Abstract
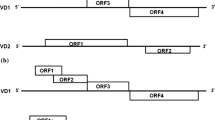
The Indian isolate of Bombyx mori bidensovirus (BmBDV) is a bipartite virus that comprises of a segmented, non-homologous, two linear single-strands of DNA molecules (VD1 and VD2). It is one of the causative agents of the fatal silkworm disease ‘Flacherie’ that causes severe crop loss for the sericulture farmers. Genome analyses of the Indian isolate of BmBDV revealed that it consists of 6 putative ORFs similar to the Japanese and Chinese isolates. VD1 consists of 4 ORFs while VD2 has 2 ORFs that code for 4 non- structural (NS) and 2 structural (VP) proteins, in total. In this study, we investigated, in detail, the impact of BmBDV pathogenesis on growth and development of the silkworm Bombyx mori, at different developmental stages. Mortality rate and weight uptake analyses were also performed on newly ecdysed 4th instar larvae. BmBDV infection was not found to be developmental stage specific and it occurred at all stages. Onset of mortality took place 8 days post infection (dpi) and 100% mortality occurred at 11 dpi. The infected larvae showed a significant difference in weight uptake wherein from 7 dpi the larvae stopped gaining weight and from 8th dpi started demonstrating the typical symptoms of flacherie. Further, the expression pattern of the 6 viral ORFs were also investigated in the newly ecdysed 4th instar BmBDV infected silkworms. Among all the six ORFs, VD2 ORF 1 and 2 revealed the highest transcript numbers, which was followed by VD1 ORF 4 that encodes for the viral DNA polymerase enzyme. This was the first ever attempt to understand the pathogenesis and the expression pattern of all the six ORF transcripts of the Indian isolate of BmBDV.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Bombyx mori bidensovirus, (BmBDV), now classified as the type species of the Bidnaviridae family of viruses, is one of the causative agents of the fatal B. mori silkworm disease ‘Flacherie’ [12]. The BmBDV infection is chronic and therefore, the symptoms appear at late stages of infection, thereby making it difficult for the farmers to detect the infection at an early larval stage. This leads to severe crop loss for the sericulture farmers. BmBDVs are identified by their unique bipartite genome with non-homologous, single stranded, linear DNA molecules (VD1 and VD2) along with a DNA polymerase motif, which differentiates them from the Parvoviridae family of viruses [8]. Three isolates of BmBDVs have been reported so far viz. the Japanese [19], the Chinese [25] and the Indian isolates [3]. The Indian isolate of BmBDV, which was recently completely sequenced, has also been reported to be highly prevalent in the Indian sericulture farms and is responsible for severe crop loss [4]. This is the first group of bipartite insect viruses discovered and is being investigated in detail for understanding its genome segregation. All the three isolates of BmBDVs have been reported to share a close homology. Studies done thus far, have reported that all the three isolates of BmBDVs share a high degree of conserved sequences. This could suggest that the minor differences in sequences might have arisen due to temperature variations, which led to the evolution of different isolates of BmBDV, adapted to different climatic conditions.
Early studies showed contradicting results regarding the number of structural protein polypeptides and also about ORF designations of BmBDV. However, the detailed investigation and complete sequencing of the Japanese and Chinese isolates confirmed that both the isolates possessed six ORFs, coding for both structural (VP) and non-structural (NS) proteins. VP as well NS proteins play key roles in a viral life cycle. The VPs are known to be associated with host cell surface receptor recognition, viral genomic encapsidation, host immune response detection, pathogenicity determination and evasion [1]. Non- structural proteins have been reported to be associated with viral replication [6]. Hence, analysing the expression pattern of viral ORF transcripts is essential for understanding the detailed pathogenesis of a virus. So far, there have been no detailed reports on the sequential investigation on the pattern of ORF transcript expression for the Indian BmBDV isolate. One of the targets of this study was to investigate the transcript expression of each of the ORFs during the course of BmBDV infection for the Indian isolate. Hence, in this study, we studied the expression pattern of all the 6 characterized ORFs during infection at the 4th instar of B. mori development.
In our earlier study, we characterized the Indian isolate of BmBDV, wherein it was found to have 6 putative ORFs similar to the Japanese and Chinese isolates [3]. Four ORFs were found to be located on the VD1 and two were found on the VD2 DNA segment. These 6 ORFs were found to code for 4 NS and 2 VP proteins, in total. However, there was a need to understand the infection pattern of this fatal pathogen. So far, there has been no such investigation regarding the expression pattern of all the 6 ORFs during the time course of silkworm development in the Indian BmBDV isolate. Hence, in this study, we studied the expression pattern of all the 6 characterized ORFs during the 4th instar of silkworm development. In addition, we also analyzed the effects of BmBDV infection on the growth and development patterns of the silkworm at various developmental stages.
Materials and methods
Silkworm and viral inoculum
The BmBDV susceptible B. mori race CSR2 was used for the infection study. The silkworms were reared at 25 °C at 75–80% humidity under controlled environment conditions. This breed is known to be the most susceptible to diseases among all the other Indian silkworm breeds and hence was used for the infection study of BmBDV. Silkworm larvae were fed with BmBDV inoculum, which was a 100 times dilution of the midgut homogenate as described by Ito et al. 2016 [10]. This virus concentration resulted in 100% infection in the CSR 2 breed.
Silkworm infection and sample collection
Newly ecdysed 1st, 2nd, 3rd and 4th instar day 0 CSR2 (n = 30, maintained in duplicates) larvae were fed with mulberry leaves smeared with BmBDV inoculum for 24 h. Each larva was fed with BmBDV only once in its life time. Post 24 h the silkworms were fed with fresh un-inoculated mulberry leaves. Simultaneously, another batch of uninfected control CSR2 (n = 30, maintained in duplicates) silkworms were maintained separately. These batches of larvae were maintained for observing the impact of BmBDV infection on silkworm growth. Fourth instar larvae from these batches were used for analyzing the weight uptake and mortality rate. In addition to this two other batches of freshly ecdysed 4th instar day 0 CSR2 larvae (n = 30, maintained in duplicates) were infected with BmBDV in a similar way as mentioned above. Samples from these two batches were collected at an interval of 24 h post infection, every day, till all the larvae died.
Isolation of total RNA and cDNA synthesis
Total RNA was isolated from the midgut of the infected larvae (n = 30, each day 2 RNA samples), every 24 h post infection till all the larvae died and Trizol (RNAiso plus, Takara) based extraction was carried out for processing the RNA. The RNA extracted was reverse transcribed as per manufacturer’s protocol (PrimeScript1st strand cDNA synthesis kit, Takara). The obtained cDNA was then diluted tenfold and used for conventional as well as quantitative Real time PCR (qPCR).
Quantification of ORF transcripts using qPCR
The Indian BmBDV isolate has been characterized with 6 ORFs in total, 4 being on the VD1 and 2 on the VD2 DNA segments [3]. Accordingly, specific primers for the 6 ORFs were designed with an average product size of 170 base pair (bp) for quantifying the ORF transcripts at different time intervals post infection. The 6 ORF specific primer sets used have been shown in Table 1. One μl of the diluted cDNA was used as template for a 10 μl qPCR reaction. Sample collected from each day of infection were tested in triplicates using the Agilent StratageneMx 3005P Real time PCR system. The transcript level of each of the ORFs was analyzed by calculating the mean SD value. The β-actin primer was used as an internal control and also a non-template control (NTC) reaction was run to detect contamination, if any. The qPCR conditions were as follows: initial denaturation step at 95° C for 8 min followed by 40 cycles of denaturation at 95° C for 30 s, annealing at 55° C for 1 min and final extension at 72° C for 30 s.
Results
Impact of BmBDV infection on growth and development of B. mori at different developmental stages
The effect of BmBDV infection was studied at different stages of silkworm development, which included the 1st, 2nd, 3rd and the 4th instars. The severity of infection was prominent at all stages of development. However, 1st, 2nd, 3rd and 4thinstar larvae did not complete their life cycle and mortality occurred between 11–20 dpi. Most of the 5th instar infected larvae (inoculated with BmBDV at the 4th instar), however, entered the spinning stage since the BmBDV infection is chronic and takes time to propagate. The most visible effects of BmBDV infection was the retardation of growth and development Fig. 1. At these four developmental stages the growth and development were completely stunted, upon BmBDV infection. The difference in sizes between the control and infected batches were clearly visible from 8 days post infection (dpi). The initial seven days of infection was, devoid of any typical ‘Flacherie’ symptoms, wherein the larval feeding was normal and size of the infected batch of larvae was at par with the control batch. Thus, it could be concluded that BmBDV infection is not developmental stage specific and occurs at all stages.
Mortality rate and weight increase upon BmBDV infection
The mortality rate and the weight uptake upon BmBDV infection was studied during the 4th instar of BmBDV infection. The infected larvae showed a significant difference in weight uptake wherein from 7 dpi the larvae stopped gaining weight and from the 8th day started demonstrating the typical symptoms of flacherie Fig. 2. The symptoms included low intake of feed, flaccidity and diarrhea, the typical symptoms as reported for the Japanese and the Chinese BmBDVs. The results confirmed the chronic nature of Indian isolate of BmBDV infection. Simultaneously, the cumulative percent mortality during the course of infection was also calculated Fig. 3. Mortality started 8 days post infection and 100% mortality occurred at 11 dpi.
Effects of Indian BmBDV isolate infection on the weight uptake of newly ecdised 4th instar B. mori larvae (n = 30). Larvae were orally infected and the weight of living larvae was recorded daily till all the larvae died. The black and the grey lines indicate the weight in infected and control samples, respectively
Cumulative percent mortality calculation of newly ecdised 4th instar B. mori larvae upon infection by Indian BmBDV isolate. Larvae (n = 30) were orally infected and the cumulative percent mortality was recorded daily till all the larvae died. The black and the grey bars indicate the mortality in infected and control samples, respectively
Expression pattern of ORFs during the course of development
The newly ecdysed 4th instar larvae were exposed to BmBDV infection. Samples were collected every 24 h post infection till there was complete mortality. Complete mortality during this study occurred at 11th dpi. The expression of ORF transcripts were confirmed initially through the conventional PCR with cDNA as template, using the ORF specific primers and β-actin as the internal control Fig. 4. The quantification of each of ORF transcripts was targeted next, during the time course of BmBDV infection. The qPCR analysis revealed that the ORF transcripts lowered during the molting phase of the infected silkworm larvae [9, 18].
The expression levels of each of the 4 ORFs were measured. All but, VD1 ORF 1 followed a similar pattern of expression, wherein the expression levels of each ORF increased gradually till day 5 post infection, followed by a drop in the level of expression on the 6th day, which coincided with the molting phase. Post molting phase, the expression levels increased once again and showed a peak on the 9th day, after, which the levels dropped down once again from the 10th day and further decreased on the last day i.e. on the 11th day, which represented complete mortality.
The order of expression level was VD2 ORF 1 and 2, followed by VD1 ORF 4, 2, 1 and 3, respectively. The expression levels of all the ORFs were detected in significant amounts post 48 h of infection Fig. 5a–f.
Discussion
The BmBDV pathogenesis is tissue specific and occurs at all developmental stages. Further, the BmBDV infection pattern is strongly affected by the molting process. This is in contrast to BmDV, which is not at all affected by the molting period of silkworms [9]. In our study, the 4th instar larvae were specifically selected for mortality, weight uptake and ORF expression studies because, the larvae at this stage were perfect in size for dissection of midgut and hence sample collection could be done with ease. We saw a significant difference in weight uptake for 4th instar larvae after 7th day of infection when infected by BmBDV. Typical ‘Flacherie’ symptoms were observed from the 8th day of infection, which included body flaccidity, brown coloration of larval body, diarrhea etc. Significant difference in growth and development was also observed eight days post infection, which included onset of mortality. This also adds to the difference between BmBDV and BmDV infection wherein in case of the latter there was no difference in weight uptake till the day the larvae died [9]. The ORF expression studies also revealed a similar pattern of expression as reported for other BmBDV isolates. The ORF expression levels clearly decreased during the molting period, which indicates that the amplified virus particles were discarded along with the old midgut cells during the molting phase. This pattern of infection was also reported by (Nakagaki et al., 1999) for the Japanese BmBDV isolate [18].
Each of the 6 BmBDV ORFs encode genes that play a vital role in viral replication. The VD1 ORF 1 has been associated with NS2. However, there have been no clear reports on the exact functions associated with NS2 in BmBDV. It has been found to have no homology with other NS2 from parvoviruses [6]. However, there have been reports on the expression of NS2 at 28 h pi in infected larvae, in very low amounts, but in high amounts at late stages of infection [24]. Also, immunofluorescence analysis has showed that NS2 ultimately gets concentrated at the nuclear membrane in Bombyx mori Nuclear (BmN) cells at late stages [24].
The result of the study also reveals NS2 transcript expression at very low amount at 24 h pi. NS2 gets concentrated at the nuclear membrane in BmN cells at later stages of infection similar to the adeno-virus death protein (ADP) [6]. ADP is an Asn-glycosylated integral membrane protein, which is expressed early but gets greatly amplified at late stages of infection [2]. Thus, it was hypothesized that NS2 also might be involved in cell lysis just like ADP [22] and be thereby associated with release of virus particles. VD1 ORF 2 encodes NS1, which has a helicase/ATPase motif homologous to the NS1 of parvoviruses [16]. NS1 in Parvoviruses is associated with functions like sequence specific DNA binding, ATP-dependent site-specific endonuclease, helicase as well as ATPase activities [11, 23]. Similar functions of NS1 associated with helicase/ATPase activities have been confirmed in BmBDVs through in-vitro experiments and also NS1 was found to interact with the viral DNA polymerase encoded by VD1 ORF 4 [16]. All these mechanisms are associated with viral DNA replication [26] and its activities indicate that NS1 is multifunctional protein, which might have a key role associated with virus replication. VD1 ORF 3 on the other hand has been reported to encode for a structural protein [17].
Lv et al. (2011) studied the structural polypeptides produced in BmBDV Z and reported 7 viral structural polypeptides named P1 to P7 [17]. Among them P5 and P6 structural polypeptides were reported to be the larger VPs and were encoded by VD1 ORF 3. MALDI-TOF/mass spectrometry analysis also confirmed that P5 and P6 were encoded by the VD1 ORF 3 [17]. However, there have been other reports wherein peptide mapping and amino acid sequencing indicated that VP1 to 4 were encoded by VD1-ORF 3 [5, 13, 20]. Lastly, the VD1 ORF 4 encodes for a protein- primed type B DNA polymerase (Pol B), which led to the establishment of Bidnaviridae family of the viruses. The evolution of Pol B in this group of viruses triggered the exclusion of bidensoviruses from the group of Parvoviruses [15].
The VD2 ORFs followed a pattern of expression similar to the VD1 ORFs. However, VD2 ORF transcript expression was the highest among all the 6 ORFs. VD2 ORF 1 has been reported to be encoding the structural protein VP6 [5]. However, another study has reported that VD2 ORF 1 is responsible for encoding a protein named p133, which has a weight of 133 kD [14]. ORF 2 of VD2 DNA segment on the other hand encodes for NS3 whose homology has been identified with NS3 of Junonia coenia densovirus (JcDNV), Galleria mellonella densovirus (GmDNV) and Mythimna loreyi densovirus (MlDNV) [27]. Further, the study also reported NS3 to be an integral part for viral DNA replication [27]. Hence, considering the close homology of BmBDV NS3 with that of JcDNV, it can be concluded that BmBDV NS3 might also have a similar role in BmBDV replication.
Our previous study had revealed that the VD1 and VD2 DNA segments of the Indian BmBDV isolate were composed of 6542 and 6023 nts, respectively, indicating that the VD2 was the shorter segment [3]. The results of the present study also revealed a higher copy number of VD2 ORF transcripts, which once again indicate towards the fact that shorter segments could have a shorter replication time and higher stability [7]. Interestingly, most of the segmented/multipartite viruses have been reported to be plant viruses like those belonging to family of Nanoviridae, Comoviridae etc. BmBDV is the only known segmented insect virus reported, so far [7]. Studies indicated that BmBDVs evolved from Parvovirus ancestors through horizontal gene transfers [21]. Hence, this unique group of viruses have evolved and radiated by gaining new genes for better survival and follow a chronic pattern of infection. This was the first ever attempt to understand the expression pattern of all six ORF transcripts and the pathogenesis of Indian BmBDV isolate.
Availability of data and material (data transparency)
Data sharing is not applicable to this article as no new data were created or analysed in this study. However, the data on qPCR analysis is available which is not required to share as the graphical representation of qPCR data is already presented in the manuscript.
References
Agbandje-McKenna M, Chapman MS. Correlating structure with function in the viral capsid. In: Kerr JR, Cotmore SF, Bloom ME, Linden M, Parrish CR, editors. Parvoviruses. London: Hodder Arnold; 2006. p. 125–39.
Doronin K, Toth K, Kuppuswamy M, Krajcsi P, Tollefson AE, Wold WSM. Over expression of the ADP (E3–11.6K) protein increases cell lysis and spread of adenovirus. Virology. 2003;305:378−87.
Gupta T, Ito K, Kadono-Okuda K, Vijaya Gowri E, Murthy GN, Ponnuvel KM. Characterization and genome comparison of an Indian isolate of bidensovirus infecting the silkworm Bombyx mori. Arch Virol. 2017;163:125–34.
Gupta T, Ramesha AR, Mishra RK, Moorthy M, Sivaprasad V, Ponnuvel KM. Functional marker assisted improvement of productive mulberry silkworm breeds conferring resistance to Bombyx mori Bidensovirus (BmBDV). Agri gene. 2019;11:1–6.
Hayakawa T, Kojima K, Nonaka K, Nakagaki M, Sahara K, Asano SI, Iizuka T, Bando H. Analysis of proteins encoded in the bipartite genome of a new type of parvo-like virus isolated from silkworm – Structural protein with DNA polymerase motif. Virus Res. 2000;66:101–8.
Hu Z, Li GH, Li GT, Yao Q, Chen KP. Bombyx mori bidensovirus: the type species of the new genus Bidensovirus in the new family Bidnaviridae. Chin Sci Bull. 2013;58:4528–32.
Hu Z, Zhang X, Liu W, Zhou Q, Zhang Q, Li G, Yao Q. Genome segments accumulate with different frequencies in Bombyx mori bidensovirus. J Basic Microbio. 2016;56:1–6.
Ito K, Fujii T, Yokoyama T, Kadono-Okuda K. Decrease in the expression level of the gene encoding the putative Bombyxmori bidensovirus receptor during virus infection. Archives of Virology. 2018;1–12.
Ito K, Kidokoro K, Shimura S, Katsuma S, Kadono-Okuda K. Detailed investigation of the sequential pathological changes in silkworm larvae infected with Bombyx densovirus type 1. J Invertebr Pathol. 2013;112:213–8.
Ito K, Shimura S, Katsuma S, Tsuda Y, Kobayashi J, Tabunoki H, Yokoyama, T, Shimada T, Kadono-Okuda K. Gene expression and localization analysis of Bombyx mori bidensovirus and its putative receptor in B. mori midgut. J Invertebr Pathol. 2016;136:50–6.
Jindal HK, Yong CB, Wilson GM, Tam P, Astell CR. Mutations in the NTP-binding motif of minute virus of mice (MVM) NS-1 protein uncouple ATPase and DNA helicase functions. J Biol Chem. 1994;269:3283–9.
Kadono-Okuda K, Ito K, Murthy GN, Sivaprasad V, Ponnuvel KM. Molecular mechanism of Densovirus resistance in silkworm, Bombyxmori. Sericologia. 2014;54:1–10.
Kawase S, Cai YM, Bando H, Seki H. Chemical properties of the Yamanashi isolate of the Bombyx densonucleosis virus. J Seric Sci Jpn. 1984;53:341–7.
Kong J, Hu Z, He Y, Li G, Cao J, Wang F, Chen K, Yao Q. Expression analysis of Bombyx mori parvo-like virus VD2-ORF 1 gene encoding a minor structural protein. Biologia. 2011;66:684–9.
Krupovic M, Koonin EV. Evolution of eukaryotic single- stranded DNA viruses of the Bidnaviridae family from genes of four other groups of widely different viruses. Sci Rep. 2014;4:5347.
Li G, Sun C, Zhang J, He Y, Chen H, Kong J, Huang G, Chen K, Yao Q. Characterization of Bombyx mori parvo-like virus non-structural protein NS1. Virus Genes. 2009;39:396–402.
Lv M, Yao Q, Wang Y, Liu X, Liu H, Huang G, Chen K, Zhang J, Li X. Identification of structural proteins of Bombyx mori parvo-like virus (China Zhenjiang isolate). Intervirology. 2011;54:37–43.
Nakagaki M, Morinaga T, Zhou C, Kajiura Z, Takei R. Increasing curves of two virus DNAs in the midgut epithelium of silkworm infected with Bombyx mori densonucleosis virus type 2 (BmDNV-2). J Seric Sci Jpn. 1999;68:173–80.
Seki H, Iwashita Y. Histopathological features and pathogenicity of a densonucleosis virus of the silkworm, Bombyxmori isolated from sericultural farms in Yamanashi prefecture. J Seric Sci Jpn. 1983;52:400–5.
Sotoshiro H, Kobayashi M. Identification of viral structural polypeptides in the midgut and feces of the silkworm, Bombyx mori infected with Bombyx densovirus type 2. J Invertebr Pathol. 1995;66:60–7.
Tijssen P, Pénzes JJ, Yu Q, Pham HT, Bergoin M. Diversity of small, single-stranded DNA viruses of invertebrates and their chaotic evolutionary past. J Invertebr Pathol. 2016;140:83–96.
Tollefson AE, Scaria A, Hermiston TW, Wold LJ, Wold WSM. The adenovirus death protein (E3–11.6K) is required at very late stages of infection for efficient cell lysis and release of adenovirus from infected cells. J Virol.1996;70:2296−2306.
Walker SL, Wonderling RS, Owens RA. Mutational analysis of the adeno-associated virus Rep68 protein: Identification of critical residues necessary for site-specific endonuclease activity. J Virol. 1997;71:2722–30.
Wang F, Hu Z, He Y, Li G, Kong J, Cao J, Chen K, Yao Q. The non-structural protein NS-2 of Bombyx mori parvo-like virus is localized to the nuclear membrane. Curr Micro biol. 2011;63:8–15.
Wang YJ, Yao Q, Chen KP, Wang Y, Lu J, Han X. Characterization of the genome structure of Bombyx mori densovirus (China isolate). Virus Genes. 2007;35:103–8.
Yang B, Zhang J, Cai D, Li D, Chen W, Jiang H, Hu Y. Biochemical characterization of Periplaneta fuliginosa densovirus non-structural protein NS1. Biochem Biophys Res Commun. 2006;342:1188–96.
Yin HJ, Yao Q, Guo ZJ, Bao F, Yu W, Li J, Chen K. Expression of non-structural protein NS3 gene of Bombyx mori densovirus (China isolate). J Genet Genom. 2008;35:239–44.
Acknowledgements
The authors gratefully acknowledged the Central Silk Board, Ministry of Textiles, Govt. of India for providing infra-structure to carry out the research work. This study was funded by Swedish Research Council, Sweden (Grant No: 2017-05463).
Funding
This study was funded by Swedish Research Council, Sweden (Grant No: 2017-05463).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Ethical approval
The authors ensured that accepted principles of ethical and professional conduct have been followed.
Consent to participate (include appropriate statements)
Not applicable.
Consent for publication (include appropriate statements)
All authors contributed significantly, and conveyed their consent for the preparation of manuscript and its publication.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Gupta, T., Raghavendar, G., Terenius, O. et al. An investigation into the effects of infection and ORF expression patterns of the Indian bidensovirus isolate (BmBDV) infecting the silkworm Bombyx mori. VirusDis. 33, 76–83 (2022). https://doi.org/10.1007/s13337-021-00750-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13337-021-00750-y