to collect different fractions eluted at different time and these fractions were used for the characterization of amino acid sequence of peptides through RP-LC/MS. For liquid chromatography, Ekspert ultra LC 100 (Eksigent, USA) was used in conjunction with ABSCIEX QTRAP® 4500 ion trap mass spectrometer via Electron Spray Ionization (ESI) interface with data acquisition and processing software Analyst (version 1.6.1). In built MASCOT script was used to characterize the unknown peptides. The Information dependent analysis (IDA) was optimized with Enhanced Mass Spectra scan (EMS) followed by three Enhanced Product Ion (EPI) scan to identify the unknown peptides. Fragmentation patterns so generated were collected and processed through Analyst software of ABSCIEX. For undigested HPLC fractions the search parameters were set as no enzymatic cleavage with zero mismatches. In addition, search parameters were modified as 1.2 Da for peptide and 0.6 Da product ions mass tolerance, and fixed modification of cysteine to carbamidomethylation and variable modification was set to carboxymethylation and methionine oxidation. Peptides with Mascot Score exceeding the threshold value corresponding to <5% false positive rate, were considered to be positively identified. Further, peptides sequence with significant score was exposed to homology search using BLAST/P online tool (NCBI) against UniProtKB/Swiss-Prot database with taxonomy ID 9838 of Camelus dromedarius. Different di/tri and tetra peptides were found matched in PIR database (Barker et al. 2001). Protein Information Resource (PIR) which is popularly used to match di, tri and tetra peptide sequences was used in present study (Tagliazucchi et al. 2016). As it has been reported by many worker that di and tri peptide are the good candidate for ACE-inhibitory activity (Liu et al. 2014; Wu et al. 2006; Hellberg et al. 1991; Cheung et al. 1983). In addition, di, tri-peptide and tetra peptides were searched in Database of Antihypertensive peptides (AHTPDB) which is an ideal platform identification of large number of antihypertensive peptides. AHTPDB provides information about 5978 peptides, out of which 3364 are with IC50 value provided and total number of unique antihypertensive peptides was 1694. Hence, it is comprehensive platform for large antihypertensive peptides with all the relevant information associated with them e.g. sequence, source, IC50, pIC50, molecular mass, purification method and therapeutic value etc. AHTPDB also provides information of bitterness/toxicity of the peptides, this is important as many antihypertensive agents have been discovered and used as food additives e.g. FDA has approved ‘VPP’ & ‘IPP’ as antihypertensive food additives. If the given peptides have been known with their bitterness/toxicity value, it would be of great use for identification of all those antihypertensive peptides which can be exploited as food additives (Kumar et al. 2015). In this context, VPP was observed in fraction J of 09 culture of 3 kDa permeates and in Y fraction of 10 kDa permeates. Similarly, IPP was observed in LBF fraction E1 of 10 kDa permeate (Tables 11, 12). A total 21 peptides were obtained from 7 fractions of 3 kDa permeate milk samples of two lactic cultures. While 26 peptides were identified from 12 fractions of 10 kDa permeates. In case of 3 kDa permeates, maximum fractions 4 were collected from 09 fermented camel milk using RP-HPLC while in case of 10 kDa permeates, maximum 16 fractions were collected from lactic culture LBF fermented camel milk. Total ion chromatogram of fraction E1 of 10 kDa permeate of LBF culture generated by EMS to EPI scan in LC-MS was presented in the Fig. 3. Presence of similar sequences such as PPPGSKSSTGT in 09 and LBF and QNVLDFHR in 09 and LBF of different fractions revealed cultures may have similar kind of proteolytic activity of among the lactic cultures. The finding was reported by previous worker (Rodríguez-Figueroa et al. 2012). All the characterized sequences from 3 and 10 kDa fractions were matched against NCBI (Database) using BLAST/P (Tables 7, 8), PIR database (Tables 9, 10) to confirm camel milk protein and matched in AHTPDB database (Tables 11, 12 to confirm ACE-inhibitory activity of characterized peptides. Presence of similar sequences such as PPPGSKSSTGT and QNVLDFHR in 09 and LBF of different fractions revealed cultures may have similar kind of proteolytic activity of among the lactic cultures. The finding was reported by previous worker (Rodríguez-Figueroa et al. 2012). Interestingly, peptide fraction E1 obtained from LBF (Lactobacillus fermentum) showed amino acid sequence QSAPGNEAIPP (Fig. 3) with Pro in the C-terminal position which reflect ACE-inhibitory activity. This finding was supported by the investigation of Pripp et al. (2004) who also specified the relationship between hydrophobicity and positively charged amino acids in the C-terminal position for ACE-inhibitory activity.
Highlighted Portion in the Chromatogram shows Spectra of QSAPGNEAIPP Peptide from E1 Fraction (10 kDa Permeate) of Lactic Culture LBF
It has been reported that α, β-, and k-CN are precursors of bioactive peptides (Mills et al., 2011). However, in the current work, casein protein as well as whey protein such as α-LA, were found to be important sources of peptides with ACE-inhibitory activity. Camel milk fermented with LBF showed QSAPGNEAIPP peptide sequence derived from Kappa-casein (119–122) which have encrypted the hypotensive tripeptide (IPP) reported by Nakamura et al. (1995). MS/MS spectrum of fraction E1 of LBF (3 kDa permeate) was presented in the Fig. 4.
Similarly, peptide sequences of 3 and 10 kDa permeates were found matched with various reported sequences and sources (Tables 11, 12, 13) like milk protein, soy protein, cereals, fungi, algae, fermented milks, cheese, wine, fish and legumes using AHTPDB.
Conclusion
This work evidenced the ACE-inhibitory effect of camel milk fermented with Lactobacillus bulgaricus NCDC (09) and Lactobacillus fermentum TDS030603 (LBF). These Lactobacillus cultures had shown maximum ACE-inhibitory activity, pepX activity and proteolytic activity during the fermentation of camel milk. Fractionation of the fermented camel milk under optimized growth conditions revealed that peptides involved in the ACE-inhibitory activity are of lower molecular weight (3 and 10 kDa permeates). This observation was confirmed by RP-LC/MS analysis, which led to the identification of many unknown ACE-inhibitory peptides and known peptides such as VPP and IPP from fermented camel milk. Moreover, these findings showed the another source of ACE-inhibitory peptides from “Camel milk (Camelus dromedarius)” in India, which has an anti-diabetic property. Preparation of functional foods and validation of ACE-inhibition through clinical studies will add another health claim of fermented camel milk.
References
Abdelgadir SW, Ahmed TK, Dirar HA (1998) The traditional fermented milk products of the Sudan. Rev. Int J Food Microbiol 44:1–13
Badkook MM (2013) Fermented Camel Milk Reduces Inflammation in Rats Fed a High-Fat Diet. Int J Health Sci Res 3:7–17
Barker WC, Garavelli JS, Hou Z, Huang H, Ledley RS, Mc Garvey PB, Mewes HW, Orcutt BC, Pfeiffer F, Tsugita A, Vinayaka CR, XIO C, Yeh LSL, WU C (2001) Protein Information Resource: a common resource for expert annotation protein data. Nucleic Acids Res 29:29–32
Chandan RC (2004) Dairy: yogurt. In: Smith JS, Hui YH (eds) Food processing: Principles and applications, Ames. Blackwell Publishing Professional, Iowa, pp 297–300
Chen Y, Wang Z, Chen X, Liu Y, Zhang H, Sun T (2010) Identification of angiotensin I-converting enzyme inhibitory peptides from koumiss, a traditional fermented mare’s milk. J Dairy Sci 93:884–892
Cheung HS, Wang FI, Ondetti MA (1983) Binding of peptide substrates and inhibitors of angiotensin-converting enzyme. Importance of the COOH-terminal dipeptide sequence. J Biological Chem 225:401–407
De Leo F, Panarese S, Gallerani R, Ceci LR (2009) Angiotensin converting enzyme (ACE) inhibitory peptides: production and implementation of functional food. Curr Pharm Des 15:3622–3643
Degraeve A, Martial G (2003) Purification and partial characterisation of X-prolyl dipeptidyl aminopeptidase of Lactobacillus helveticus ITG LH1 P. Int Dairy J 13:497–507
Donkor ON, Henriksson A, Vasiljevic T, Shah NP (2007) Proteolytic activity of dairy lactic acid bacteria and probiotics as determinant of growth and in vitro angiotensin-converting enzyme inhibitory activity in fermented milk. Lait 87:21–38
El-Agamy EI (2006) Camel milk. In: Park YW, Haenlein GF (eds) Handbook of milk of non-bovine mammals, Ames. Blackwell Publishing Professional, Iowa, pp 297–344
FitzGerald R, Murray BA (2006) Bioactive peptides and lactic fermentations. Int J Dairy Technol 59:118–125
Hartmann R, Meisel H (2007) Food-derived peptides with biological activity: from research to food applications. Curr Opin Biotechnol 18:163–169
Hati S, Sreeja V, Solanki J and Prajapati JB (2015) Significance of proteolytic microorganisms on ACE-inhibitory activity and release of bioactive peptides during fermentation of milk. Indian J Dairy Sci 68:584–591
Hati S, Sakure A, Mandal S (2016) Impact of proteolytic Lactobacillus helveticus MTCC5463 on Production of bioactive peptides derived from honey based fermented milk. Int J Pept Res Ther DOI:10.1007/s10989-016-9561-5
Hayes M, Ross RP, Fitzgerald GF, Stanton C (2007) Putting microbes to work: dairy fermentation, cell factories and bioactive peptides. Part I: overview. Biotechnol J 2:426–434
Hellberg S, Eriksson L, Jonsson J (1991) Minimum analogue peptide sets (MAPS) for quantitative structure-activity relationships. Int J Pept Protein Res 37(5):414–424
Jäkälä P, Vapaatalo H (2010) Antihypertensive peptides from milk proteins. Pharmaceuticals 3:251–272
Jakubczyk A and Baraniak B (2014) Angiotensin I converting enzyme inhibitory peptides obtained after in vitro hydrolysis of Pea (Pisum sativum var. Bajka) Globulins. BioMed Res Int 2014:1–8
Korhonen H, Pihlanto A (2006) Bioactive peptides: production and functionality. Int Dairy J 16:945–960
Kumar R, Chaudhary K, Sharma M, Nagpal G, Chauhan JS, Singh S, Gautam A, Raghava GPS (2015) AHTPDB: a comprehensive platform for analysis and presentation of antihypertensive peptides. Nucleic Acids Res 43:956–962
Kunji ERS, Mierau I, Hagting A, Poolman B, Konings WN (1996) The proteotytic systems of lactic acid bacteria. Antonie Van Leeuwenhoek 70:187–221
Liu Y, ZhangL, Guo M, Wu H, Xie J, Wei D (2014) Virtual screening for angiotensin I-converting enzyme inhibitory peptides from Phascolosoma esculenta. Bioresour Bioprocess 1:1–9
López-Expósito R, Recio I (2006) Antibacterial activity of peptides and folding variants from milk proteins. Int Dairy J 16:1294–1305
Mehari Y, Mekuriaw Z and Gebru Z (2007) Potentials of camel production in Babilie and Kebribeyah woredas of the Jijiga Zone, Somali Region, Ethiopia. LRRD 19(4):1–10
Moslehishad M, Ehsani MR, Salami M, Mirdamadi S, Ezzatpanah H, Naslaji AN, Moosavi-Movahedi AA (2013) The comparative assessment of ACE-inhibitory and antioxidant activities of peptide fractions obtained from fermented camel and bovine milk by Lactobacillus rhamnosus PTCC 1637. Int Dairy J 29:82–87
Nakamura YM, Yamamoto N, Sakai K, Okubo A, Yamazaki S, Takano T (1995) Purification and characterization of angiotensin I-converting enzyme inhibitors from sour milk. J Dairy Sci 78(4):777–783
Padghan PV, Mann B, Sharma R, Bajaj R, Saini P (2016) Production of angiotensin-I-converting-enzyme-inhibitory peptides in fermented milks (Lassi) fermented by Lactobacillus acidophillus with consideration of incubation period and simmering treatment. Int J Pept Res Ther pp: 1–11. DOI:10.1007/s10989-016-9540-x
Pan D, Luo Y, Tanokura M (2005) Antihypertensive peptides from skimmed milk hydrolysate digested by cell-free extract of Lactobacillus helveticus JCM1004. Food Chem 91:123–129
Papadimitriou CG, Vafopoulo-Mastrojiannaki A, Viera Silva S, Gomes AM, Malcata FX, Alichanidis E (2007) Identification of peptides in traditional and probiotic sheep milk yoghurt with angiotensin I-converting enzyme (ACE)-inhibitory activity. Food Chem 15:647–656
Park YW (2009) Overview of bioactive components in milk and dairy products. In: Park YW (ed) Bioactive components in milk and dairy products, Ames. Wiley-Blackwell, Iowa, pp 3–5
Pastar I, Tonic I, Golic N, Kojic M, van Kranenburg R, Kleerebezem M (2003) Identification and genetic characterisation of a novel proteinase, PrtR, from the human isolate Lactobacillus rhamnosus BGT10. Appl Environ Microbiol 69:5802–5811
Pripp HA, Isaksson T, Stepaniak L, Sørhaug T (2004) Quantitative structure-activity relationship modelling of ACE-inhibitory peptides derived from milk proteins. Eur Food Res Technol 219:579–583
Qian B, Xing M, Cui L, Deng Y, Xu Y, Huang M (2011) Antioxidant, antihypertensive, and immunomodulatory activities of peptide fractions from fermented skim milk with Lactobacillus delbrueckii ssp. bulgaricus LB340. J Dairy R 78:72–79
Rahman IE, Dirar HA, Osman MA (2009) Microbiological and biochemical changes and sensory evaluation of camel milk fermented by selected bacterial starter cultures. Afr J Food Sci 3:398–405
Rodríguez-Figueroa JC, González-Córdova AF, Torres-Llanez MJ, Garcia HS, Vallejo-Cordoba B (2012) Novel angiotensin I-converting enzyme inhibitory peptides produced in fermented milk by specific wild Lactococcus lactis strains. J Dairy Sci 95:5536–5543
Savijoki K, Ingmer H, Varmanen P (2006) Proteolytic systems of lactic acid bacteria. Appl Microbiol Biotechnol 71:394–406
Seifu E (2007) Handling, preservation and utilization of camel milk and camel milk products in Shinile and Jijiga Zones, eastern Ethiopia. LRRD 19(6):86
Tagliazucchi D, Martini S, Bellesia S, Conte A (2015) Identification of ACE-inhibitory peptides from Phaseolus vulgaris after in vitro gastrointestinal digestion. Int J Food Sci Nutr 66(7):774–782
Tagliazucchi D, Shamsia S, Conte A (2016) Release of angiotensin converting enzyme-inhibitory peptides during in vitro gastrointestinal digestion of camel milk. Int Dairy J 56:119–128
Vasiljevic T, Jelen P (2002) Lactose hydrolysis in milk as affected by neutralizers used for the preparation of crude β-galactosidase extracts from Lactobacillus bulgaricus 11842. Innov Food Sci Emerg Technol 3:175–184
Vegarud GE, Langsrud T, Svenning C (2000) Mineral-binding milk proteins and peptides; occurrence biochemical and technological characteristics. Br J Nutr 84:91–98
Vermeirssen V, Van CJ, Verstraete W (2004) Bioavailability of angiotensin I converting enzyme inhibitory peptides. Brazilian. J Nutr 92:357–366
Wu J, Aluko RE, Nakai S (2006) Structural requirements of angiotensin I-converting enzyme inhibitory peptides: quantitative structure-activity relationship modelling of peptides containing 4–10 amino acid residues. QSAR Comb Sci 25:873–880
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Solanki, D., Hati, S. & Sakure, A. In Silico and In vitro Analysis of Novel Angiotensin I-Converting Enzyme (ACE) inhibitory Bioactive Peptides Derived from Fermented Camel Milk (Camelus dromedarius). Int J Pept Res Ther 23, 441–459 (2017). https://doi.org/10.1007/s10989-017-9577-5
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10989-017-9577-5