Abstract
The bipartite genome of an Indian isolate of Bombyx mori bidensovirus (BmBDV), one of the causative agents of the fatal silkworm disease ‘Flacherie’, was cloned and completely sequenced. Nucleotide sequence analysis of this Indian isolate of BmBDV revealed two viral DNA segments, VD1 and VD2 as well as a DNA polymerase motif which supports its taxonomical status as the type species of a new family of Bidnaviridae. The Indian isolate of BmBDV was found to have a total of six putative ORFs four of which were located on the VD1 with the other two being on the VD2 DNA segment. The VD1 DNA segment was found to code for three non-structural proteins including a viral DNA polymerase as well as one structural protein, while the VD2 DNA segment was found to code for one structural and one non-structural protein, similar to that of the Japanese and Zhenjiang isolates of BmBDV. A BmBDV ORF expression study was done through real time qPCR wherein the VD2 ORF 1 and 2 showed the maximum transcript levels. This is the first report of the genome characterization of an Indian isolate of BmBDV, infecting silkworm B. mori.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Bombyx mori bidensovirus (BmBDV) previously known as B. mori densovirus type 2 (BmDNV-2) [1] was previously taxonomically grouped under the subfamily Densovirinae of the family Parvoviridae [2]. BmBDVs are one of the causative agents of the silkworm disease, ‘flacherie’ which causes immense economic losses to the sericulture industry [3]. BmBDVs are small DNA viruses and can exclusively infect the columnar cells of midgut epithelium [4]. Structurally, BmBDVs have resemblance with B. mori densovirus type 1 (BmDNV-1) and other parvoviruses wherein BmBDVs exhibit a similar non-enveloped, spherical, icosahedral structure of approximately 22 nm diameter [5]. However, unlike the monopartite genomes of BmDNV-1 and other parvoviruses, BmBDVs have a bipartite genome with two non-homologous, single stranded, linear DNA molecules (VD1 and VD2) encapsulated in separate virions. The VD1 and VD2 DNA segments of BmBDVs have inverted terminal repeats (ITRs) and share a common terminal sequence (CTS) [1]. However, unlike BmDV and other parvoviruses, BmBDVs do not possess any common terminal palindromic sequences that can form terminal hairpins and the ITRs form a ‘panhandle structure’ [6]. In addition, BmBDV is the only virus that possesses a single stranded DNA that encodes for the DNA polymerase enzyme [7, 8]. Considering all these individual specificities, the International Committee on the Taxonomy of Viruses (ICTV), in 2011, established a new family of viruses, Bidnaviridae and assigned BmBDV as the type species in the new genus, Bidensovirus [1].
Three isolates of BmBDVs have been reported, thus far: the Japanese (Yamanashi isolate), the Chinese (BmBDV-3) and the Zhenjiang (BmBDV-Z) isolates [5]. The Chinese isolate (accession nos. VD1: DQ017268 and VD2: DQ017269), the Japanese isolate (accession nos. VD1: AB033596 and VD2: S78547) as well as the Zhenjiang isolate (accession nos. VD1: EU623082 and VD2: EU623083), all have the same host species; however, their virulence and the symptoms of infection are different [6, 9]. In India, BmBDV is also a major factor for crop loss for sericulture farmers. Preliminary studies and the partial cloning of the Indian isolate of BmBDV revealed a close homology between the Indian isolate of BmBDV and the Japanese isolate [10]. Neighborhood joining analysis revealed two major clusters, wherein the Indian and the Japanese isolates of BmBDV formed separate groups under one cluster, while, the Chinese isolate formed a second cluster. However, the full length genome sequence of the Indian isolate of BmBDV and its ORFs were not known at that stage. Hence, it was essential to clone and sequence the complete genome of the Indian isolate of BmBDV and compare its sequence with the other reported isolates. This need initiated this study towards the characterization of the complete genome of the Indian isolate of BmBDV. In this study, the complete genome of the Indian isolate of BmBDV has been cloned, sequenced, compared and analyzed with that of the Chinese, Japanese and Zhenjiang isolates. We have also investigated the expression pattern of the genes through Real time q-PCR.
Materials and methods
Collection of viral sample
BmBDV was propagated in the fifth instar larvae of BmBDV-susceptible B. mori silkworm race CSR 2, under a controlled environment in the laboratory. The midgut was dissected out from the infected larvae and 100mg of midgut sample was homogenized in 150ml of 0.05M Tris- HCl buffer having a pH of 7.2 along with 0.01M EDTA. The homogenate was filtered through three layers of gauze cloth and the filtrate was aliquoted into 50 ml falcon tubes and centrifuged at 3000rpm at 4°C for 10 minutes to remove the debris. The supernatant was mixed with half a volume of chloroform, centrifuged at 6000rpm at 4°C for 10 minutes and the aqueous phase was collected for purification of the viral particles. Purification was done using a 40% (w/w) Sucrose density-gradient ultracentrifugation, performed at 40,000 rpm, at 4°C for 3.5 hours using a Beckman 70Ti rotor. The pellet containing the viral particles were resuspended and carefully overlaid on a sucrose gradient and kept for centrifugation at 25,000 rpm at 4°C, for 3.5 h using a swing rotor, Beckman SW28. The fractions containing the viral particles were identified through PCR, using BmBDV specific forward/reverse primers. The pooled fractions containing the BmBDV bands were digested using proteinase K in Tris Buffer (100mM NaCl, 15mM MgCl2, 0.5% SDS, 0.5mg/ml, Proteinase K) and were incubated at 56°C for 4 hours. The digested viral sample was then extracted successively with an equal volume of phenol, phenol/ chloroform (1:1) and finally with chloroform solution. The resultant DNA was precipitated out using twice the volume of ethanol and one tenth the volume of 3M sodium acetate. The DNA was dried and reconstituted in 100 μl of TE solution after a 70% ethanol wash.
Cloning of the viral DNA
Earlier reports had indicated a close homology between the Indian isolate of BmBDV and the Japanese isolate [10]. Based on these reports several primer sets were designed as per the sequence of the Japanese isolate of BmBDV, using the Primer3 software. The purified viral genomic DNA was first checked for the presence of BmBDV with the primer sets VD1 and VD2, used in previous studies [10]. Once a sample was confirmed to be BmBDV-positive, the designed primer sets were used for amplifying the target regions. The DNA polymerase gene located on the VD1 fragment was targeted first for cloning and sequencing. This was followed by cloning and sequencing of the other regions. PCR was carried out in an Eppendorf AG 22331 Hamburg Thermocycler and the products were resolved on 1.2% agarose gels in Tris boric acid/EDTA buffer with a constant voltage of 80 in parallel using a standard DNA marker (Mass Ruler). The PCR amplified DNA samples were purified using a Promega wizard SV Gel and PCR clean up kit followed by cloning in a TA cloning vector (pGEM-TEasy Vector– Promega) as per the protocols indicated by the respective manufacturers. The recombinant plasmids were transformed into the E.coli JM109 strain. The cloned column purified plasmid DNA samples were PCR amplified with gene-specific, as well as M13, primers to confirm the insertion of DNA in the vector.
Sequence analysis
Multiple clones containing the VD1 and VD2 fragments were sequenced using the M13 primer at Eurofins Genomics India Pvt. Ltd, Bangalore in order to verify the sequences. Different sets of overlapping primers were designed to clone the complete VD1 and VD2 fragments. The obtained sequences were used for multiple sequence alignment using ClustalW, while, the phylogenetic relationship of the Indian isolate of BmBDV with that of the Chinese, Japanese and BmBDV-Z isolates was analyzed using the MEGA 5.1 tool [11]. ORF characterization and amino acid deduction were carried out using NCBI tools. Transcriptional control signals were assessed by Neural Network Promoter Prediction (NNPP) software.
Quantification of ORF transcripts by qPCR
The putative open reading frames (ORFs) identified within the Indian isolate of BmBDV through sequence analysis were confirmed using q-PCR. A total of six ORFs were identified for the Indian isolate of BmBDV and accordingly, six ORF specific q-PCR based primers were designed with an average product size of ~200bp. The template used for the q-PCR analysis was cDNA synthesized from the Densovirus-infected midgut RNA samples. The RNA was extracted from the dissected midguts using Trizol reagent (Invitrogen) and quantified by measuring the UV absorbance. The RNA samples were then purified by DNAse treatment. This was followed by first strand cDNA synthesis as per a standard protocol (using M-MLV Superscript III reverse transcriptase [Invitrogen]) and subsequent q-PCR analysis. One μl of first strand cDNA was used in a 10 μl reaction mixture using primers specifically designed to amplify the DNA of each of the six ORFs. The reactions were carried out on a STRATAGENEMx 3005 P Realtime PCR system with each sample tested in triplicate; the mean value ± SD was used for analysis of relative transcript levels. A non-template control (NTC) sample was also run to detect contamination, if any.
Results
Analysis of the viral genome
The complete nucleotide sequence of the Indian isolate of BmBDV has been determined and submitted to NCBI. The sequence data was assembled into two contiguous sequences named VD1 (6542 nts) and VD2 (6023 nts). The DNA polymerase gene located on the VD1 fragment was cloned and sequenced first followed by cloning and sequencing of the other regions on the VD1 fragment. The Genbank accession numbers assigned for the VD1 and VD2 DNA segments are KX760110 and KX779526, respectively, while the Genbank accession number assigned for the DNA polymerase gene of the Indian isolate of BmBDV is KP886818. The base composition of VD1 was found to be 71.12% A+T, while, that of VD2 was 67.92% A+T. Both VD1 and VD2 DNA fragments were found to possess high A+T content. A complete linear genomic text map of the VD1 and VD2 DNA segments of the Indian isolate of BmBDV is provided in online resource 1 and 2, respectively. As per previous reports, the terminal palindromic sequences commonly present in other densoviruses were not found in the Indian isolate of BmBDV, thereby confirming a typical feature of bidnaviruses. VD1 was found to have inverted terminal repeats (ITRs) of 224 nts, while, VD2 had ITRs of 499 nts. The ‘panhandle structure’ formation by the single stranded viral DNA molecule can be inferred through structural analysis of both VD1 and VD2 ITRs. The 5’ terminal end sequence from 1-224 nts of VD1 was found to be complimentary with that of nts 6318-6542 in the 3’ end terminal sequence. The ITRs GC content in the Indian isolate of BmBDV (VD1:42.41%; VD2:43.34%) was found to be much higher than that of the entire viral genome (VD1: 32.06%, VD2: 28.86%).
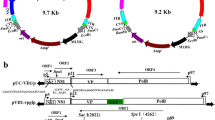
Putative ORFs present in the genome of the Indian isolate of BmBDV were identified using the NCBI ORF finder software. Methionine-initiated codon (ATG), stop codons (TAA, TAG and TGA) along with ORFs with a minimum of 100aa in length and showing minimal overlap were some of the criteria used for ORF searching within the Indian isolate of BmBDV. Six putative ORFs were identified in the genome of the Indian isolate of BmBDV. The VD1 DNA segment of the Indian isolate of BmBDV was found to have four ORFs while the VD2 DNA segment contained two ORFs, similar to the Yamanashi and BmBDV-Z isolates. The orientation, size and location of the predicted ORFs in the VD1 and VD2 DNA segments of the Indian isolate of BmBDV and the BmBDV-3 (Chinese isolate) is shown in Fig. 1a and b, respectively. The VD1 segment of DNA was found to contain four ORFs (VD1 ORF1, VD1 ORF2, VD1 ORF3 and VD1 ORF4), while, VD2 had two ORFs (VD2 ORF1, VD2 ORF2). The orientation and the number of putative ORFs identified in the Indian isolate of BmBDV resembled that of the Japanese and the Zhenjiang isolates, which have also been reported to have six ORFs in total.
To gain a better understanding of the genomic structure of the Indian isolate of BmBDV, the distribution of initiation and polyadenylation signals was also analyzed using the NNPP algorithm. The genomic organization of the VD1 and VD2 DNA segments along with the putative promoter signals are shown in Figures 2a and 2b, respectively. The first putative TATA box (TATATAA), on VD1, was found to be located between nts 259 and 265. This putative promoter might control transcription of VD1 ORF1 and VD1 ORF2. Another TATA box was found to be located between nt 1401 and 1407, which possibly controls the transcription of VD1 ORF3. Further, a third TATA box was found located on the minus strand between nt 6300 and 6306. VD1 ORF4 transcription might be controlled by this promoter. Polyadenylation signals (AATAAA) were also found between nt 1406–1411 and nt 2911- 2916. This signal was also found located on the complementary strand at nt 2943–2938. In the case of the VD2 DNA sequence, a putative TATA box was found between nt 644 and 650 which could possibly control the transcription of VD2 ORF1. Also, a polyadenylation signal was noted between nts 650-655. However, no promoter signal was detected for the VD2 ORF2 gene sequence.
a: The genome organization of the VD1 DNA segment of the Indian isolate of BmBDV. The horizontal line represents the VD1 DNA segment. The colored bars above the line represent the ORFs encoded by the positive strand and those below the line represent the ORFs encoded by the complement sequence. The TATA boxes indicate the functional promoters. Figure 2b: The genome organization of the VD2 DNA segment of the Indian isolate of BmBDV. The horizontal line represents the VD2 DNA segment. The colored bars above the line represent the ORF encoded by the positive strand and the one below the line represents the ORF encoded by the complementary sequence. The TATA boxes indicate the functional promoters
Transcript analysis through q-PCR
The identification of 6 putative ORFs within the Indian isolate of BmBDV was confirmed through q-PCR wherein the expression level of each of the ORFs was also analyzed and compared. All the putative ORFs showed significant transcription; however, VD2 ORF1 and ORF 2 showed the maximum levels of transcription. Among the six ORFs, VD2 ORF 1 was found to exhibit the maximum level of transcripts while VD1 ORF4 had the least mRNA expression levels (Fig. 3).
Comparison of the Indian isolate of BmBDV with other isolates
Comparison of the genome length, ORF length and ITRs of the Indian isolate of BmBDV with that of other BmBDV isolates is shown in Table 1. The VD1 DNA segment of the Indian and the Japanese isolates are of the same length i.e. 6542bp, while the Chinese and the Zhenjiang isolates are 1bp larger than the Indian isolate (6543bp). However, the VD2 DNA segment of all four isolates varies in length, with the Japanese isolate having the maximum length of 6031bp, while, the Chinese isolate has the shortest (6022bp). The Indian BmBDV and BmBDV-Z differ by only one nucleotide: the Indian isolate is 6023bp in length, while, BmBDV-Z is 6024bp. Further, ORF characterization revealed that the Indian, Japanese and BmBDV-Z isolates have a total of 6 ORFs; four on the VD1 segment and two on the VD2 DNA segment. However, the Chinese isolate has been reported to have seven ORFs; four on the VD1 segment and three on the VD2 DNA segment. A single nucleotide “A” deletion at position nt 1583 in the VD2 ORF1 of the Chinese isolate resulted in a frameshift mutation that brought about a premature stop codon. Consequently, the Chinese isolate has three ORFs on VD2 viz., VD2 ORF1-a and VD2 ORF1-b [6]. However, this mutation has not been observed in the other isolates. The Indian isolate was individually compared with each of the previously reported isolates. Tables 2, 3 and 4 show the comparison of the Indian isolate of BmBDV with that of the Japanese, Chinese and Zhenjiang isolates, respectively. The comparison of the Indian and Japanese isolate revealed a total of 221 bp differences, of which 159 were bp substitutions found to occur in the coding regions of the six ORFs. Consequently, 58 amino acid changes were observed in all 6 ORFs of the Indian isolate of BmBDV. Among the 159 substitutions in the coding regions, 101 bp substitutions were silent. Similarly, a comparison between BmBDV Z and the Indian isolate revealed a total of 310 bp substitutions, of which 159 occurred in the 6 ORF sequences just like the Japanese isolate. However, the overall number of substitutions was higher in this case because the substitutions mostly occurred in the non-coding region of the sequence. The comparison between the Chinese and the Indian isolates of BmBDV revealed a total of 271 bp substitutions, of which 153 substitutions occurred in the coding regions. The 153 substitutions brought about only 46 amino acid changes, suggesting that 118 nt changes were silent. Comparison of the ITR of the Indian isolate with that of the other isolates revealed the VD2 DNA segment of the Indian isolate to possess a 26 bp nucleotide addition at the 5’ terminal end.
Phylogenetic analysis of the Indian isolate of BmBDV
Phylogenetic analysis of all the reported isolates of BmBDV was carried out using the Mega6 software to study homology between the VD1 and VD2 DNA segments. The scale range for analysis of the VD1 DNA segment was from 0.001 to 0.005 (Fig. 4), while, in the case of the VD2 DNA segment the scale range was from 0.002 to 0.012 (Fig. 5). The scale range clearly indicates that both VD1 and VD2 DNA segments share a close homology without any significant differences. However, despite the close homology between the VD1 and VD2 DNA segments, VD1 clearly has a higher degree of conservation than the VD2 DNA segment, as indicated by the scale range. Further, the phylogenetic tree constructed revealed two separate clusters wherein the first cluster was formed by the Indian and the Yamanashi isolates, while, the Chinese isolate and BmBDV-Z isolates formed the second cluster. Phylogenetic analysis of each of the ORFs was also carried out separately (online resource 3) wherein each of the ORFs revealed a close homology, which indicated that the ORF sequences are highly conserved in all the reported isolates of BmBDV. However, among all the ORFs, the VD1 ORF 2 showed the maximum degree of conservation, while, VD2 ORF 1 and 2 exhibited the maximum variability. Among the two DNA segments the VD1 ORFs seem to be more highly conserved.
Discussion
The present study was carried out with the purpose of characterizing an Indian isolate of BmBDV, a causative agent of the deadly silkworm disease ‘Flacherie’ [3]. In this study, the whole genome and the ORFs of an Indian isolate of BmBDV have been sequenced and characterized. The Indian isolate, like the other reported isolates of BmBDV, has been found to have Bidnaviridae-like characteristics; it possesses a bipartite genome (VD1 and VD2) [4]. Six putative ORFs were identified within the genome of the Indian isolate of BmBDV which resembled that of the Zhenjiang and the Japanese isolates [12]. The analysis of the DNA genome of the Indian isolate revealed that it bore a close resemblance to that of the Japanese isolate. The VD1 nucleotide sequence shared 98% identity with the Japanese and Zhenjiang isolates and 97% with the VD1 segment from the Chinese isolate. In the case of the VD2 nucleotide sequence, the Indian isolate shared 98% identity with the other BmBDV isolates. Phylogenetic analysis of the VD1 and VD2 DNA segments of the Indian isolate of BmBDV with the other reported isolates, revealed a genetic divergence ranging from 0.001 to 0.005 for the VD1 DNA segment, while in the case of the VD2 DNA segment the range was from 0.002 to 0.012. Thus, our data confirms that there is a high degree of conservation among all reported isolates of BmBDV. The comparison of all the BmBDV isolates indicates that they share a similar genetic makeup yet there are differences in the number and orientation of the ORFs. This difference in the genome structure may be attributed to different environmental conditions which might have resulted in the evolution of different isolates of BmBDV [12].
The BmBDV genome encodes structural proteins (VP) as well as non-structural proteins (NS) which are encoded by the various ORFs. Previous studies have been carried out wherein the NS and VP expression pattern in the reported isolates has been analysed [13,14,15,16,17]. The prediction of ORF number in the BmBDV genome varies between the reported isolates. The Chinese isolate, a variant of the Zhenjiang isolate, has been reported to have seven ORFs wherein the VD2 ORF 1 is split into two, VD2 ORF1a and VD2 ORF 1b, due to a point mutation which leads to the production of a premature stop codon [12]. On the contrary, the Zhenjiang isolate has been reported to have six ORFs like the Indian isolate. Similarly, the Japanese isolate has also been reported to have six ORFs (NCBI nucleotide database). For the Japanese and Zhenjiang isolates, both of which have been reported to have six ORFs in total wherein, the VD1 ORF 4 has been assigned as a non-structural protein in the case of the Japanese (AB033596) isolate while for the Zhenjiang isolate (EU623082) VD1 ORF 4 is referred to as a structural protein. The differences in these reports in the database might have arose due to the early nature of such studies. However, recent and advanced VD1 ORF 4 sequence analysis has revealed that the sequences share a common homology with the family B DNA polymerase enzymes (PolB) and should hence be considered as a non-structural protein. In our report we have predicted six ORFs in total, four on the VD1 DNA segment and two on the VD2 DNA segment. Further, we have also proposed VD1 ORF4 (KX760110), encoding the DNA polymerase enzyme, as a non-structural protein as it is theoretically involved in the replication of BmBDV. The Indian isolate of BmBDV is hence predicted to have three NS proteins including the DNA polymerase enzyme, encoded by the VD1 ORFs 1, 2 and 4 respectively, while one NS protein is from VD2 ORF2. Also, our study indicates the presence of two VP proteins encoded by VD1 ORF3 and VD2 ORF1, respectively. Further, for BmBDV-Z, VD2 ORF 1 has been reported to code for a structural protein P133 [18]. There have been previous reports on the expression pattern as well as the localization of various NS and VP proteins. The NS2 protein of the Chinese isolate of BmBDV has been reported to be localized in the nuclear membrane [19]. Equivalent characterization for the NS and VP proteins from the Indian isolate of BmBDV is yet to be performed. The bidnaviruses are not only unique but have also revealed an interesting pattern of evolution wherein they have acquired genes from other viruses; in fact they have been referred to as kleptomaniacs in the viral kingdom [20]. Horizontal gene transfer has been the key source for the evolution of the bidnaviruses wherein genes have been acquired and lost to equip better survival [21]. Phylogenetic and sequence analysis performed by Krupovic and Koonin [21] have revealed the inheritance of bidnavirus genes from various other virus species. Their studies have suggested that the bidnaviruses might have acquired genes from parvoviruses, reoviruses and polintiviruses thereby having a blended ancestry [20, 21]. The characterization of the Indian isolate of BmBDV also revealed the presence of putative ORFs, similar to the Japanese and the Zhenjiang isolates.
Reports have indicated that BmBDV undergoes a leaky scanning mechanism of translation which has led to the investigation of whether multiple proteins might be encoded by a single ORF [19, 22]. Li et al [22] analysed the characteristics and functions of the NS1 protein of BmBDV-Z. ATPase and ATPase coupled helicase activity was demonstrated through enzymatic assays. The study also reported the co-precipitation of a DNA polymerase motif encoded by the VD1 ORF4, identified by mass spectrometry, indicating a leaky scanning mechanism for translation [22]. Wang et al, [6] have also highlighted the role of leaky scanning in generating two transcriptional products from a single ORF. The distribution of strong promoter and polyadenylation signals in the genome of the Indian isolate and its similarities with the other reported isolates of BmBDV indicates that the Indian isolate might also adapt the leaky scanning mechanism for translation. However, further experiments are required to prove such a hypothesis.
BmBDV, which is one of the causative agents of the fatal flacherie disease in silkworms, replicates only in the midgut columnar cells, in contrast to B. mori densoviruses (BmDV) that replicates in both the midgut columnar as well as in the goblet cells [2]. B. mandarina and B. mori are considered to have a common ancestor and both are susceptible to BmBDV [23]. Many Japanese strains acquired BmBDV resistance through improved breeding. These resistant strains became susceptible to the parvovirus-like BmDV [24]. BmBDV is therefore likely to be the native disease of the Bombyx, whereas the origin of BmDV BmDNV-1is likely to be from other insects [25]. The pyralid Glyphodes pyloalis, a pest of mulberry plantations of sericultutal farms is also susceptible to BmDV and may be the real host of BmDV [26, 27].
As mentioned earlier, BmBDVs are one of the causative agents of the viral silkworm disease Flacherie which goes unnoticed until the late larval stages, since there is a dearth of early detection techniques for this disease. As a result it is essential to detect pathogens at a very early stage, in order to prevent crop loss. With this backdrop, the characterization of the whole genome sequence of the Indian isolate of BmBDV will pave the way towards the development of diagnostics and control measures for this disease. Early diagnosis and control measures against the disease are being developed in our laboratory, using simple and precise techniques such as loop-mediated isothermal amplification (LAMP). Accordingly, the genome sequence of the Indian isolate is being put to use in developing diagnostic measures against this disease since detection techniques require accurate primer design, pertaining to the correct amplification of the appropriate target region of the virus.
References
Bando H, Choi H, Ito Y, Nakagaki M, Kawase S (1992) Structural analysis on the single-stranded genomic DNAs of the virus newly isolated from silkworm: the DNA molecules share a common terminal sequence. Arch Virol 124(1):187–193. doi:10.1007/BF01314637
Adams MJ, Carstens EB (2012) Ratification vote on taxonomic proposals to the international committee on taxonomy of viruses. Arch Virol 157(7):1411–1422. doi:10.1007/s00705-012-1299-6
Tijssen P, Bergoin M (1995) Densonucleosis viruses constitute an increasingly diversified subfamily among the parvoriruses. Virology 6(5):347–355. doi:10.1006/smvy.1995.0041
Hu Z, Li GH, Li GT, Yao Q, Chen KP (2013) Bombyx mori bidensovirus: the type species of the new genus Bidensovirus in the new family Bidnaviridae. Chin Sci Bull 58(36):4528–4532. doi:10.1007/s11434-013-5876-1
Zhang P, Miao D, Zhang Y, Wang M, Hu Z, Lü P, Yao Q (2016) Cloning and rescue of the genome of Bombyx mori bidensovirus, and characterization of a recombinant virus. Virol J 13:126. doi:10.1186/s12985-016-0576-5
Wang YJ, Yao Q, Chen KP et al (2006) Organization and transcription strategy of genome of Bombyx mori bidensovirus (China isolate) VD1.Chin. J Biotechnol 22(5):707–712. doi:10.1016/S1872-2075(06)60052-6
Zhang J, Li G, Chen H, Li X, Lv M, Chen K, Yao Q (2010) Molecular cloning and expression of key gene encoding hypothetical DNA polymerase from B. mori parvo-like virus. Genet Mol Biol 33(4):739–744. doi:10.1590/S1415-47572010005000083
Hayakawa T, Kojima K, Nonaka K, Nakagaki M, Sahara K, Asano SI, Iizuka T, Bando H (2000) Analysis of proteins encoded in the bipartite genome of a new type of parvo-like virus isolated from silkworm—structural protein with DNA polymerase motif. Virus Res 66(1):101–108. doi:10.1016/S0168-1702(99)00129-x
Watanabe H, Kurihara Y (1988) Comparative histopathology of two densonucleoses in the silkworm, Bombyx mori. J Invertebr Pathol 51(3):287–290. doi:10.1016/0022-2011(88)90038-9
Murthy GN, Ponnuvel KM, Awasthi AK, Rao CGP, Chandrasekhar Sagar BK (2014) The Indian isolate of Densovirus-2-Impact of infection and mechanism of resistance in Bombyx mori. J Invertebr Pathol 115:48–50. doi:10.1016/j.jip.2013.10.010
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi:10.1093/molbev/msr121
Wang YJ, Yao Q, Chen KP, Wang Y, Lu J, Han X (2007) Characterization of the genome structure of Bombyx mori densovirus (China isolate). Virus Genes 35(1):103–108. doi:10.1007/s11262-006-0034-3
Lü M, Yao Q, Wang Y et al (2011) Identification of structural proteins of Bombyx mori parvo like virus (China Zhenjiang isolate). Intervirology 54:37–43. doi:10.1159/000318888
Li G, Hu Z, Guo X et al (2013) Identification of Bombyx mori bidensovirus VD1-ORF4 reveals a novel protein associated with viral structural component. Curr Microbiol 66(6):527–534. doi:10.1007/s00284-013-0306-9
Zhu S, Li G, Hu Z et al (2012) Characterization of the promoter elements of Bombyx mori bidensovirus nonstructural gene 1. Curr Microbiol 65(5):643–648. doi:10.1007/s00284-012-0199-z
Sotoshiro H, Kobayashi M (1995) Identification of viral structural polypeptides in the Midgut and feces of the silkworm, Bombyx mori, infected with Bombyx densovirus type 2. J Invertebr Pathol 66:60–67. doi:10.1006/jipa.1995.1061
Yin HJ, Yao Q, Guo ZJ, Bao F, Yu W, Li J et al (2008) Expression of non-structural protein NS3 gene of Bombyx mori densovirus (China isolate). J Genet Genom 35(4):239–244. doi:10.1016/S1673-8527(08)60033-8
Kong J, Hu Z, He Y et al (2011) Expression analysis of Bombyx mori parvo-like virus VD2-ORF1 gene encoding a minor structural protein. Biologia 66(4):684–689. doi:10.2478/s11756-011-0074-6
Wang F, Hu Z, He Y et al (2011) The non-structural protein NS-2 of Bombyx mori parvo-like virus is localized to the nuclear membrane. Curr Microbiol 63(1):8–15. doi:10.1007/s00284-011-9933-1
Tijssen P, Pénzes JJ, Yu Q, Pham HT, Bergoin M (2016) Diversity of small, single-stranded DNA viruses of invertebrates and their chaotic evolutionary past. J Invertebr Pathol 140:83–96. doi:10.1016/j.jip.2016.09.005
Krupovic M, Koonin EV (2014) Evolution of eukaryotic single-stranded DNA viruses of the Bidnaviridae family from genes of four other groups of widely different viruses. Sci Rep 4:5347. doi:10.1038/srep05347
Li GH, Sun C, Zhang JH, He Y et al (2009) Characterization of Bombyx mori parvo-like virus non-structural protein NS1. Virus Genes 39(3):396–402. doi:10.1007/s11262-009-0402-x
Goldsmith MR, Shimada T, Abe H (2005) The genetics and genomics of the silkworm, Bombyx mori. Ann Rev Entomol 50:71–100. doi:10.1146/annurev.ento.50.071803.130456
Furuta Y (1994) Susceptibility of Indian races of the silkworm, Bombyx mori to the nuclear polyhedrosis virus and densonucleosis viruses. Acta Seric Entomol 8:1–10
Furuta Y (1995) Susceptibility of the races of the silkworm, Bombyx mori, preserved in NISES to the nuclear polyhedrosis virus and densonucleosis viruses. Bull Natl Inst Seric Entomol Sci 15:119–145
Watanabe H, Maeda S (1981) Genetically determined nonsusceptibility of silkworm Bombyx mori to infection with a densonucleosis virus (Densovirus). J Invertebr Pathol 38:370–373
Bergoin M, Tijssen P (2000) Molecular biology of Densovirinae. Contrib Microbiol 4:12–32
Acknowledgements
This research work has been supported through collaborative DST-JSPS project jointly funded by Department of Science and Technology (DST), Ministry of Science and Technology, Government of India as well as Japan Society for the Promotion of Science (JSPS), Government of Japan (Grant no: DST/INT/JSPS/P-187/2014, 16th June 2014 to 15th June 2016).
Author information
Authors and Affiliations
Corresponding author
Additional information
Handling Editor: T. K. Frey.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Gupta, T., Ito, K., Kadono-Okuda, K. et al. Characterization and genome comparison of an Indian isolate of bidensovirus infecting the silkworm Bombyx mori . Arch Virol 163, 125–134 (2018). https://doi.org/10.1007/s00705-017-3584-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-017-3584-x