Abstract
The complete genome sequences of four low-temperature Escherichia coli-specific tevenviruses, vb_EcoM-VR5, vb_EcoM-VR20, vb_EcoM-VR25 and vb_EcoM-VR26, were determined. Genomic comparisons including recently described genomes of vb_EcoM-VR7 and JS98 as well as phage T4 allowed the identification of two genetic groups that were consistent with defined host-range phenotypes. Group A included the broad-host-range phages vb_EcoM-VR5 and JS98, while group B included vb_EcoM-VR7, vb_EcoM-VR20, vb_EcoM-VR25 and vb_EcoM-VR26, which all had somewhat limited host ranges. All four sequenced phages had genomes that were similar in length (~170 kb) and GC content (~40 %), and, with the exception of vb_EcoM-VR5, at the nucleotide level, they were much more closely related to each other than either was to any other tevenvirus currently characterized. Nevertheless, the overall genome organization of vb_EcoM-VR5, vb_EcoM-VR20, vb_EcoM-VR25 and vb_EcoM-VR26 was comparable to that seen in tevenviruses.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Foodborne illnesses resulting from the consumption of produce commodities contaminated with enteric pathogens (e.g., Escherichia coli or Salmonella) is an increasing problem around the world [11, 12]. In this regard, bacteriophages (phages) may provide a safe and effective intervention against bacterial contamination. A number of reports in the literature have described encouraging results, with the use of phages to reduce foodborne pathogens on a variety of fresh and fresh-cut produce. However, one of the main barriers to the success of such applications is the restrictive temperature used for the storage of many fresh and minimally processed produce, since at 4 °C, most pathogenic bacteria will not be metabolically active and the cycle of phage infection cannot be completed [11].
In 1980, three physiological types of phages were recognized by N.D. Seeley and S.B. Primrose: high-temperature (HT) phages plating at or above 25 °C, low-temperature (LT) phages at or below 30 °C, and mid-temperature (MT) phages in the range of 15 to 42 °C [10]. Tevenviruses – phages that share an evolutionary history with bacteriophage T4 – comprise one of the most diverse and common groups of bacteriophages found in nature [2]. About 90 % of such viruses are MT phages that grow on E. coli or related enterobacteria and have an optimum temperature for plating about 37–40 °C, which correlates closely with the optimum growth temperature of the host. To date, only three E. coli-specific tevenviruses, vB_EcoM-VR5, vB_EcoM-VR7 and vB_EcoM-VR20 (described in our previous paper [3]), have been assigned to the LT group. We have successfully expanded this group by isolating two additional low-temperature phages, vb_EcoM-VR25 and vb_EcoM-VR26. However, the complete genome sequence of only vB_EcoM-VR7 has been published thus far [4].
In this paper, we present a brief analysis of the complete genome sequences of four coliphages, vb_EcoM-VR5, vb_EcoM-VR20, vb_EcoM-VR25 and vb_EcoM-VR26 (subsequently referred to by their shorter common laboratory names VR5, VR20, VR25 and VR26), that all show the unique low-temperature plating profile and can replicate at 4 °C.
Tevenviruses VR20, VR25 and VR26 were originally isolated from Lithuanian municipal wastewater, whereas phage VR5 was isolated from sewage. E. coli BE (VR5 and VR20) and MH1 (VR25 and VR26) were used as the host for phage propagation, and all bacteria were cultivated at 30 °C. Phage isolation, further purification, plating and enumeration were carried out as described previously [3]. To determine the nature of host receptors, bacteriophages VR were tested against a set of E. coli K-12 strains (Table S2) that have deletions of the genes encoding outer membrane proteins or LPS components [1]. Initially, a quantitative spot dilution test was employed as described in ref. [14]. When clearly modified infections were observed, determination of the efficiency of plating (EOP) was performed as described in ref. [3]. Briefly, high-titer phage stocks were diluted and plated in duplicate. Plates incubated at 30 °C were read after 18–24 h, those at 10 °C after 5 days. The number of plaques formed on the lawn of E. coli K-12 BW25113 (at the appropriate temperature) was taken as the standard for the EOP calculation. All phage experiments, host range determination and the isolation of phage DNA were performed as described previously [3, 4]. Phage genomes were sequenced using Illumina DNA sequencing technology at BaseClear (The Netherlands). Genome sequence analysis and bioinformatics were conducted as described in [13].
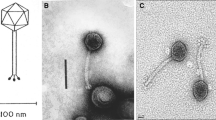
The phages presented here have a dsDNA genome with sizes ranging from 170,336 bp (VR20) to 171,541 bp (VR26). The genomic G+C content ranged from 39.3 % (VR5) to 40.4 % (VR20 and VR25), which is close to the G+C content found in VR7 [3]. Genome alignment comparison (Fig. 1A) revealed that three out of four phages presented here, namely VR20, VR25 and VR26, are more closely related to VR7 and each other (~96 % identity/~86 % coverage) than to other sequenced tevenviruses. In contrast, the genome sequence of VR5 is more similar to that of JS98 (89 % of the genome sequence shares 92 % sequence identity) than to the genome of VR7 (50/97) (Table S1). Phylogenetic analysis using whole-genome alignment allowed the identification of two evolutionary groups (Fig. 1 B) that correlate well with distinctive lytic phenotypes. As seen in Table 1, phage VR5 (group A) has the capacity to infect a broad range of laboratory E. coli strains. In contrast, phages from group B show a clear preference for either E. coli BE derivatives (VR20 and VR7) or K-12 strains (VR25 and VR26). In spite of the aforementioned differences, VR phages have a similar genome organization and share between 34 and 100 percent amino acid sequence identity to each other among 225 conserved open reading frames (ORFs). The majority of the ORFs in the genomes of VR phages presented here appear to originate within the T4 superfamily. Comparative genome analysis indicates that VR phages and phage T4 share a conserved core genome [8], particularly within the regions coding for DNA replication/recombination and virion structural proteins. Nonconserved modules are primarily composed of genes of unknown origin as well as those coding for DNA modification enzymes, homing nucleases, tRNAs, distal tail fibers and internal head proteins. Interestingly, the majority of T4 genes that are absent from VR7 [4] appear to be also missing from the genomes of VR phages presented here, whereas the gene for a head vertex protein is duplicated in all VR genomes. Out of four bacteriophages presented here, only phage VR20 encodes both for anaerobic ribonucleotide reductase (NrdD) and NrdD-activating enzyme (NrdG), suggesting that VR5, VR25 and VR26 are less adapted to anaerobic growth conditions. As was mentioned above, the tail fiber module is the most plastic structural region in the VR genomes, and the long distal fiber locus is particularly diverged. In VR7 [4] and VR25, the long tail fiber adhesin is located in the C-terminal domain of a protein that is somewhat similar to T4 gp37. In the case of other VR phages, the adhesin is a separate protein. Phylogenetic analysis based on the alignment of the amino acid sequences of the tail fiber adhesins from various tailed viruses (Fig. S1) suggests that bacteriophages VR25 and VR26 recognize either OmpC (not produced by E. coli B [7]) or OmpA of the E. coli K-12 type. Interestingly, apart from E. coli K-12 derivatives, phage VR25 is also capable of infecting a number of STEC strains (data not shown), including E. coli O157:H7, O111:H8 and O26:H11, that also harbour OmpC [5]. Nevertheless, the results presented in Table S2 suggest that, unlike bacteriophage VR26, which clearly requires OmpA but is also sensitive to the LPS composition, phage VR25 can recognize more than one host receptor, including, but not limited to, OmpC. In addition, our results indicate that VR phages likely have more complex temperature-dependent behavior, and at different temperatures may utilize different host cell receptors. However, since the change in temperature has marked effects on the cell surface structure/composition [6], further studies are required to test this hypothesis.
Comparative whole-genome analysis. (A) Linear pairwise genome comparison performed using the mVISTA tool (available at http://genome.lbl.gov/vista/index.shtml) with a threshold of 70 % similarity over 100 bp. The genome of bacteriophage VR7 (#HM563683) was used a reference sequence. (B) Neighbor-joining tree based on the whole-genome sequence alignment. The tree was constructed using Geneious v5.5, available from http://www.geneious.com. The numbers at the nodes indicate the bootstrap probabilities
Several changes take place inside the cell when it is exposed to low temperature. These include (1) loss of membrane flexibility, which affects the overall membrane structure and membrane-associated functions such as transport; (2) stabilization of secondary structures in nucleic acids, which leads to impaired transcription and translation due to hindered movement of RNA polymerase and ribosomes, respectively; (3) increase in negative supercoiling of DNA, which affects DNA replication, (4) unfolding or improper folding and methylation of some proteins, and (5) transient induction of proteins involved in cold response [9]. This, in conjunction with our previously published findings [3], suggests that LT phages must be adapted to alterations within the host cell at all stages of infection, starting from adsorption and ending with the virion release.
Unfortunately, as is the case with cold-active viruses [15], it is not possible to demonstrate cold-adaptation of LT phages solely from sequence data. As was mentioned previously, the vast majority of the ORFs in the genomes of VR phages presented here appear to be derived from tevenviruses and other MT phages, and no exclusively VR-specific genes may be found. Consequently, it would appear that the adaptation of LT coliphages to replicate within the mesophilic host at low temperatures consists of a collection of synergistic changes in the overall genome content and composition (e.g., changes in the amino acid composition that may lead to fine-tuning of protein activity, flexibility and stability balance) rather than the presence of a unique set of particular genes.
Finally, while the genome sequence analysis did not provide us with a straightforward answer as to why the viruses presented here belong to the low-temperature class of coliphages, the differential host specificity observed among VR phages can most likely be attributed to the differences within the tail fiber genes coding for the receptor-recognizing proteins. Thus, VR bacteriophages, as well as their long tail fiber adhesins, are an attractive tool for application in the selective detection and/or the control of bacterial contamination in produce at low temperatures – approaches that are being extensively researched in our laboratory.
Nucleotide sequence accession numbers
The complete genome sequences of VR5, VR20, VR25 and VR26 are available in the GenBank database under the accession numbers KP007359, KP007360, KP007361 and KP007362, respectively.
References
Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H (2006) Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2(2006):0008
Comeau AM, Bertrand C, Letarov A, Tetart F, Krish HM (2007) Modular architecture of the T4 phage superfamily: a conserved core genome and a plastic periphery. Virology 362:384–396
Kaliniene L, Klausa V, Truncaite L (2010) Low-temperature T4-like coliphages vB_EcoM-VR5, vB_EcoM-VR7 and vB_EcoM-VR20. Arch Virol 155:871–880
Kaliniene L, Klausa V, Zajanckauskaite A, Nivinskas R, Truncaite L (2011) Genome of low-temperature T4-related bacteriophage vB_EcoMVR7. Arch Virol 156:1913–1916
Liao WC, Ng WV, Lin IH, Syu WJ, Liu TT, Chang CH (2011) T4-Like genome organization of the Escherichia coli O157:H7 lytic phage AR1. J Virol 85:6567–6578
Mihoub F, Mistou MY, Guillot A, Leveau JY, Boubetra A, Billaux F (2003) Cold adaptation of Escherichia coli: microbiological and proteomic approaches. Int J Food Microbiol 89:171–184
Mizuno T, Shinkai A, Matsui K, Mizushima S (1990) Osmoregulatory expression of porin genes in Escherichia coli: a comparative study on strains B and K-12. FEMS Microbiol Lett 56:289–293
Petrov VM, Ratnayaka S, Nolan JM, Miller ES, Karam JD (2010) Genomes of the T4-related bacteriophages as windows on microbial genome evolution. Virol J 7:292
Phadtare S, Inouye M (2008) Cold-shock proteins. In: Margesin R, Schinner F, Marx J-C, Gerday C (eds) Psychrophiles: from biodiversity to biotechnology. Springer, Berlin, pp 191–210
Seeley ND, Primrose SB (1980) The effect of temperature on the ecology of aquatic bacteriophages. J Gen Virol 46:87–95
Sharma M (2013) Lytic bacteriophages: potential interventions against enteric bacterial pathogens on produce. Bacteriophage 3:e25518
Scharff RL (2012) Economic burden from health losses due to foodborne illness in the United States. J Food Prot 75:123–131
Šimoliūnas E, Kaliniene L, Truncaitė L, Zajančkauskaitė A, Staniulis J, Kaupinis A, Ger M, Valius M, Meškys R (2013) Klebsiella phage vB_KleM-RaK2—a giant singleton virus of the family Myoviridae. PLoS ONE 8:e60716
Trojet SN, Caumont-Sarcos A, Perrody E, Comeau AM, Krisch HM (2011) The gp38 adhesins of the T4 superfamily: a complex modular determinant of the phage’s host specificity. Genome Biol Evol 2011(3):674–686. doi:10.1093/gbe/evr059
Wells LE (2008) Cold-active viruses. In: Margesin R, Schinner F, Marx J-C, Gerday C (eds) Psychrophiles: from biodiversity to biotechnology. Springer, Berlin, pp 157–173
Acknowledgments
This research was funded by a grant (No. SVE-04/2012) from the Research Council of Lithuania.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kaliniene, L., Zajančkauskaitė, A., Šimoliūnas, E. et al. Low-temperature bacterial viruses VR – a small but diverse group of E. coli phages. Arch Virol 160, 1367–1370 (2015). https://doi.org/10.1007/s00705-015-2388-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-015-2388-0