Abstract
In this study, after the expression of a pyruvate carboxylase gene (PYC) cloned from Meyerozyma guilliermondii in a marine-derived yeast Yarrowia lipolytica SWJ-1b, a transformant PG86 obtained had much higher PYC activity than Y. lipolytica SWJ-1b. At the same time, the PYC gene expression and citric acid (CA) production by the transformant PG86 were also greatly enhanced. When glucose concentration in the medium was 60.0 g L−1, CA concentration formed by the transformant PG86 was 34.02 g L−1, leading to a CA yield of 0.57 g g−1 of glucose. During a 10-L fed-batch fermentation, the final concentration of CA was 101.0 ± 1.3 g L−1, the yield was 0.89 g g−1 of glucose, the productivity was 0.42 g L−1 h−1 and only 5.93 g L−1 reducing sugar was left in the fermented medium within 240 h of the fed-batch fermentation. HPLC analysis showed that most of the fermentation products were CA.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Citric acid (CA) is a symmetric tricarboxylic acid, a natural constituent of a variety of citrus fruits, pineapple, pear, peach and fig, and today is a most important food acidulant. It has a variety of uses in food, pharmaceuticals and other industrial fields, e.g. as a flavor acidifying and a preservative additive in the food and pharmaceutical industry, as a stabilizer for vegetable oils and fats or as a complex-forming and bleaching component in many washing detergents [1–3].
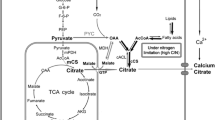
CA, on the other hand, is the product of citrate synthase, an enzyme of the mitochondrial oxidative TCA cycle, while the cytosolic l-malic acid is an intermediate in CA biosynthesis. Currently, A. niger is almost exclusively used to produce CA. However, the yeast Yarrowia lipolytica as the producer has certain advantages over the filamentous fungi (e.g. A. niger) [4]. For example, it has broader substrate spectrum, higher maximal product formation rate, higher substrate concentrations and yield, greater tolerance to metal ions and lower oxygen requirement and simpler process control than A. niger. It also can be easily genetically modified [5]. However, the disadvantage of Y. lipolytica is to produce isocitric acid [3]. Therefore, the reduction in isocitric acid and fatty acid biosynthesis in Y. lipolytica can enhance CA production [3, 6]. Although it has been known that CA production of Y. lipolytica is triggered by an excess of carbon source and the nitrogen limitation conditions, the understanding of the events relevant to CA biosynthesis, accumulation and regulation is not completely understood. Therefore, for an economical aspect, it becomes important to further improve CA production by genetic engineering of Y. lipolytica [6].
Pyruvate carboxylase (PYC: EC6.4.1.1) is a biotin-dependent tetrameric enzyme that catalyzes the carboxylation of pyruvic acid to oxaloacetic acid. It is a key enzyme in the cytosolic reductive TCA pathway rather than in the mitochondrial oxidative TCA cycle [7]. Since it is situated at the branch point of pyruvate metabolism in the cytosol, thus, it is expected that the expression of the PYC gene would be strictly regulated. Only in certain filamentous fungi and in the yeast Saccharomyces cerevisiae, the enzyme including a cytosolic malate dehydrogenase (MDH), and a fumarase (FUM) is situated exclusively in the cytosol because of lack of a mitochondrial-targeting peptide [8]. The cytosolic localization appears to be important for the ability of these organisms to accumulate high concentrations of organic acids by scavenging of unwanted acids in the cytosol. It has been well documented that PYC is associated with malic acid, polymalate, succinic acid, fumaric acid, the aspartate family of amino acids and α-ketoglutaric acid production [9–14]. However, it is still a little known whether it can play a role in CA biosynthesis or not [15].
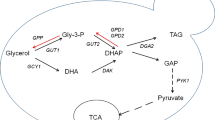
It has been known that pyruvate from glucose can be converted into oxaloacetate by fixing 1 mol of carbon dioxide onto pyruvate under the catalysis of PYC. The formed oxaloacetate and acetyl-CoA are condensed to form CA (Fig. 1). When A. niger was grown in a high-glucose concentration (140.0 g L−1) medium, CA formation is closely related to the PYC reaction [16]. In our previous studies [4–6], it was found that Y. lipolytica SWJ-1b could produce a large amount of CA and after an inulinase gene was over-expressed in Y. lipolytica SWJ-1b, a recombinant yeast strain 30 produced 84.0 g L−1 of CA from 100.0 g L−1 of inulin within 214 h. It also has been reported that the biosynthesis of malic acid and lipid in M. guilliermondii Pcla22 is highly associated with a pyruvate carboxylase [10, 17]. After the PYC gene cloned from M. guilliermondii Pcla22 was over-expressed in Y. lipolytica ACA-DC 50109, the amount of lipid and the secreted citric acid by the recombinant strains was greatly enhanced [18]. Thus, we infer that the PYC gene from the marine yeast M. guilliermondii Pcla22 may play an important role in citric acid biosynthesis. In this study, after the PYC gene from the marine yeast M. guilliermondii Pcla22 which can produce malic acid and lipid was over-expressed in Y. lipolytica SWJ-1b, a recombinant yeast strain was used to produce CA from glucose in the presence of CaCO3.
Materials and methods
Strains and media
The CA producer used in this study was Y. lipolytica SWJ-1b which was isolated from a gut of the marine fish at Bohai Sea [4]. A uracil mutant was isolated from Y. lipolytica SWJ-1b using 5′-FOA (5′-fluororotic acid) [19]. The marine yeast M. guilliermondii Pcla22 that could produce both high level of lipid and malate was used as the source of the PYC gene encoding a cytolic pyruvate carboxylase [12]. Yeast strains were grown in a yeast peptone dextrose (YPD) medium [4]. Yeast transformants were selected on a YNB-N5000 medium [4]. An Escherichia coli strain used in this study was DH5α [F − endA1 hsdR17 (rK_/mK +) supE44 thi − λ − recA1gyr96DlacU169 (j80lacZDM15)] and was grown in a Luria–Bertani broth (LB). The E. coli transformants were grown in the LB medium with 100 μg mL−1 of ampicillin or 30 μg mL−1of kanamycin. A CA production medium (g L−1) contained glucose 60.0, K2HPO4 1.0, KH2PO4 1.0, MgSO4∙ H2O 1.2, yeast extract 0.25, corn steep liquor 0.5, CaCO3 20.0 [4].
Plasmids
A genetically modified expression vector pINA1312-GY which did not contain a DNA fragment encoding the signal sequence was used to express an intracellular enzyme. A pMD-19 T simple vector was purchased from TaKaRa (Japan).
Isolation of DNA, restriction digestions, and transformation
A genomic DNA was isolated from the marine yeast M. guilliermondii Pcla22 and the DNA manipulations were carried out using the standard methods [20]. A bacterial plasmid DNA was purified using a PerfectPrep plasmid minikit (TIANGEN). Restriction endo-nuclease digestions and DNA ligations were performed according to the manufacturer’s recommendations (New England Biolabs, UK). E. coli was transformed with a plasmid DNA according to the E. coli transformation methods described by Sambrook et al. [20]. Y. lipolytica was transformed according to the methods described by Xuan et al. [21].
Construction of a vector for expression of the PYC gene in Y. lipolytica
To express the PYC gene from M. guilliermondii Pcla22 in Y. lipolytica SWJ-1b, the primers for amplification of the PYC gene were designed according to the sequence of the gene (Accession No. XM_001484326.1). The forward primer was PG1 and the reverse primer was PG2 (Table 1 and Supplementary file 1). The genomic DNA of M. guilliermondii Pcla22 was used as the template for PCR. The reaction system contained 1.0 μL of a La Taq Polymerase, 5.0 μL of a 10× La PCR buffer II (Mg2+ Plus), 8.0 μL of 2.5 mM dNTPs, 1.0 μL of 20.0 μM each primer, 1.0 μL of 10 ng mL−1 of the genomic DNA. The final volume was 50.0 μL by supplementation with the sterile distilled water. The conditions for the PCR amplification were initial denaturation at 94 °C for 10 min, denaturation at 94 °C for 1 min, annealing temperature at 55 °C for 1 min, extension at 72 °C for 4 min, final extension at 72 °C for 10 min. PCR was run for 30 cycles. The PCR products (3.6 kb) were separated by an agarose gel electrophoresis and ligated into the plasmid pMD19-T simple vector. The recombinant vector was transformed into E. coli DH5α. The recombinant vectors carrying the PYC gene were extracted from the E. coli transformants and purified. The purified recombinant vectors carrying the whole PYC gene were digested with SacII and SpeI, and the digests were ligated into the plasmid pINA1312-GY digested with the same enzymes (Supplementary file 1). The resulting plasmid carrying the PYC gene was designated as pINA1312-GY-PYC (Supplementary file 1).
Transformation and isolation of a high CA producing transformant
The plasmid pINA1312-GY-PYC obtained above was digested with the enzyme NotI. The fragments carrying the PYC gene were separated in an agarose gel and recovered using a TaKaRa Agarose Gel DNA Purification Kit Ver.3.0. The recovered fragments (6.8 kb) carrying the PYC gene were transformed into the uracil mutant of Y. lipolytica SWJ-1b mentioned above by lithium acetate methods [21]. The transformants obtained were grown on the YNB-N5000 plates without uracil. The positive transformants were cultivated in the CA production medium at 28 °C for 96 h and CA produced by the cells of different positive transformants were determined as described below, respectively, and Y. lipolytica SWJ-1b was used as a control. After determination of CA yields produced by the cells of over 128 positive transformants, it was found that CA yield produced by a transformant PG86 among them was the highest. Therefore, the transformant PG86 was used as the CA producer subsequently.
Confirmation of the integrated PYC gene
The genomic DNAs in the transformant PG86 and Y. lipolytica SWJ-1b were extracted as described above and used as the templates for PCR. The DNA fragments (the PYC gene) were PCR amplified using the primers MGPYC-s/MGPYC-a (Table 1). The sizes of the PCR products were estimated using the Automated Documentation and Analysis System (Gene-Genius, USA). The PCR products were sequenced by the Nanjing Genscript Company [22].
CA production at flask level
The effects of different concentrations (40.0, 50.0, 60.0, 70.0, 80.0, 90.0, 100.0, 110.0 and 120.0 g L−1) of glucose on CA production and changes in reducing sugar by the transformant PG86 were performed by incubating the culture in the CA production medium. The cells of the transformant PG86 were transferred to 50.0 mL of the medium for the seed culture and cultivated at a shaking speed of 180 rpm and 28 °C for 24 h. Two milliliters of the culture (2.5 × 108 cells mL−1) were transferred to 50.0 mL of the CA production medium and the yeast cells were cultivated at 180 rpm and 28 °C for 72 h. The CA concentration and the amount of reducing sugar in the cultures were determined as described below.
Preparation of intracellular extracts
The intracellular extracts of the transformant PG86 and Y. lipolytica SWJ-1b were prepared as described by Zhang et al. [23]. The disrupted cells were centrifuged at 12,000×g and 4 °C for 20 min and the supernatants (the crude intracellular enzymes) obtained were used for the determination of a pyruvate carboxylase activity. Total protein quantity in the supernatant was determined using a coomassie brilliant blue assay [24]. The crude intracellular enzymes heated at 100 °C for 5 min were used as the inactivated intracellular enzymes.
Measurement of the pyruvate carboxylase (PYC) activity
The pyruvate carboxylase (PYC) activity was measured according to the methods described by Bologna [25]. One unit of the PYC activity was defined as the amount of enzyme that catalyzed the oxidization of 1 μmol of NADH per minute.
Fluorescent real-time PCR
A fluorescent real-time PCR was performed according to the methods described by Liu et al. [26]. All the primers used for the fluorescent real-time PCR were designed according to the corresponding gene sequences of M. guilliermondii Pcla22 and Y. lipolytica SWJ-1b. The primers pc1 and pc2 were designed according to the PYC gene sequence in M. guilliermondii, and the primers 26s1 and 26s2 were designed according to the 26S rRNA gene sequence (GenBank Accession No. JQ690257.1) in Y. lipolytica (Table 1). The transcriptional level of the PYC gene in Y. lipolytica SWJ-1b was regarded as 100 %.
CA production by a fed-batch fermentation
CA production by a fed-batch fermentation was carried out in a 10-L fermentor [BIOQ-6005-6010B, Huihetang Bio-Engineering Equipment (Shanghai) Co., Ltd]. The seed culture of the transformant PG86 was prepared as described above. Seven hundred milliliter of the seed culture (OD600nm = 18.0) were transferred to 7.0 L of the CA production medium with initial 60.0 g L−1 of glucose and 20.0 g L−1 of CaCO3. The fermentation was performed under the conditions of an agitation speed of 200 rpm, an aeration rate of 7 L min−1, a temperature of 28 °C and a fermentation period of 288 h. At 96 h of the fermentation, additional 60.0 g L−1 of glucose was supplemented to the fermentation medium. Only 10.0 mL of the culture was collected in the interval of 12 h and was centrifuged at 5000×g and 4 °C for 5 min. The cell mass, reducing sugar and CA in the supernatant obtained were determined as described above. The cell dry weight in 5.0 mL of the culture during the 10-L fermentation was also measured.
Determination of CA
CA was estimated using the methods described by Camp and Farmer [27].
Determination of reducing sugar in the fermented media
Reducing sugar in the fermented media was determined by the Nelson-Somogyi method [28].
Measurement of cell dry weight
Cell dry weight was measured according to the methods described by Chi et al. [29].
HPLC analysis of citric acid
The culture obtained during the fermentation was treated with 1.0 M H2SO4 and the precipitates (CaSO4) formed were removed by centrifugation. The supernatant obtained was suitably diluted. The dilute was analyzed using HPLC (Agilent 1100 series, Germany). First, the CA in the dilute was separated on an Agilent 5 TC-C18 (2) column (5.0 μM, 150 mm × 4.6 mm). The HPLC conditions were that the mobile phase was 0.01 M (NH4)2HPO4 in 10.0 % methanol solution whose pH was adjusted to 2.7 using 1.0 M phosphoric acid and degassed by a microwave; the flow rate was 1.0 mL min−1; the column temperature was 30 °C; the sample volume was 5.0 μL; the detector was a waters 996 Diode-Array Detector; the detection wavelength was 210 nm; the sensitivity was 0.02 AUFS. The pure l-malic acid and CA (Sigma, USA) were used as the standards.
Results
Expression of the PYC gene in the CA-producing yeast Y. lipolytica
The PYC gene from the marine yeast M. guilliermondii Pcla22 which can produce both high level of malate and lipid was cloned and expressed in Y. lipolytica SWJ-1b [4, 17] (Supplementary file 1). After determination of CA produced by the 128 positive transformants grown in the CA production medium at the flask level, it was found that a transformant PG86 among all the transformants and Y. lipolytica SWJ-1b could produce the highest amount of CA (Fig. 2). For example, the concentration of CA secreted by the transformant PG86 in the presence of CaCO3 reached to 54.2 g L−1, resulting in conversion of 45 % of the added glucose to citric acid, while the concentration of CA yielded by Y. lipolytica SWJ-1b under the same conditions was only 27.3 g L−1, leading to the conversion of 22.7 % of the added glucose to citric acid (Fig. 2). This confirmed that the expression of the PYC gene indeed could play an important role in CA biosynthesis in Y. lipolytica SWJ-1b used in this study.
Confirmation of the integrated PYC gene in the genomic DNA of the transformant PG86
It can be observed from the results in Fig. 3 that the PYC gene was amplified from the genomic DNA in the transformant PG86. However, no such PCR products were amplified from the genomic DNA of Y. lipolytica SWJ-1b (Fig. 3). This showed that the PYC gene was indeed integrated into the genomic DNA in the transformant PG86, leading to stable occurrence of the gene in the transformant PG86.
Activity of pyruvate carboxylase and expression of the PYC gene
After the determination of the pyruvate carboxylase activity in the transformant PG86 and Y. lipolytica SWJ-1b, the results in Fig. 4a show that the specific activity of pyruvate carboxylase in the transformant PG86 was 0.38 U mg−1 while that of pyruvate carboxylase in Y. lipolytica SWJ-1b was only 0.07 U mg−1. This meant that the specific activity of pyruvate carboxylase in the transformant PG86 carrying the heterologous PYC gene was much higher than that of pyruvate carboxylase in Y. lipolytica SWJ-1b. At the same time, the transcriptional level of the PYC gene in the transformant PG86 was also much higher than that of the PYC gene in Y. lipolytica SWJ-1b (Fig. 4b).
Optimization of glucose concentration at the flask level
As CaCO3 plays an important role in organic acid biosynthesis, it is important to optimize the concentration of it in the fermentation media. Our results showed that 20.0 g L−1 of CaCO3 in the medium was the most suitable for CA production by the transformant PG86 (data not shown). So, the effects of different concentrations of glucose on citric acid production in the presence of 20.0 g L−1 of CaCO3 by the transformant PG86 were tested. The results in Fig. 5 show that when the glucose concentration in the medium was 60.0 g L−1, CA concentration formed in the fermented medium was 34.02 g L−1, leading to conversion of 56.7 % of the added glucose into citric acid, the yield was 0.57 g g−1 of glucose and when the glucose concentration in the medium was 120.0 g L−1, the CA concentration formed in the fermented medium was 52.85 g L−1, leading to conversion of 44.0 % of glucose into CA, the yield was 0.44 g g−1 of glucose. This meant that the more the added glucose, the more the residual reducing sugar left in the fermented medium. To make more reducing sugar be used by the transformant PG86, initial 60.0 g L−1 of glucose was applied in the following fed-batch fermentation.
Ten-liter fed-batch fermentation
The results in Fig. 5 had revealed that the more the added glucose, the more the residual reducing sugar left in the fermented medium. Therefore, the fed-batch cultivation mode was used for CA production in this study. When the residual glucose concentration reduced to 4.66 g L−1 at 96 h of the fermentation, additional 60.0 g L−1 of glucose was added to the fermentation broth and the fermentation was continued to last for another 192 h (Fig. 6). In this case, a final concentration of CA was 101.0 ± 1.3 g L−1, a yield was 0.89 g g−1 of glucose and a productivity was 0.42 g L−1 h−1 within 240 h of the fed-batch fermentation (Fig. 6). Furthermore, only a trace amount of isocitric acid was detected in the fermented medium (data not shown). The results above demonstrated that the molecular engineering of Y. lipolytica with the PYC gene indeed could enhance production of CA and other organic acids.
HPLC analysis of the fermentation products
The culture obtained during the fed-batch fermentation was treated with H2SO4 and CaSO4 formed was removed by centrifugation. The supernatant obtained was analyzed using HPLC as described in Materials and methods. The results in Supplementary file 2C revealed that most (65.61 %) of the fermentation products were citric acid. However, a considerable amount of malic acid (12.38 %) and other unknown products also appeared in the fermentation broth (Supplementary file 2A, B and C).
Discussion
After the PYC gene from P. guilliermondii Pcla22 was integrated into the genomic DNA and expressed in Y. lipolytica SWJ-1b, the transformant PG86 obtained could produce more CA than Y. lipolytica SWJ-1b. It also has been reported that the addition of 20.0 g L−1 CaCO3 and 0.8 mg L−1 biotin to the culture of Y. lipolytica led to an increase in productivity from 16.6 to 39.2 g L−1 of α-ketoglutaric acid (KGA) [15]. Concentrations of the by-products fumarate (FA), malate (MA), succinic acid (SA) and pyruvic acid (PA) decreased significantly by overproduction of fumarase (FUM), but increased by overproduction of PYC and also of FUM and PYC simultaneously in a recombinant yeast Y. lipolytica. In contrast, 137–147 g L−1 of KGA was produced by a multicopy strain H355A (over-production of FUM1-PYC1) after 93–114 h of cultivation [30]. This meant that in addition to CA biosynthesis, PYC also played an important role in biosynthesis of other organic acids appeared in the TCA cycle. The specific activity of the PYC and the transcriptional level of the PYC gene in the transformant PG86 were much higher than that of the PYC and that of the PYC gene in Y. lipolytica SWJ-1b. It also has been reported that overexpression of the fumarase (FUM) or pyruvate carboxylase (PYC) genes in Y. lipolytica resulted in strongly increased specific enzyme activities during cultivation of these strains on raw glycerol [30].
It can be seen from Fig. 1 that CO2 is required for carboxylation of pyruvate to oxaloacetate under catalysis of PYC. It has been reported that during organic acid biosynthesis, CaCO3 is the best CO2 source among the carbonates because CaCO3 plays an important role in the organic acid biosynthesis by keeping pH constant of around 6.5 and providing CO2 as a substrate for efficient production of organic acid [7]. The results also have shown that calcium is involved in cellular signaling pathways and influences pyruvate carboxylase activity [31]. After optimization of the concentrations of CaCO3 and glucose, the results showed that 20.0 g L−1 CaCO3 and 60.0 g L−1 glucose were the most suitable for CA production by the transformant PG86. During the fed-batch fermentation, the final concentration of CA, the CA yield and the CA productivity were 101.0 ± 1.3 g L−1, 0.89 g g−1 and 0.42 g L−1 h−1 within 240 h, respectively. It has been reported that 68.9 g L−1 of CA and 4.1 g L−1 of iso-citric acid were produced by the yeast strain expressing an inulinase within 312 h of the 2-L fermentation [4]. During a 2-L fermentation, 84.0 g L−1 of CA and 1.8 g L−1 of iso-citric acid were attained from 100.0 g L−1 of inulin within 214 h by the transformant 30 in which the inulinase gene and the iso-citrate lyase gene (ICL1) were expressed and some of the ATP-citrate lyase genes (ACL1) were deleted [6]. The maximal CA amount of 127–140 g L−1 with a yield of 0.75–0.82 g g−1 were reached in a fed-batch cultivation process on sucrose with a recombinant yeast Y. lipolytica H222-S4 in which the invertase encoding ScSUC2 gene was expressed [2]. A recombinant Y. lipolytica strain H222-S4(p67ICL1)T5, harboring an invertase encoding ScSUC2 gene from Saccharomyce cerevisiae and the multiple ICL1 gene copies (10–15) could produce 127–140 g L−1 CA with a yield of 0.75–0.82 g g−1 within 200 h of the cultivation [3]. Yin et al. [11] reported that a recombinant yeast Y. lipolytica-RoPYC2 overexpressing a heterologous pyruvate carboxylase gene could yield a maximum concentration of 62.5 g L−1 KGA in a pH-controlled 3-L fermenter in which an evident decrease in pyruvate yield from 35.2 to 13.5 g L−1 [11] took place. Furthermore, citrate production through gene insertion in A. niger was also enhanced [32]. This demonstrated that after overexpression of the PYC gene, CA production by the recombinant yeast strain PG86 was also greatly enhanced.
The fermentation products produced by the transformant PG86 included citric acid (major), malic acid (minor) and other unknown organic acids (minor). This suggested that the pathway shown in Fig. 1 is correct and some malic acid formed during the CA biosynthesis in the transformant PG86 overexpressing the PYC gene could not be transformed into CA. Because the pyruvate formed during the glycolysis can be actively converted into oxaloacetate by fixing 1 mol of carbon dioxide onto pyruvate under the catalysis of the enhanced PYC activity in the transformant PG86, the oxaloacetate formed was reduced to l-malic acid. After the l-malic acid entered the TCA cycle, it was oxidized to oxaloacetate with which acetyl-CoA react to be condensed to form CA. In fact, in the presence of CaCO3, the mixture of calcium malate and calcium citrate was formed during the fermentation. Therefore, the transformant PG86 may have highly potential applications in food and pharmaceutical industries.
References
Karaffa L, Kubicek CP (2003) Aspergillus niger citric acid accumulation: do we understand this well working black box? Appl Microbiol Biotechnol 61:189–196
Förster A, Aurich A, Mauersberger S, Barth G (2007) Citric acid production from sucrose using a recombinant strain of the yeast Yarrowia lipolytica. Appl Microbiol Biotechnol 75(6):1409–1417
Förster A, Jacobs K, Juretzek T, Mauersberger S, Barth G (2007) Overexpression of the ICL1 gene changes the product ratio of citric acid production by Yarrowia lipolytica. Appl Microbiol Biotechnol 77(4):861–869
Liu XY, Chi Z, Liu GL, Wang F, Madzak C, Chi ZM (2010) Inulin hydrolysis and citric acid production from inulin using the surface-engineered Yarrowia lipolytica displaying inulinase. Metab Eng 12:469–476
Stottmeister U, Hoppe K, Ruttloff H (1991) Organische Genußsäuren. Lebensm Biotechnol Entwickl Asp 1:516–547
Liu XY, Chi Z, Liu GL, Madzak C, Chi ZM (2013) Both decrease in ACL1 gene expression and increase in ICL1 gene expression in marine-derived yeast Yarrowia lipolytica expressing INU1 gene enhance citric acid production from inulin. Mar Biotechnol 15(1):26–36
Chi Z, Wang ZP, Wang GY, Khan I, Chi ZM (2015) Microbial biosynthesis and secretion of l-malic acid and its applications. Crit Rev Biotechnol 41(2):228–237
Jitrapakdee S, St Maurice M, Rayment I, Wallace Cleland WW, John C, Wallace JC, Attwood PV (2008) Structure, mechanism and regulation of pyruvate carboxylase. Biochem J 413:369–387
Li Y, Chi Z, Wang GY, Wang ZP, Liu GL, Lee CF, Ma ZC, Chi ZM (2015) Taxonomy of Aureobasidium spp. and biosynthesis and regulation of their extracellular polymers. Crit Rev Microbiol 41(2):228–237
Brown SH, Bashkirova L, Berka R, Chandler T, Doty T, McCall K, McCulloch M, McFarland S, Thompson S, Yaver D, Berry A (2013) Metabolic engineering of Aspergillus oryzae NRRL 3488 for increased production of l-malic acid. Appl Microbiol Biotechnol 97(20):8903–8912
Yin X, Madzak C, Du G, Zhou J, Chen J (2012) Enhanced alpha-ketoglutaric acid production in Yarrowia lipolytica WSH-Z06 by regulation of the pyruvate carboxylation pathway. Appl Microbiol Biotechnol 96(6):1527–1537
Peleg Y, Battat E, Scrutton MC, Goldberg I (1989) Isoenzyme pattern and subcellular localisation of enzymes involved in fumaric acid accumulation by Rhizopus oryzae. Appl Microbiol Biotechnol 32(3):334–339
Lin H, San KY, Bennett GN (2005) Effect of Sorghum vulgare phosphoenolpyruvate carboxylase and Lactococcus lactis pyruvate carboxylase coexpression on succinate production in mutant strains of Escherichia coli. Appl Microbiol Biotechnol 67(4):515–523
Peters-Wendisch PG, Schiel B, Wendisch VF, Katsoulidis E, Mockel B, Sahm H, Eikmanns BJ (2001) Pyruvate carboxylase is a major bottleneck for glutamate and lysine production by Corynebacterium glutamicum. J Mole Microbiol Biotechnol 3(2):295–300
Zhou J, Zhou H, Du G, Liu L, Chen J (2010) Screening of a thiamine-auxotrophic yeast for α-ketoglutaric acid overproduction. Lett Appl Microbiol 51(3):264–271
Peksel A, Torres N, Liu J, Juneau G (2002) 13C-NMR analysis of glucose metabolism during citric acid production by Aspergillus niger. Appl Microbiol Biotechnol 58(2):157–163
Wang GY, Chi Z, Song B, Wang ZP, Chi ZM (2012) High level lipid production by a novel inulinase-producing yeast Pichia guilliermondii Pcla22. Bioresour Technol 124:77–82
Wang GY, Zhang Y, Chi Z, Liu GL, Wang ZP, Chi ZM (2015) Role of pyruvate carboxylase in accumulation of intracellular lipid of the oleaginous yeast Yarrowia lipolytica ACA-DC 50109. Appl Microbiol Biotechnol 99(4):1637–1645
Wang F, Yue L, Wang L, Madzak C, Li J, Wang XH, Chi ZM (2009) Genetic modification of the marine-derived yeast Yarrowia lipolytica with high-protein content using a GPI-anchor-fusion expression system. Biotechnol Prog 25(5):1297–1303
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory, Beijing, pp 367–370 (Chinese translated edn)
Xuan JW, Fournier P, Gaillardin C (1988) Cloning of the LYS5 gene encoding saccharopine dehydrogenase from the yeast Yarrowia lipolytica by target integration. Curr Genet 14(1):15–21
Zhao CH, Cui W, Liu XY, Chi ZM, Madzak C (2010) Expression of inulinase gene in the oleaginous yeast Yarrowia lipolytica and single cell oil production from inulin-containingmaterials. Metab Eng 12(6):510–517
Zhang F, Wang ZP, Chi Z, Raoufi Z, Abdollahi S, Chi ZM (2013) The changes in Tps1 activity, trehalose content and expression of TPS1 gene in the psychrotolerant yeast Guehomyces pullulans 17-1 grown at different temperatures. Extremophiles 17(2):241–249
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72(1):248–254
Bologna FP, Andreo CS, Drincovich MF (2007) Escherichia coli malic enzymes: two isoforms with substantial differences in kinetic properties, metabolic regulation, and structure. J Bacteriol 189(16):5937–5946
Liu GL, Wang DS, Wang LF, Zhao SF, Chi ZM (2011) Mig1 is involved in mycelial formation and expression of the genes encoding extracellular enzymes in Saccharomycopsis fibuligera A11. Fungal Genet Biol 48(9):904–913
Camp BJ, Farmer L (1967) A rapid spectrophotometric method for the determination of citric acid in blood. Clin Chem 13(6):501–505
Spiro RG (2001) Analysis of sugars found in glycoproteins. Methods Enzymol 8:3–26
Chi Z, Liu J, Zhang W (2001) Trehalose accumulation from soluble starch by Saccharomycopsis fibuligera sdu. Enzyme Microb Technol 28(2):240–245
Otto C, Yovkova V, Aurich A, Mauersberger S, Barth G (2012) Variation of the by-product spectrum during α-ketoglutaric acid production from raw glycerol by overexpression of fumarase and pyruvate carboxylase genes in Yarrowia lipolytica. Appl Microbiol Biotechnol 95(4):905–917
Zelle RM, de Hulster E, van Winden WA (2008) Malic acid production by Saccharomyces cerevisiae: engineering of pyruvate carboxylation, oxaloacetate reduction, and malate export. Appl Environ Microbiol 74(9):2766–2777
de Jongh WA, Nielsen J (2008) Enhanced citrate production through gene insertion in Aspergillus niger. Metab Eng 10:87–96
Acknowledgments
This study was supported by Natural Science Foundation of Shandong Province, the Grant Number is ZR2014CZ001.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
449_2016_1607_MOESM2_ESM.doc
Supplementary material 2: Analysis of the fermentation products (C), standard CA (B) and l-malic acid (A) by using HPLC. (DOC 291 kb)
Rights and permissions
About this article
Cite this article
Tan, MJ., Chen, X., Wang, YK. et al. Enhanced citric acid production by a yeast Yarrowia lipolytica over-expressing a pyruvate carboxylase gene. Bioprocess Biosyst Eng 39, 1289–1296 (2016). https://doi.org/10.1007/s00449-016-1607-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00449-016-1607-8