Abstract
Transportation of Cryptosporidium oocysts in river type source water is of great concern in an area where extensive human activities exist. In this study, a total of 47 samples were collected from Tongxiang, China, where drinking source water was taken from a complicated river network system, by three sampling campaigns over a rainy season in 2009, to reveal the presence, genotypes, and likely source of Cryptosporidium oocysts in river water. Immunofluorescence microscopy analyses show that 37 (78.7%) were Cryptosporidium positive, with a mean concentration of 0.51 oocysts per liter. These results suggest that the protozoa were commonly distributed in the river network type source water of Tongxiang with a relatively low concentration level. PCR analysis was used to determine the species/genotypes of Cryptosporidium, which revealed the presence of the animal related species/genotypes including Cryptosporidium suis, Cryptosporidium fragile, and the avian III, pig II, cervine genotypes. Three of them were also detected in wastewater samples taken from neighboring animal farms, showing that farm animals rather than human might be the major pollution sources. This is the first report on simultaneous detection and genotyping of Cryptosporidium oocysts from surface water in China.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Cryptosporidium spp. are common protozoa that have caused many waterborne outbreaks which affected hundreds of thousands of people (Karanis et al. 2007a). Until now, at least 12 outbreaks of cryptosporidiosis associated with contamination of river-type source water have been reported (Karanis et al. 2007a). In Zhejiang Province southern-eastern China, a relatively high occurrence of Cryptosporidium in diarrheal children has been found in this region (Lu et al. 2000), which is known for its high-population density and prosperous livestock and poultry husbandry industries, possesses abundant of rivers connecting to each other, used for multipurposes including transportation canals, drinking water source, and receiving water bodies of wastewater.
In fact, river systems, which have long been utilized as an important water source in many countries, are in general affected by human activities most intensively, and are difficult to protect due to their openness. Especially in regions with a complicated river network system, all of the rivers are connected together, making it very difficult to establish a barrier to protect source water effectively. Moreover, oocysts cannot be completely removed/inactivated by conventional water treatment practices in waterworks (Karanis et al. 2007a; Hijnen et al. 2007; Montemayor et al. 2008). Therefore, it is urgent for the protection of drinking water resource to survey on the occurrence of Cryptosporidium in the river network.
In addition, the species/genotype of Cryptosporidium is another important factor to affect the outbreak of cryptosporidiosis, which has been often ignored in the previous study about the risk assessment of Cryptosporidium in drinking water. In fact, not all species/genotypes of Cryptosporidium can infect human (Smith and Nichols 2010). Consequently, understanding the source and species/genotypes of Cryptosporidium in source water is necessary to identify public health risk and take appropriate preventative measures.
To provide some information on contamination of Cryptosporidium in a complicated river network type source water region, we performed this study to determine the occurrence and genotypes of Cryptosporidium in drinking water source over a rainy season in 2009 (three sampling campaigns in June, August, and October, respectively). Our study is believed to be the first attempt to simultaneous detection and genotyping of Cryptosporidium oocyst from source water in China and will be helpful for the management of source water where human activities are intensive.
Materials and methods
Water sampling sites
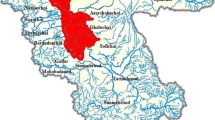
Tongxiang (30°28' to 30°47′ N, 120°37′ to 120°39′ W), located in Zhejiang Province, is intersected by the Beijing-Hangzhou Grand Canal, and 6.73% of its surface is covered by water. Besides the Canal, there are 46 main rivers running through the region, most of which are intersecting with the Canal, forming a complicated water network. The livestock and poultry breeding industry, particular the sheep, pig, and duck breeding, is one of the important industries in the region. The population density was 1,383 inhabitants per square kilometer in 2008 (http://www.txsz.gov.cn/nianjian/). The Canal and some other rivers, which are still used for transportation, are used as major source waters for approximately one million people in Tongxiang.
A total of 47 water samples were collected from the sites shown in Fig. 1 for the detection of Cryptosporidium oocysts. Firstly, 25 10-L water samples were collected in June 2009. Based on the detection results of the samples in June, 22 20-L samples were then collected for both microscopy and molecular analyses (10 L for each) in August and October 2009. Besides the river water samples, three 50-mL wastewater samples were collected from nearby duck, pig, and sheep farms, respectively, for molecular analyses in the last campaign.
Sample collection and filtration
Clean plastic tanks were used for collection of water from approximately 30 cm below the water's surface at the middle of the rivers. According to the method described by Aldom and Chagla (1995), samples were filtered through a filter holder with a mixed cellulose ester membrane filter (diameter, 142 mm; pore size, 1 μM; Advantec MFS, Inc. Japan) with a peristaltic pump on the inlet side of the filter holder according to the recommendation of the manufacturer. The typical flow rate is 0.3 L/min. After filtration, the membrane filter was transferred to a fresh screw-capped conical 50-mL centrifuge tube and transported on ice to the laboratory for further processing within 72 h (US Environmental Protection Agency (USEPA) 2005).
Sample elution, concentration, and purification
The membrane filters were dissolved in acetone solutions to recover oocysts/cysts as described previously (Aldom and Chagla 1995). Briefly, the membrane filter was dissolved completely in 40 mL of acetone with vigorous shaking and successively centrifuged at 1,050×g for 15 min at 4°C and resuspended in acetone, 10% ethanol, and phosphate-buffered saline (PBS, pH 7.4). The packed pellets were resuspended in a suitable volume of PBS buffer containing 0.01% Tween 80 (PBST).
The concentrated oocysts were separated from other particulate materials by flotation on Percoll-sucrose gradients, as described previously (Hashimoto et al. 2002). Briefly, the particulate suspension (3 mL) was transferred to a 15-mL conical centrifuge tube and underlaid with approximately 5 mL of Percoll-sucrose solution (specific gravity, 1.10), using a capillary pipette. The Percoll-sucrose gradients were centrifuged at 20°C in a swinging bucket rotor at 1,050×g for 10 min. The entire fluid layer above the pellet in each flotation tube was harvested by pipette and transferred into a fresh 50-mL conical centrifuge tube. To avoid losses that might occur, the pellet was resuspended in 3 mL of PBST, repurified, harvested, and transferred into the same conical centrifuge tube, and the volumes were adjusted to 40 mL with additional PBS buffer. The harvests were pelleted by centrifugation at 1,050×g for 10 min at 20°C. Supernatants were aspirated off, leaving a 3-mL concentrated sample solution, including the pellets.
Sample staining, examination, and quality assurance
The method for sample staining was according to a previous study (Nieminski et al. 1995). Twenty-five-millimeter diameter, 3.0-μm-pore-size cellulose acetate membrane filters (Advantec MFS, Inc. Japan), which were marked in the middle with a circle, using a DAKO pen (Dako North America, Inc. California, USA) to designate the area of inoculation, were wetted in PBS, and then, placed onto a glass filter holder. The purified sample was inoculated within the encircled area for filtration and washed twice by passing 1 mL of PBS for each. Then 0.5 mL of 1% bovine serum albumin (BSA) in PBS was applied and incubated for 1 min. The membrane filter was transferred on a fresh plane glass slide and stained with a combined fluorescein isothiocyanate (FITC) conjugated anti-Cryptosporidium monoclonal antibody (Waterborne, Inc., New Orleans, LA). An aliquot (100 μL) of antibody was applied to the filter and the slide was placed in a humid chamber in the dark. After incubation at room temperature for 30 min, 50 μL of 4′,6′-diamidino-2-phenylindole (DAPI) was applied and incubated for 10 min. The membrane filter was transferred back onto the filter holder, rinsed with 2 mL of PBS, and then treated sequentially with 1 mL of 30%, 70%, and 90% ethanol solutions containing 5% glycerol. Positive and negative staining controls were routinely included according to the recommendations of the manufacturer.
For microscopic observation, the membranes were mounted on glass microscope slides with 75 μL of 1,4-diazabicyclo[2.2.2]octane (DABCO)-glycerol mounting medium and sealed with coverslips and nail varnish. The entire encircled area of the membrane was systematically examined using fluorescence microscopy (Olympus BX51, USA) for the FITC, DAPI fluorescence and differential interference contrast (DIC) examination. Cryptosporidium spp. oocysts were identified on the basis of their size, shape, and internal structures. Only those meeting the morphological criteria of oocysts, as described in method 1623 (USEPA 2005), were regarded as positive and recorded. The calculated numbers were used to extrapolate to concentrations of oocysts per liter of sample.
To assure acceptable method performance, initial precision and recovery (IPR) tests were included prior to the initiation of field sampling and method blanks were included with each sampling campaign as recommended in method 1623 (USEPA 2005). For the IPR tests, four replicate trials were performed, 10 L of reagent water was spiked directly with Cryptosporidium parvum oocyst (Iowa isolate, approximately 1 month old; supplied by Waterborne Inc., New Orleans, LA, USA), and then the seeded reagent water was further treated using the method described above.
Data of the prevalence of the parasites in three campaigns were tested using χ 2 test, whereas the concentration in campaigns and different sections of the Beijing-Hangzhou Grand Canal were tested using Kruskal–Wallis test. Differences with P values of <0.05 were defined as being statistically significant.
Preparation of DNA templates
Molecular analysis was performed on all water samples collected in August and October 2009. Genomic DNA was extracted from Percoll-sucrose flotation-purified oocysts of concentrated water samples using the FastDNA SPIN kit for soil (MP Biomedicals, France), according to the manufacturer's instructions, with the addition of six freeze-thaw cycles (liquid nitrogen 2 min/56°C 5 min) after resuspension in lysis solution in order to rupture the Cryptosporidium oocysts. DNA was eluted in 50–100 μL buffers and stored at −20°C for further processing (Jiang et al. 2005a).
After concentration by centrifugation, the three wastewater samples from animal farms were directly submitted for DNA extraction as described above.
PCR assay
A nested PCR was performed using the primers and PCR conditions published by Jiang et al. (2005b) to amplify an 840-bp fragment of the small subunit (SSU) rRNA gene of Cryptosporidium. Briefly, primary PCR amplifications were performed in 25-μL volumes using Taq polymerase with 1× Taq buffer (TaKaRa, Dalian, China) and containing 1.5 mM MgCl2, 200 μM of each diethylnitrophenyl thiophosphate (dNTP), 0.4 mg/mL BSA, 0.1 μM primers of SSU-F2 (5′-TTC TAG AGC TAA TAC ATG CG-3′) and SSU-R2 (5′-CCC ATT TCC TTC GAA ACA GGA-3′). After an initial denaturation at 94°C for 7 min, the cycling program consisted of 35 cycles of denaturation at 94°C for 45 s, annealing at 55°C for 45 s, and primer extension at 72°C for 1 min. A final extension cycle at 72°C for 7 min was followed by soaking at 4°C. Three PCR replicates per sample, using 1–3 μL of extracted DNA per PCR, were performed to increase the accuracy of Cryptosporidium oocyst detection (Xiao et al. 2006). The secondary nested PCR was performed in a total volume of 50 μL using Taq polymerase with 1× Taq buffer, 1.5 mM MgCl2, 200 μM of each dNTP, and 0.2 μM of each primer SSU-F3 (5′-GGA AGG GTT GTA TTT ATT AGA TAA AG-3′) and SSU-R4 (5′-CTC ATA AGG TGC TGA AGG AGT A-3′). One microliter of primary PCR product was used as the template for the secondary nested PCR. Cycling parameters were identical to the first round amplification with the exception of the annealing temperature, which was increased to 58°C. The nested PCR products were analyzed after electrophoresis in 1.5% agarose gels and staining with 0.2 μg/mL of ethidium bromide and visualized on a UV transilluminator.
Sequence and phylogenetic analysis
The secondary PCR products positive for Cryptosporidium were purified with DNA UNIQ-5 spin column purification kit (Tiangen, China) and cloned into the pMD18-T plasmid vector by using a cloning kit (TaKaRa, China), according to the manufacturer's recommendations. Clones were sent to Beijing Augct Co., Ltd. for sequencing using ABI 3730 automated DNA sequencer (BigDye Terminator Chemistry). Nucleotide sequences were read using the software Chromas (http://www.technelysium.com.au). The obtained sequences, with other reference sequences downloaded from the GenBank database, were aligned using the Clustal W (http://www.clustal.org/) programs. Phylogenetic trees were constructed using MEGA version 4.0 (MEGA4.0: Molecular Evolutionary Genetics Analysis software, Arizona State University, Tempe, Arizona, USA). Bootstrap analyses were conducted using 1,000 replicates to assess the reliability of inferred tree topologies. Neighbor-joining algorithms were conducted using the Kimura 2 parameter distance analysis.
Results and discussion
Initial precision and recovery of Cryptosporidium parvum oocysts
In general, the detection of Cryptosporidium oocysts in water includes several sequential steps: sample collection and concentration, separation of oocyst from contaminating debris, staining, and microscopic examination. The cartridge/foam filter and immunomagnetic separation, for their quickly filtration and specificity, were recommended for Cryptosporidium detection by the method 1623 of the USEPA, but their high cost makes it difficult to conduct environmental survey in large scale. Meanwhile, many studies have noted declines in Cryptosporidium and Giardia recovery levels to near or below minimum acceptable levels in waters of high turbidity or high total suspended solid (Krometis et al. 2009; Feng et al. 2003; Hu et al. 2004). It is possible that Cryptosporidium oocysts, which could be adhered to particles, were still retained in the filter after elution process (Feng et al. 2003). So in this study, we employed a concentration and purification method adopted by Japan including cellulose ester membrane filtration, acetone dissolution, and Percoll-sucrose flotation in succession (Japanese Ministry of Health Labor and Welfare 2007). As shown in Table 1, this method permitted a mean recovery and relative standard deviation (RSD) of 41.25% and 21.54% for initial precision and recovery tests, respectively, which could meet the acceptance criteria of method 1623 (24% to 100% recovery with a maximum RSD of 55%).
Occurrence of Cryptosporidium species oocysts in river water samples
A total of 47 river water samples were examined by microscopy in this study, with the parasites detected in 37 (78.7%) samples (Table 2). In this study, the oocyst concentrations were calculated without correction using the recovery efficiency of the detection method (World Health Organization (WHO) 2009). This was because the recovery efficiencies for different samples could vary greatly due to the difference of water quality in different sites (Keeley and Faulkner 2008). The detection percentages of the oocysts of Cryptosporidium did not change much (63.6–84.0%) for the three sampling campaigns (χ 2 = 1.97; P > 0.05). The geometric mean of Cryptosporidium was 0.51 oocysts per liter. The prevalence and concentrations of Cryptosporidium oocysts in different sections of the Beijing-Hangzhou Grand Canal (Tongxiang section) are shown in Table 3. Of the 25 samples in the first campaign, 100% of the upstream and midstream samples and 73.3% of the downstream ones were contaminated by oocyst. The oocyst concentrations of the midstream and upstream samples were much higher than those of downstream ones (P < 0.05).
Cryptosporidium oocysts were detected with a relatively high percentage during the three sampling campaigns, suggesting that these parasites had been discharged into the rivers continuously. High prevalence of Cryptosporidium oocysts in river water was also obtained in some other studies conducted in the United States (99%) (Keeley and Faulkner 2008), Spain (63.5%) (Carmena et al. 2007), Netherlands (100%) (Schets et al. 2008), Brazil (100%) (Franco et al. 2001), Taiwan (72.2%) (Hsu et al. 1999), and Japan (100%) (Hashimoto et al. 2002). According to our field investigation, animal breeding activities are extensive near the rivers which flow into the midstream section of the Beijing-Hangzhou Grand Canal (Fig. 1), and the use of animal feces as natural fertilizer is common in this area. So maybe that's why the abundance of Cryptosporidium in the midstream section was much higher than the other sections along the Grand Canal.
Previous studies have reported that heavy rainfall is likely to cause short periods of heavy contamination of oocysts in surface waters (Hansen and Ongerth 1991). In the studied area, the rainy season in 2009 lasted from June to September (http://www.jxwater.gov.cn/). The stronger washing effects of rainfalls in the beginning of the rainy season might be a reason for the higher Cryptosporidium concentrations observed in the first campaign. Nevertheless, the concentrations of Cryptosporidium oocysts were in a relatively low level for the three campaigns with 20/37 (54.1%) positive samples having less than five oocysts in 10-L samples. These concentration levels of the parasites were in accordance with some previous studies in China (Cai et al. 2007; Sun et al. 2007) and Japan (Hashimoto et al. 2002), but much lower than other studies (Srisuphanunt et al. 2010; Castro-Hermida et al. 2008).
Molecular identification of parasites
PCR amplification was performed for Cryptosporidium on all water samples collected in the last two campaigns, and the expected size of amplicon at the SSU rRNA gene locus was produced in 27.3% (3 out of 11) and 36.4% (4 out of 11) water samples collected in August and October, respectively. All of the seven positive amplicons were successfully cloned, and at least two clones from each sample were sequenced. According to the sequence analysis result, the presence of the following five species/genotypes of Cryptosporidium was confirmed: Cryptosporidium suis (samples LIULIGANG and JINFENGQIAO), Cryptosporidium fragile (sample CONGCHANGGANG), and the avian III (samples ZHUANGQIANQIAO and DAGAOYANGGANG), pig II (sample ZHUCHANGFUJIN), and cervine genotype (sample YANGCHANGRUKOU) (Fig. 2). In light of the host-specific nature of Cryptosporidium species, all the above Cryptosporidium species/genotypes were animal related. At the same time, all of the three wastewater samples were successfully amplified, cloned, and sequenced. The avian III and pig II genotypes were detected in the duck and pig farm, respectively, while the cervine genotype and Cryptosporidium bovis were present at the sheep farm.
Phylogenetic position of Cryptosporidium isolates inferred by a neighbor-joining analysis of the partial SSU rDNA sequences. The evolutionary distances between sequences were calculated by the Kimura two-parameter model. A sequence of Eimeria tenella (AF026388) was used as the out-group. Values on the branches are percent bootstrapping values (>50%) using 1,000 replicates. GenBank accession numbers of the sequences used in the construction of the phylogenetic tree are listed following species or genotypes
PCR has been considered as a sensitive method for detecting oocysts in environmental samples (Xiao et al. 2001). However, nine samples were microscopically positive but negative by PCR in this study. Similar results were also obtained in some other studies conducted in Hungary (Plutzer et al. 2008), Spain (Castro-Hermida et al. 2008), France (Coupe et al. 2006), and Finland (Hanninen et al. 2005). This low ratio of positive PCR result may be related to the persistence of potent inhibitor or other interfering compounds in the DNA eluate or to the heterogenous distribution of low oocyst numbers in the samples.
Among the 13 Cryptosporidium species/genotypes reported to be responsible for human cryptosporidiosis cases, five species including Cryptosporidium hominis, C. parvum, Cryptosporidium meleagridis, Cryptosporidium felis, and Cryptosporidium canis have most often been found to cause human infections, while the others are occasionally found in humans (Xiao 2010). However, none of the detected Cryptosporidium in river water in this study belongs to the five human infective species. Similar results have also been reported in previous studies (Ruecker et al. 2007). At the same time, as shown in Fig. 2, three of the five species/genotypes detected in the river water samples could be found in duck, pig, and/or sheep farms (the avian III, pig II, and cervine genotypes). Consequently, it is concluded that the animal breeding industry should be the major contamination sources of Cryptosporidium oocysts in the studied region. However, it should be noted that among the five animal originated species/genotypes detected in this study, the Cryptosporidium suis has been reported in patients in Peru and England (Leoni et al. 2006) and in HIV-infected patients in China (Wang et al. 2010a), and the cervine genotype Cryptosporidium has been reported in patients in Canada (Trotz-Williams et al. 2006), the United Kingdom (Ong et al. 2002), the United States (Feltus et al. 2006), and Slovenia (Soba et al. 2006). Besides humans, the cervine genotype has also been found in a wide host range including wildlife, zoo, and farm animals (Karanis et al. 2007b) and could possibly emerge as an important human pathogen with increasing contact between humans and animals. In China, this genotype was firstly found in ibex (Karanis et al. 2007b) and was later demonstrated to be the major Cryptosporidium genotype in sheep (Wang et al. 2010b). So, the impacts of these Cryptosporidium species/genotypes to human health should not be neglected, and it is recommended to improve the management of wastes and wastewater from the animal farms.
Conclusions
Results of this study show that Cryptosporidium oocysts were widely discharged into the rivers in Tongxiang continuously, with a relatively low concentration level. Species/genotypes analysis revealed that animal husbandries were probably the major pollution sources. Cryptosporidium infection is not likely to transmit by the drinking water in the studied region. Nevertheless, these results also can be serving to an alert for other areas where source water maybe contaminated by oocysts from human contamination. Consequently, continued environmental, epidemiological, and molecular studies of Cryptosporidium are necessary to develop appropriate source water protection plans in China.
References
Aldom JE, Chagla AH (1995) Recovery of Cryptosporidium oocysts from water by a membrane filter dissolution method. Lett Appl Microbiol 20:186–187
Cai J, Ye J, Du HL, Hu XW, Liu J, Huang W, Cui X (2007) Investigation of Cryptosporidium and Giardia contaminating drinking-water in Chengdu. Chin J Health Lab Tech 17:2165–2167
Carmena D, Aguinagalde X, Zigorraga C, Fernandez-Crespo JC, Ocio JA (2007) Presence of Giardia cysts and Cryptosporidium oocysts in drinking water supplies in northern Spain. J Appl Microbiol 102:619–629
Castro-Hermida JA, Garcia-Presedo I, Almeida A, Gonzalez-Warleta M, Correia Da Costa JM, Mezo M (2008) Presence of Cryptosporidium spp. and Giardia duodenalis through drinking water. Sci Total Environ 405:45–53
Coupe S, Delabre K, Pouillot R, Houdart S, Santillana-Hayat M, Derouin F (2006) Detection of Cryptosporidium, Giardia and Enterocytozoon bieneusi in surface water, including recreational areas: a one-year prospective study. FEMS Immunol Med Microbiol 47:351–359
Feltus DC, Giddings CW, Schneck BL, Monson T, Warshauer D, McEvoy JM (2006) Evidence supporting zoonotic transmission of Cryptosporidium spp. in Wisconsin. J Clin Microbiol 44:4303–4308
Feng YY, Ong SL, Hu JY, Song LF, Tan XL, Ng WJ (2003) Effect of particles on the recovery of Cryptosporidium oocysts from source water samples of various turbidities. Appl Environ Microbiol 69:1898–1903
Franco RM, Rocha-Eberhardt R, Cantusio Neto R (2001) Occurrence of Cryptosporidium oocysts and Giardia cysts in raw water from the Atibaia River, Campinas, Brazil. Rev Inst Med Trop Sao Paulo 43:109–111
Hanninen MJ, Hormann A, Rimhanen-Finne R, Vahtera H, Malmberg S, Herve S, Lahti K (2005) Monitoring of Cryptosporidium and Giardia in the Vantaa river basin, southern Finland. Int J Hyg Environ Health 208:163–171
Hansen JS, Ongerth JE (1991) Effects of time and watershed characteristics on the concentration of Cryptosporidium oocysts in river water. Appl Environ Microbiol 57:2790–2795
Hashimoto A, Kunikane S, Hirata T (2002) Prevalence of Cryptosporidium oocysts and Giardia cysts in the drinking water supply in Japan. Water Res 36:519–526
Hijnen WA, Dullemont YJ, Schijven JF, Hanzens-Brouwer AJ, Rosielle M, Medema G (2007) Removal and fate of Cryptosporidium parvum, Clostridium perfringens and small-sized centric diatoms (Stephanodiscus hantzschii) in slow sand filters. Water Res 41:2151–2162
Hsu BM, Huang C, Jiang GY, Hsu CL (1999) The prevalence of Giardia and Cryptosporidium in Taiwan water supplies. J Toxicol Environ Health A 57:149–160
Hu J, Feng Y, Ong SL, Ng WJ, Song L, Tan X, Chu X (2004) Improvement of recoveries for the determination of protozoa Cryptosporidium and Giardia in water using method 1623. J Microbiol Methods 58:321–325
Japanese Ministry of Health Labor and Welfare (2007) Methods for the detection of Cryptosporidium, Giardia and indicator microorganisms in water supply systems. http://www.mhlw.go.jp/topics/bukyoku/kenkou/suido/kikikanri/dl/ks-0330006.pdf. Accessed Mar. 29 2011
Jiang J, Alderisio KA, Singh A, Xiao L (2005a) Development of procedures for direct extraction of Cryptosporidium DNA from water concentrates and for relief of PCR inhibitors. Appl Environ Microbiol 71:1135–1141
Jiang J, Alderisio KA, Xiao L (2005b) Distribution of Cryptosporidium genotypes in storm event water samples from three watersheds in New York. Appl Environ Microbiol 71:4446–4454
Karanis P, Kourenti C, Smith H (2007a) Waterborne transmission of protozoan parasites: a worldwide review of outbreaks and lessons learnt. J Water Health 5:1–38
Karanis P, Plutzer J, Halim NA, Igori K, Nagasawa H, Ongerth J, Liqing M (2007b) Molecular characterization of Cryptosporidium from animal sources in Qinghai province of China. Parasitol Res 101:1575–1580
Keeley A, Faulkner BR (2008) Influence of land use and watershed characteristics on protozoa contamination in a potential drinking water resources reservoir. Water Res 42:2803–2813
Krometis LAH, Characklis GW, Sobsey MD (2009) Identification of particle size classes inhibiting protozoan recovery from surface water samples via US Environmental Protection Agency Method 1623. Appl Environ Microbiol 75:6619–6621
Leoni F, Amar C, Nichols G, Pedraza-Diaz S, McLauchlin J (2006) Genetic analysis of Cryptosporidium from 2414 humans with diarrhoea in England between 1985 and 2000. J Med Microbiol 55:703–707
Lu S, Lin A, Chen R, Wen L, Cheng Y, Chen X, Zhu G (2000) The diagnosis and analysis of Cryptosporidium infection in children of Zhejiang Province. Chin J Zoonoses 16:42–44
Montemayor M, Costan A, Lucena F, Jofre J, Munoz J, Dalmau E, Mujeriego R, Sala L (2008) The combined performance of UV light and chlorine during reclaimed water disinfection. Water Sci Technol 57:935–940
Nieminski EC, Schaefer FW 3rd, Ongerth JE (1995) Comparison of two methods for detection of Giardia cysts and Cryptosporidium oocysts in water. Appl Environ Microbiol 61:1714–1719
Ong CS, Eisler DL, Alikhani A, Fung VW, Tomblin J, Bowie WR, Isaac-Renton JL (2002) Novel Cryptosporidium genotypes in sporadic cryptosporidiosis cases: first report of human infections with a cervine genotype. Emerg Infect Dis 8:263–268
Plutzer J, Karanis P, Domokos K, Torokne A, Marialigeti K (2008) Detection and characterisation of Giardia and Cryptosporidium in Hungarian raw, surface and sewage water samples by IFT, PCR and sequence analysis of the SSUrRNA and GDH genes. Int J Hyg Environ Health 211:524–533
Ruecker NJ, Braithwaite SL, Topp E, Edge T, Lapen DR, Wilkes G, Robertson W, Medeiros D, Sensen CW, Neumann NF (2007) Tracking host sources of Cryptosporidium spp. in raw water for improved health risk assessment. Appl Environ Microbiol 73:3945–3957
Schets FM, van Wijnen JH, Schijven JF, Schoon H, de Roda Husman AM (2008) Monitoring of waterborne pathogens in surface waters in Amsterdam, the Netherlands, and the potential health risk associated with exposure to Cryptosporidium and Giardia in these waters. Appl Environ Microbiol 74:2069–2078
Smith HV, Nichols RA (2010) Cryptosporidium: detection in water and food. Exp Parasitol 124:61–79
Soba B, Petrovec M, Mioc V, Logar J (2006) Molecular characterisation of Cryptosporidium isolates from humans in Slovenia. Clin Microbiol Infect 12:918–921
Srisuphanunt M, Karanis P, Charoenca N, Boonkhao N, Ongerth JE (2010) Cryptosporidium and Giardia detection in environmental waters of southwest coastal areas of Thailand. Parasitol Res 106:1299–1306
Sun L, Lv J, He Y, Li C, Jin Q, Zhou Y, Rong Y (2007) Investigation on removal effect of Giardia and Cryptosporidium in purification process in Shanghai Pudong waterworks. China Water Wastewater 23:5–7
Trotz-Williams LA, Martin DS, Gatei W, Cama V, Peregrine AS, Martin SW, Nydam DV, Jamieson F, Xiao L (2006) Genotype and subtype analyses of Cryptosporidium isolates from dairy calves and humans in Ontario. Parasitol Res 99:346–352
US Environmental Protection Agency (USEPA) (2005) Method 1623: Cryptosporidium and Giardia in water by filtration/IMS/FA. EPA 815-R-05-002, Office of Water. US Government Printing Office, Washington DC, USA
Wang L, Zhao XD, Zhang HW, Zhang LX, Feng YY (2010a) Molecular epidemiology of Cryptosporidium in HIV patients in Henan, China. Paper presented at the Proceedings of the 10th International Coccidiosis Conference, Guangzhou, China.
Wang YL, Feng YY, Cui B, Jian FC, Ning CS, Wang RJ, Zhang LX, Xiao LH (2010b) Cervine genotype is the major Cryptosporidium genotype in sheep in China. Parasitol Res 106:341–347
World Health Organization (WHO) (2009) Risk assessment of Cryptosporidium in drinking water. WHO/HSE/WSH/09.04. WHO Press, Geneva, Switzerland
Xiao L (2010) Molecular epidemiology of cryptosporidiosis: an update. Exp Parasitol 124:80–89
Xiao L, Alderisio KA, Jiang J (2006) Detection of Cryptosporidium oocysts in water: effect of the number of samples and analytic replicates on test results. Appl Environ Microbiol 72:5942–5947
Xiao L, Singh A, Limor J, Graczyk TK, Gradus S, Lal A (2001) Molecular characterization of Cryptosporidium oocysts in samples of raw surface water and wastewater. Appl Environ Microbiol 67:1097–1101
Acknowledgements
This work was supported by grants from National Natural Science Foundation of China (grant no. 50778171, 20807013).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Xiao, S., An, W., Chen, Z. et al. Occurrences and genotypes of Cryptosporidium oocysts in river network of southern-eastern China. Parasitol Res 110, 1701–1709 (2012). https://doi.org/10.1007/s00436-011-2688-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-011-2688-6