Abstract
Bacteriophages vB_EcoM-VR5, vB_EcoM-VR7 and vB_EcoM-VR20, showing an unusual low-temperature plating profile and producing constantly growing plaques, were isolated from aquatic environments of Lithuania. Although vB_EcoM-VR5, vB_EcoM-VR7 and vB_EcoM-VR20 resembled phage T4 both in their genome size and in their major structural protein (gp23) pattern, physiological properties of all three phages tested differed significantly from those of T4. With an optimum temperature for plating around 24°C and a high efficiency of plating in the range 7–30°C, bacteriophages vB_EcoM-VR7 and vB_EcoM-VR20 failed to plate at 37°C, whereas phage vB_EcoM-VR5 could not be plated at 40°C. Sequence analysis of diagnostic g23 PCR products revealed that g23 of vB_EcoM-VR5, vB_EcoM-VR7 and vB_EcoM-VR20 differed from the corresponding T4 g23 DNA sequence by 21, 21 and 20%, respectively.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Bacteriophages are the most abundant entities in the biosphere, exceeding the number of bacteria by a factor of 5:25 [1, 2]. It has been estimated that in the biosphere, there are >1030 tailed phages, among which the myoviruses, phages with contractile tails, are widespread and diverse. Bacteriophage T4, the most researched archetype of the myovirus family, can be shown by phylogenetic analysis of the head and tail genes to belong to the monophyletic group of T4-type bacteriophages. Among more than 160 bacteriophages that have been classified as T4-type exclusively based on morphological criteria, nearly 130 are morphologically indistinguishable from T4 [3]. The strong conservation of T4-type virion morphology suggests that the portion of the phage genome encoding structural components may be the most tenaciously fixed in evolution.
Based on a sequence comparison of three major structural proteins (gp18, gp19 and gp23), the T4-type phages have been classified into four subgroups with increasing divergence from T4: the T-evens, pseudo-T-evens, shizo-T-evens and exo-T-evens [4–6]. The designation mezo-T-even has also been proposed in order to classify so called “chimeric” T4-type bacteriophages (e.g. RB69, TuIA), which appear to occupy an intermediate position between T-even and pseudo-T-even phages (Krisch, personal communications). Moreover, culture-independent sequence analysis of PCR-amplified environmental DNA [7, 8] revealed that the T4-type myoviruses are much more diverse than was previously suspected and indicated that the T4-type phage superfamily may comprise at least 15 distinct subgroups.
About 90% of the known T4-related phages grow on Escherichia coli or other enterobacteria, and the remaining 10% grow on phylogenetically more distant bacteria, e.g., representatives of such genera as Aeromonas, Vibrio, Synechococcus etc. [9]. The vast majority of enterobacterial T4-type phages are from municipal wastewater or sewage, indicating that their natural habitat is the human or animal intestine [3]. Consequently, the optimum temperature for coliphage development is about 37–40°C, and this correlates closely with the optimum growth temperature of the host—the thermotrophic cells of E. coli [10]. Based on the effect of temperature on the efficiency of plating, three physiological types of bacteriophages have been recognized—high-temperature (HT) phages plating at or above 25°C, low-temperature (LT) phages plating at or below 30°C, and mid-temperature (MT) phages plating in the range 15–42°C [11]. E. coli phage T4, the archetype of T4-type superfamily, is the typical representative of MT bacteriophages. According to Yanagida et al. [12], certain steps in T4 assembly seems to be inhibited even at 19°C, leading to an accumulation of capsids, preheads, partially sheathed tails and naked cores. No T4-type E. coli-specific bacteriophages have been reported to plate efficiently at low temperatures, and, among characterized bacteriophages, only Aeromonas hydrophyla-specific phages have been assigned to the LT group to date [11, 13].
In this paper, we present three T4-like bacteriophages, vB_EcoM-VR5, vB_EcoM-VR7 and vB_EcoM-VR20, with physiological properties that have never been observed before among the characterized T4-related E. coli phages. Our aim was to introduce the main characteristics of the newly identified bacteriophages, as it is usually recommended prior to publication of the whole genome sequence. Following the nomenclature of viruses proposed by Kropinski et al. [14], we named our newly isolated bacteriophages vB_EcoM-VR5, vB_EcoM-VR7 and vB_EcoM-VR20, although they are subsequently referred to by their shorter common laboratory names VR5, VR7 and VR20.
Materials and methods
Phages and bacterial strains
T4-like phages VR5, VR7 and VR20 were originally isolated from Lithuanian municipal wastewater (VR20) and sewage (VR5 and VR7) as described previously [15]. Wild-type phage T4D was kindly provided by Dr. W. B. Wood. E. coli strain BE (sup 0), a gift from Dr. L. W. Black, was used for phage propagation and phage growth experiments. For host range determination, a double agar overlay plaque assay method [16] was employed, and the list of E. coli strains used is presented in Table 1.
For all phage experiments, bacteria were cultivated in Luria–Bertani broth (LB) or LB agar. Bacterial growth was monitored turbidimetrically by reading OD600. An OD600 of 0.8 corresponded to 2.0 × 108 cells/ml.
Phage techniques
Phage isolation, plating and titering were carried out as described in Ref. [15], and adsorption tests and one-step growth experiments were carried out as described by Khusainov et al. [17] and Carlson and Miller [18], respectively.
Determination of the efficiency of plating (e.o.p.) was performed as described by Seeley and Primrose [11]. High-titer phage stocks were diluted and plated in duplicate. Plates incubated at 17, 24, 30, 35, 37, 39 and 40°C were read after 18–24 h, those at 12°C after 48 h, and those at 7°C after 5 days. The temperature at which the largest number of plaques were formed was taken as the standard for the e.o.p. calculation.
Viral DNA isolation and restriction analysis
Aliquots of phage suspension (1011–1012 pfu/ml) were subjected to phenol/chloroform extraction and ethanol precipitation as described in Ref. [18]. Isolated phage DNA was subsequently used in restriction analysis. Restriction digestion was performed with EcoRV, DraI, VspI, NdeI, SmiI, Bsp1107I, AluI, Bst1170I, Bsp143I, HinfI, BglII and MunI restriction endonucleases (Fermentas) according to the supplier’s recommendations. DNA fragments were separated by electrophoresis in a 0.8% agarose gel containing ethidium bromide. Restriction digestion was repeated at least three times to confirm the results.
Pulsed-field gel electrophoresis (PFGE)
Purified phage particles (approximately 1.0 × 1011 pfu/ml) were used as the source for the preparation of intact genomic DNA. After overnight incubation with proteinase K, samples were mixed with 1.2% low-melting agarose and loaded into a 1.2% agarose gel. PFGE was performed for 24 h at 6 V/cm and 8°C with a pulse time of 10 (E/W) to 10 s (S/N) using the Gene Navigator system from Pharmacia Biotech. The gel was stained for 30 min with ethidium bromide (0.5 μg/ml) and visualized on a UV box.
PCR and oligonucleotide primers
PCR with Pfu DNA polymerase (Fermentas), using denaturated phage particles as a template, g23-targeting degenerate primers MZIA1bis (5′-GAT ATT TGI GGI GTT CAG CCI ATG A-3′) and MZIA6 (5′-CGC GGT TGA TTT CCA GCA TGA TTT C-3′) was performed following the method described by Filée et al. [7]. PCR products were cloned using a CloneJET™ PCR Cloning Kit (Fermentas) and sequenced.
Sequence analysis
Sequence analysis was performed using the Transeq (DNA sequence translation), FASTA3 (nucleotide and deduced amino acid [aa] sequence comparison) and ClustalW2 (multiple sequence alignment) programs, which are available on the EMBL-EBI website. A neighbor-joining tree was constructed using MEGA 4.0 [19]. DNA sequences were deposited in the EMBL database under the accession numbers FN641800, FN641801, and FN641802.
Protein techniques
Purified virus particles were dissolved in the sample buffer, heated at 95°C for 5 min, and subjected to 12% SDS-PAGE gel electrophoresis. Protein bands were visualized by staining the gel with Coomassie brilliant blue.
Results
Phage morphology
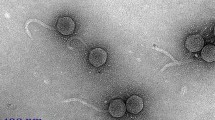
Electron microscopy of VR5, VR7 and VR20 phage particles showed that the virion morphology of all three phages tested resembled that of members of the family Myoviridae, A2 morphotype (indistinguishable from phage T4) [15]. SDS-PAGE of VR5, VR7 and VR20 virion proteins confirmed their close phylogenetic relatedness to the T4-type bacteriophages. Comparison of the structural protein profiles showed that in all three phages tested, the most abundant protein band (usually corresponding to the major head protein gp23 in T4-related phages) displayed a protein electrophoretic migration profile similar to that of gp23 of bacteriophage T4 (Fig. 1). Meanwhile, the bands of the other virion proteins of VR5, VR7 and VR20 clearly exhibited distinct electrophoretic migration profiles compared with those of T4.
The host range of VR5, VR7 and VR20
Bacteriophages VR5, VR7 (sewage sample isolates) and VR20 (wastewater sample origin) were isolated on E. coli strain BE as the indicator host [15]. Unfortunately, in the course of the investigation of the abundance of T4-type bacteriophages in municipal wastewater and sewage, no natural bacterial hosts were collected. Subsequently, in order to investigate the host range of VR5, VR7 and VR20, they were tested for their ability to infect different laboratory strains of E. coli (Table 2). In the plaque assay test, only the infectivity of bacteriophage VR5 was shown to be similar to that of T4 against all E. coli strains used. Meanwhile, phages VR7 and VR20 showed a clear preference for E. coli BE and BL21(DE3) over B40, K12 (or K12 derivatives). These results suggest that VR7 and VR20 have narrower host range specificity than VR5 or T4.
VR5, VR7 and VR20 were also tested on several laboratory isolates of Klebsiella, Aeromonas, Pseudomonas and Salmonella (data not shown). None of these bacteria were susceptible to VR5, VR7 or VR20, indicating that these phages have known host range limited only to E. coli.
The effect of temperature on the efficiency of plating (e.o.p.) and adsorption
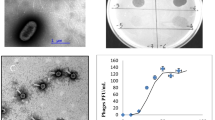
The e.o.p. of VR5, VR7, VR20 and T4 was examined in the temperature range of 7–48°C. Unlike phage T4, all three VR phages tested had an optimum temperature for plating around 24°C, but the e.o.p. dropped sharply to 0 between 30 and 37°C in the case of VR7 and VR20, or between 30 and 40°C in the case of VR5 (Fig. 2). These results indicate that VR5, VR7 and VR20 are adapted to grow at the lower temperatures of extracolonic environments rather than in the range of mammalian gut temperatures.
There are at least two possible reasons for the phage to fail to form plaques at a certain temperature: failure to adsorb to the host cell, or failure to multiply [11]. In order to test the effect of temperature on the ability of phages VR5, VR7 and VR20 to adsorb the cells of E. coli BE, adsorption tests were performed at 24, 30 and 35°C (in the case of VR7 and VR20) or 24, 30 and 37°C (in the case of VR5). During the adsorption tests, we also calculated the final number of viable phages formed by plating chloroform-treated cells at the end of the rise period, deduced experimentally (the rates are given in square brackets within the legend of adsorption charts). Figure 3a shows that the decrease of the temperature from 37°C to 24°C negatively affected both adsorption and multiplication rates of phage T4. The adsorption rates of VR5, VR7 and VR20 (Fig. 3b–d, respectively) showed that there was also a certain increase in the adsorption rates of VR5, VR7 and VR20 as the temperature was raised from 24 to 35°C (or 37°C). Meanwhile, the burst sizes were reduced by 97–98% (in the case of VR7 and VR5) and by 92% (in the case of VR20). These results suggest that multiplication (and/or the injection step) of VR5, VR7 and VR20 were affected at temperatures above 30°C. On the other hand, a slight abortive adsorption profile displayed by VR20 at 35°C (Fig. 3d) implies that the adsorption step at restrictive temperatures in the case of this phage may be also disturbed.
Effect of temperature on the ability of T4 (a) (control), VR5 (b), VR7 (c) and VR20 (d) to adsorb to E. coli BE cells. Each point represents the mean of three individual experiments. Numbers in square brackets represent values of viable phage particles formed calculated after plating chloroform-treated samples at the end of the rise period. The temperature at which the largest number of viable phage was formed was taken as the standard (1.00)
Phage growth characteristics
Single-step growth experiments were performed at 24°C as described in “Materials and methods”. The curves obtained (Fig. 4a–c) indicated that the latent periods of VR5, VR7 and VR20 were 37, 40 and 42 min, respectively. The eclipse periods were 31, 36 and 32 min, respectively, and average burst sizes of 50, 70 and 48 PFU per cell were obtained. As was expected, phage T4, with an optimum temperature for development around 37°C [20], had a substantially longer latent period at 24°C, 75 min. The eclipse period was 50 min, and the average burst size was 136 PFU per cell (Fig. 4d).
Single-step growth curves of bacteriophages VR5 (a), VR7 (b), VR20 (c), T4 (d) (control) at 24°C. Shown are PFU per infected E. coli BE cell in chloroform-treated cultures (circle) and untreated cultures (filled circle) at different time points. Each point represents the mean of two to four experiments
It is worth mentioning that in the case of VR5, VR7 and VR20, increasing the temperature to 30°C resulted in a decrease in both the latent (30, 31 and 30 min, respectively) and eclipse (25, 27 and 25 min, respectively) periods. Meanwhile the burst sizes decreased by ~40% (data not shown).
The size of genomic DNA
Usually it is impossible to determine the genome size of relatively large (>100 kb) genomic DNA-possessing bacteriophages by summing the restriction fragment lengths. Therefore, pulse-field gel electrophoresis was performed to mobilize the intact genomic DNA of bacteriophages VR5, VR7 and VR20 (Fig. 5). Like phage T4, bacteriophages VR5 and VR7 were found to possess a genome of ~170 kb in length. Meanwhile, the genome of bacteriophage VR20 appeared slightly smaller than that of T4. On the other hand, these non-significant differences in gel migration could be caused by unknown individual characteristics of the genomic DNA of phage VR20.
Restriction analysis
Restriction analysis is a valuable method for characterizing and differentiating various phages. The DNA of T4 contains hydroxymethyl cytosine (Hm-Cyt) instead of cytosine, and these residues are generally glycosylated, which makes T4 DNA resistant to digestion with most restriction enzymes, and only few of them are known to cut the DNA of T4. However, nucleotide modifications are neither invariably present among T-evens nor absolute among T4-related phages in general [3].
To test for the digestibility of VR5, VR7 and VR20 DNA, 12 type II restriction endonucleases (DraI, VspI, NdeI, SmiI, Bsp1107I, AluI, Bst1170I, Bsp143I, HinfI, BglII and MunI) were used. Restriction analysis revealed that the DNA of all three phages tested can only be digested by the restriction enzymes that are known to recognize Hm-Cyt-DNA of T4 [EcoRV (Fig. 6a), DraI, VspI, NdeI, SmiI, Bsp1107I], suggesting that phages T4, VR5, VR7 and VR20 may contain similar DNA modifications. However, clearly distinct restriction patterns visualized after digestion with SmiI (whose recognition sequence is 5′ ATTTAAAT 3′) imply that DNA of bacteriophages VR5, VR7 and VR20 (Fig. 6b) may contain a limited number of ATTTAAAT octamers compared with DNA of T4. This feature may be related to the adaptation of phages to a particular host and is quite common among other T4-type bacteriophages, e.g. a limited number of SmiI recognition sequences can be found within the genomic DNA of T-even phage RB69, shizo-T-even phage Aeh1 and pseudo-T-even phage RB49 (4, 4 and 0, respectively).
Phylogenetic relatedness
In the absence of a complete genomic sequence, a phylogenic classification of T4-related phages based on gp23 is most commonly employed [4–9, 21–24]. In this study, we used degenerate PCR primers that had been reported to amplify a homologous segment of the g23 sequence in all of the subgroups of T4-type phages [7]. PCR products of predicted size (~0.6 kb) were detected for all three bacteriophages tested. The PCR-generated VR5, VR7 and VR20 DNA fragments were subsequently cloned and sequenced, and the corresponding amino acid sequences were deduced. As was expected, all of the nucleotide sequences obtained were similar to an internal region of g23 of T4. The ClustalW2 sequence alignment revealed that PCR-generated fragments of VR5, VR7 and VR20 g23 were closely related to each other (97–98% nucleotide and amino acid sequence identity). The FASTA-Protein search within the NCBI database revealed that the deduced amino acid sequences of the internal part of gp23 of VR5, VR7 and VR20 were most closely related to the corresponding part of gp23 in JS98 (95, 94, and 94% identical aa, respectively). Meanwhile, the amino acid sequence identities shared with the corresponding part of gp23 of T4 were only 84, 84 and 83%, respectively. To determine their phylogenetic relatedness, the deduced sequences of gp23 fragments of VR5, VR7 and VR20 were aligned with the corresponding fragments of the major capsid proteins from 14 T4-type phages available in databases, and a neighbor-joining tree was constructed (Fig. 7). Based on phylogenetic analysis, bacteriophages VR5, VR7 and VR20, together with JS98, fall into the distinct subgroup of T4-type bacteriophages.
Neighbor-joining tree based on the alignment of the central part of gp23 (corresponding to 110–303 aa of gp 23 of T4) of VR5, VR7, VR20 and fourteen T4-type bacteriophages available at http://phage.bioc.tulane.edu or NCBI databases. The bootstrap values are indicated
Discussion
As a consequence of increasing antibiotic resistance in bacteria, interest in the ability of phages to control bacterial populations has extended from medical applications into the fields of agriculture, aquaculture, the food industry, and wastewater treatment. However promising, non-clinical applications of phage-mediated bioprocessing have many limitations, most of which are related to the physiology of the phage applied (e.g. the inability of the phage to survive or replicate under the conditions desired) or the lack of the characterized phages available [25, 26]. In this paper, we present the characterization of three low-temperature E. coli phages, VR5, VR7 and VR20, that could be useful in the areas where bacteriophages adapted to grow at the lower temperatures of extracolonic waters are required (e.g. wastewater treatment). In order to apply the phage to any of the areas mentioned: (1) the phage must be characterized, (2) it must be classified and (3) its complete genome sequence must be determined. Although basic morphological and physiological properties of VR5, VR7 and VR20 have been described in the previous section of this paper and the whole genome sequencing data are on the way, we still find some difficulties in classifying these newly isolated bacteriophages.
Phylogenetic analysis based on the alignment of the central third of gp23 classified phage JS98 and all of the VR phages tested into a discrete subgroup of T4-type bacteriophages. Since bacteriophage JS98 was classified into the group of T-evens [22, 23], one could assume that VR5, VR7 and VR20 should naturally fall into this group of T4-type bacteriophages as well. However, in order to be characterized as T-even, the phage should meet several determined requirements: (1) the phage should display similar physiological properties with T4, and (2) it should possess the orthologue of the major structural protein gp23 of T4, exhibiting amino acid sequence identity greater than 90% [6, 27].
The single-step growth characteristics of VR5, VR7 and VR20 were hardly comparable with those of the T-evens, since no T-even phage had been reported to have an optimum temperature around 24°C. Meanwhile, the adsorption test results suggested, as was observed with T-evens, that bacteriophages VR5, VR7 and VR20 adsorbed more efficiently at temperatures above 30°C. On the other hand, the same effect was observed with numerous different representatives of other T4-type subgroups. Although there was a significant decrease in the burst size at temperatures above 30°C in the case of all three phages tested, it would be advantageous for the virus to have maximal adsorption under conditions that favor the growth of the host [28] to survive unfavorable conditions while remaining suspended within the infected cell in an early stage of infection, and commence growth when the host cell enters more favorable surroundings.
Another feature of phage physiology that can be compared is plaque formation. T-evens generally display small, rough, cloudy-bordered plaques [29, 30]. Bacteriophage T4 (as well as T-evens in general) cannot produce bursts on stationary-phase cells, and that is why T4 plaques are limited in size, rather than continuing to grow indefinitely as do those of phiX174 and T7 [20, 31]. Meanwhile, in the range of permissive temperatures, bacteriophages VR5, VR7 and VR20 produced constantly growing plaques, e.g. after 24 h of incubation at 24°C, bacteriophages VR5, VR7 and VR20 formed plaques of 1.6 (±0.3), 1.5 (±0.5), and 0.9 (±0.3) mm in diameter respectively; after 48 h, 1.8 (±0.4), 2.1 (±0.6), and 1.75 (±0.2) mm; after 72 h, 2.3 (±0.3), 2.5 (±0.5), and 2.3 (±0.3) mm; and the plaques reached 4.3 (±0.7), 4.8 (±0.7), 3.6 (±0.4) mm in diameter within a period of 6 days (Fig. 8). In the case of T4, the growth of plaques at 24°C could only be observed for 42–48 h, reaching the size limit of 1.15 (±0.35) mm in diameter.
More surprising was the fact that the plaques formed by all three VR phages tested continued to spread even after the plates were stored at 4°C; e.g., after 24 h of incubation at 30°C, phages VR5, VR7, VR20 and T4 formed plaques of 1.4 (±0.4), 1.35 (±0.25), 1.2 (±0.3) and 0.75 (±0.15) mm in diameter, respectively. However, after an additional 120 h of incubation at 4°C, plaques of VR5, VR7 and VR20 reached 1.8 (±0.2), 1.5 (±0.3) and 1.4 (±0.4) mm in diameter respectively, while plaques of T4 maintained their initial size (Fig. 9).
As was mentioned previously, the amino acid sequence of the major structural protein gp23 of typical T-even differs from gp23 of T4 by less than 10%. Meanwhile, gp23 of VR5, VR7 and VR20 differed from gp23 of T4 at a level that would be expected for pseudo-T-evens. Moreover, the amino acid sequences of gp18 and gp19 of VR7, deduced in the course of the whole-genome sequencing project (data in preparation for publishing), showed greater relatedness with JS98 [22, 23] (90 and 91% aa identity, respectively) or Klebsiella phage Kpp95 [24] (76 and 83% aa identity, respectively) than with gp18 and gp19 of T4 (71 and 70% aa identity, respectively).
Based on all of these considerations, bacteriophages VR5, VR7 and VR20 cannot be classified into the group of T-evens. In fact, based on the classification requirements mentioned, phages VR5, VR7 and VR20 could hardly fall into any established subgroup of T4-type bacteriophages. However, a new publication regarding the classification of bacteriophages of the family Myoviridae was published very recently [32]. According to Lavigne et al. [32], three subfamilies Peduovirinae, Teequatrovirinae and Spounavirinae should be established within the family Myoviridae, and the subfamily Teequatrovirinae would contain two genera: T4-like viruses (includes previously termed T-even and pseudo-T-even) and KVP40-like viruses (consists of two former members of the shizo-T-evens). The genus of T4-like viruses should be subdivided into four subgroups sharing >70% proteins: T4-type, 44RR-type, RB43-type, and RB49-type. Since the whole-genome sequence must be analyzed in order to classify the phage into any newly established subgroup, for now, we can classify VR5, VR7 and VR20 phages as E. coli bacteriophages vB_EcoM-VR5, vB_EcoM-VR7 and vB_EcoM-VR20, members of the family Myoviridae, proposed subfamily Teequatrovirinae, genus T4-like viruses and will specify the more detailed classification when the whole-genome sequencing is finished.
References
Balter M (2000) Evolution on life’s fringes. Science 289:1866–1867
Fuhrman JA (1999) Marine viruses and their biogeochemical and ecological effects. Nature 399:541–548
Ackermann H-W, Krisch HM (1997) A catalogue of T4-type bacteriophages. Arch Virol 142:2329–2345
Desplats C, Krisch HM (2003) The diversity and evolution of the T4-type bacteriophages. Res Microbiol 154:259–267. doi:10.1016/S0923-2508(03)00069-X
Hambly E, Tétart F, Desplats C, Wilson WH, Krisch HM, Mann NH (2001) A conserved genetic module that encodes the major virion components in both the coliphage T4 and the marine cyanophage S-PM2. Proc Natl Acad Sci USA 98:11411–11416
Tétart F, Desplats C, Kutateladze M, Monod C, Ackermann H-W, Krisch HM (2001) Phylogeny of the major head and tail genes of the wide-ranging T4-type bacteriophages. J Bacteriol 183:358–366
Filée J, Tétart F, Suttle CA, Krisch HM (2005) Marine T4-type bacteriophages, a ubiquitous component of the dark matter of the biosphere. Proc Natl Acad Sci USA 102:12471–12476. doi:10.1073/pnas.0503404102
Fujii T, Nakayama N, Nishida M, Sekiya H, Kato N, Asakawa S, Kimura M (2008) Novel capsid genes (g23) of T4-type bacteriophages in a Japanese paddy field. Soil Biol Biochem 40:1049–1058. doi:10.1016/j.soilbio.2007.11.025
Comeau AM, Bertrand C, Letarov A, Tétart F, Krisch HM (2007) Modular architecture of the T4 phage superfamily: a conserved core genome and a plastic periphery. Virology 362:384–396. doi:10.1016/j.virol.2006.12.031
Leclerc H, Mossel DAA, Edberg SC, Struijk CB (2001) Advances in the bacteriology of the coliform group: their suitability as markers of microbial water safety. Annu Rev Microbiol 55:201–234
Seeley ND, Primrose SB (1980) The effect of temperature on the ecology of aquatic bacteriophages. J Gen Virol 46:87–95
Yanagida M, Suzuki Y, Toda T (1984) Molecular organization of the head of bacteriophage T-even: underlying design principles. Adv Biophys 17:97–146
Chow MS, Rouf MA (1983) Isolation and partial characterisation of two Aeromonas hydrophyla bacteriophages. Appl Environ Microbiol 45(5):1670–1676
Kropinski AM, Prangishvill D, Lavigne R (2009) Position paper: the creation of a rational scheme for the nomenclature of viruses of bacteria and archea. Env Microbiol 11:2775–2777. doi:10.1111/j.1462-2920.2009.01970.x
Klausa V, Piešinienė L, Staniulis J, Nivinskas R (2003) Abundance of T4-type bacteriophages in municipal wastewater and sewage. Ekologija 1:47–50
Kropinski AM, Mazzocco A, Waddell T, Lingohr E, Johnson RP (2009) Enumeration of bacteriophages by double agar overlay plaque assay. In: Clokie MRJ, Kropinski AM (eds) Bacteriophages: methods and protocols. Humana Press, New York, pp 69–76
Khusainov AA, Shilnikov GV, Emelyanenko VE, Ivantsky GR (1992) Effect of thermoinduced changes in T4 bacteriophage structure on the process of molecular recognition of host cells. Biochim Biophys Acta 737:51–115
Carlson K, Miller E (1994) Experiments in T4 genetics. In: Karam JD (ed) Bacteriophage T4. ASM Press, Washington DC, pp 419–483
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Kutter E, Kellenberg E, Carlson K, Eddy S, Neitzel J, Messinger L, North J, Guttman B (1994) Effects of bacterial growth conditions and physiology on T4 infection. In: Karam JD (ed) Bacteriophage T4. ASM Press, Washington DC, pp 406–418
Chen CR, Lin CH, Lin JW, Chang CI, Tseng YH, Weng SF (2007) Characterization of a novel T4-type Stenotrophomonas maltophilia virulent phage Smp14. Arch Microbiol 188:191–197. doi:10.1007/s00203-007-0238-5
Chibani-Chennoufi S, Canchaya C, Bruttin A, Brüssow H (2004) Comparative genomics of the T4-like Escherichia coli phage JS98: implications for the evolution of T4 phages. J Bacteriol 186(24):8276–8286. doi:10.1128/JB.186.24.8276-8286.2004
Zuber S, Ngom-Bru C, Barretto C, Bruttin A, Brüssow H, Denou E (2007) Genome analysis of phage JS98 defines a fourth major subgroup of T4-like phages in Escherichia coli. J Bacteriol 189(22):8206–8214. doi:10.1128/JB.00838-07
Wu LT, Chang SY, Yen MR, Yang TC, Tseng YH (2007) Characterization of extended-host-range pseudo-T-even bacteriophage Kpp95 isolated on Klebsiella pneumoniae. Appl Environ Microbiol 73(8):2532–2540. doi:10.1128/AEM.02113-06
Withey S, Cartmell E, Avery LM, Stephenson T (2005) Bacteriophages-potential for application in wastewater treatment processes. Sci Tot Env 339:1–18. doi:10.1016/j.scitotenv.2004.09.021
Verma V, Harjai K, Chhibber S (2009) Characterization of a T7-Like Lytic Bacteriophage of Klebsiella pneumoniae. Curr Microbiol doi:10.1007/soo284-009-9430-y
Kutter E, Sulakvelidze A (2005) Bacteriophages: biology and applications. CRC Press, Boca Raton
Kukkaro P, Dennis HB (2009) Virus-host interactions in environments with a wide range of ionic strengths. Environ Microbiol Rep 1:71–77. doi:10.1111/j.1758-2229.2008.00007.x
Abedon ST, Hyman P, Thomas C (2003) Experimental examination of bacteriophage latent-period evolution as a response to bacterial availability. Appl Environ Microbiol 69(12):7499–7506. doi:10.1128/AEM.69.12.7499-7506.2003
Dressman HK, Drake JW (1999) Lysis and lysis inhibition in bacteriophage T4: rV mutations reside in the holin t gene. J Bacteriol 181:4391–4396
Fort J, Méndez V (2002) Time-delayed spread of viruses in growing plaques. Phys Rev Lett 89(17):178101-1–178101-4. doi:10.1103/PhysRevLett.89.178101
Lavigne R, Darius P, Summer EJ, Seto D, Mahadevan P, Nilsson A, Ackerman HW, Kropinski AM (2009) Classification of Myoviridae bacteriophages using protein sequence similarity. BMC Microbiology 9:224. http://www.biomedcentral.com/1471-2180/9/224. Accessed 26 October 2009
Acknowledgments
We thank Dr. W. Black and Dr. Kenneth N. Kreuzer for E. coli strains and Dr. William B. Wood for phage T4. This work was supported in part by the Lithuanian State Science and Studies Foundation.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kaliniene, L., Klausa, V. & Truncaite, L. Low-temperature T4-like coliphages vB_EcoM-VR5, vB_EcoM-VR7 and vB_EcoM-VR20. Arch Virol 155, 871–880 (2010). https://doi.org/10.1007/s00705-010-0656-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-010-0656-6