Abstract
Cryptosporidium is an obligate intracellular parasite reported from all over the world. This protozoan infects a wide range of animals as well as humans. Cryptosporidium parvum and Cryptosporidium hominis are the most prevalent infecting species with mild and self-limiting infection in healthy people. The protozoan oocyst is resistant to common water purifiers. Based on emerging evidence, Cryptosporidium is one of waterborne parasites considered a major public health problem in developing and developed countries. In this study, 42 samples were collected from 14 rivers in the catchment area of Lake Urmia in northwest of Iran. Moreover, amplification of SSU rRNA gene was performed, and polymerase chain reaction products were sequenced. The results of sequencing and comparing the sequences with those in the GenBank revealed that all the 17 positive samples were C. parvum, a zoonotic species and one of the most frequent human-infecting species. Considering these data, it is highly important to inhibit the spread of this protozoan by treating livestock and preventing human and animal effluents from entering the water.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Cryptosporidium is a mandatory intracellular protozoan parasite reported in more than 90 countries around the world. Person to person (fecal–oral), animal to animal, and animal to human are the ways of transmission of this parasite mainly spreading through water, food, air, and sexual contact, among which water plays a very essential role in transmitting this protozoan via drinking or recreational water. Cryptosporidium transmission to humans occurs by eating food or drinking water contaminated with oocysts (Berahmat et al., 2017; Cunha et al., 2019).
Cryptosporidium, intestinal protozoan parasites responsible for Cryptosporidiosis, are among the most common waterborne parasites stated globally. In humans, diarrhea, dehydration, and malabsorption are the primary clinical signs of Cryptosporidiosis infection. Children under 5 years of age and people with immunodeficiency, especially AIDS patients, are at risk for Cryptosporidiosis. However, infection with this parasite in healthy individuals is mild and self-limiting. In addition to humans, wild and domestic animals are contaminated with Cryptosporidium. One of the main concerns for public health is the resistance of the parasite’s oocyst (contaminant form) to usual disinfectants used to treat water (Cunha et al., 2019). For laboratory diagnosis of Cryptosporidium, methods such as acid-fast staining, indirect hemagglutination (IHA), indirect immunofluorescence assay (IFA), enzyme-linked immunosorbent assay (ELISA), polymerase chain reaction (PCR), loop-mediated isothermal amplification (LAMP), and Western blot are used (Berahmat et al., 2017).
Numerous countries have reported the prevalence of Cryptosporidium in drinking water, indicating that water is the main cause of Cryptosporidium transmission, and this protozoan is a waterborne zoonotic parasite (Fayer, 2004; Rouhani et al., 2017). Of 40 species of Cryptosporidium identified, about 20 species are detected in humans (Xu et al., 2019). Molecular epidemiological research has shown that several Cryptosporidium species, particularly Cryptosporidium (C.) hominis, C. parvum, C. meleagridis, C. felis, C. canis, C. baileyi, and C. muric, infect nonhuman vertebrate hosts in addition to humans, the main host for C. hominis. The most common zoonotic Cryptosporidium species, C. parvum, is also more prevalent among nonhuman hosts (Fayer, 2004). C. meleagridis, C. felis, C. canis, C. ubiquitum, C. cuniculus, C. muris, and C. viatorum are other species frequently reported in immunocompromised humans and infrequently in humans (Fayer, 2004; Xu et al., 2019). In contrast, a high percentage (90%) of human Cryptosporidiosis is caused by C. parvum and C. hominis, two major species that infect humans and are mostly involved in human infections (Cunha et al., 2019).
Cryptosporidium species are a prime public health problem in both developing and developed countries, as well as the leading cause of morbimortality in immunocompromised individuals (Mahmoudi et al., 2015). Outbreaks of Cryptosporidiosis have been detected in homes, healthcare and daycare centers, swimming pools and reservoirs, districts with polluted public drinking water resources, and people using private drinking water supplies (Bartram et al., 2002). Given the increased risk of the prevalence of Cryptosporidium spp. due to waterborne transmission, the possible multiplication of this parasite in biofilms, water, or sewage treatment plants has created new challenges for the proper management of these tools. Furthermore, microbial biofilms can grow in the interior of the human intestine, thereby enhancing the host’s vulnerability both before and during the infection (Clode et al., 2015). In 1984, the first incidence of waterborne human Cryptosporidiosis was recorded in the USA. The most widespread outbreak happened in Milwaukee, WI, USA in 1993 when 400,000 people were contaminated with Cryptosporidium oocysts after drinking water from a water treatment facility (Mac Kenzie et al., 1994). In India, the prevalence of this protozoan has been reported to be 4–13%, but in south India, its prevalence among children living in poor health conditions is 40%. In addition, Africa is the first continent to have a large number of HIV cases, with a Cryptosporidium prevalence ranging from 3.8 to 73.6% (Vanathy et al., 2017).
Iran is a country with six main catchments. As mentioned above, Cryptosporidium is mostly transmitted through contaminated water and water resources. Therefore, the study of catchments for identifying this parasite seems to be necessary. One of Iran’s catchments is Lake Urmia, situated in the northwest of the country. The lake has a total area of 51,440 km2 and is fed primarily by 21 permanent and seasonal rivers. Until now, no studies have been conducted to identify parasites in catchment rivers. As the most important waterborne zoonotic parasite is Cryptosporidium, the present study investigated the prevalence and species of this protozoan in the catchment area of Lake Urmia.
Methods and materials
Sampling sites
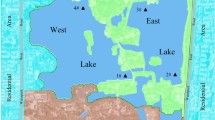
To collect water samples, we first identified sampling sites comprising Ajichai, Ojanchai, Ghalechai, Sofichai, and Azarshahrchai rivers in east Azarbaijan province and Zarinehrood, Siminehrood, Mahabadrood, Gadar, Nazloochai, Rozachai, Shaharchai, Barandozchai, and Zola rivers in west Azerbaijan province (Fig. 1).
Sample collection
Sampling was performed from late summer (September 8, 2020) to mid autumn (November 6, 2020), the rainy and wet months. Samples were obtained from 14 rivers connecting to the catchment area of Lake Urmia using a 1.5-L container. To this end, 30 L of water was collected from three parts of each river, including the beginning, the middle before entering the cities and farming centers, and the middle before entering the lake, 10 L for each part. Sampling was performed seven times at 5-min intervals. Overall, 42 samples (10-L containers) were collected. Water samples were transferred to the laboratory of the Department of Parasitology and Mycology at Urmia University of Medical Sciences for further experiments.
Isolation of oocysts from water samples
Water samples collected from rivers were filtered by ordinary filter paper with large pores to separate large particles from the water. Next, the filtrated samples were passed through a 47-mm diameter cellulose acetate membrane filter of 0.45-μm pore size (Millipore Co., USA) with a vacuum device. The filter paper of the second stage was put inside a plate containing physiological serum. The process was continued by the filtration and separation of particles from water samples by a 0.45-μm filter paper, which was then washed with physiological saline. A slide or scalpel was used to scratch and wash the surface of the filter paper in four directions. To obtain particles and protozoa, we poured the resulting liquid into a test tube and then centrifuged it at 3000 rpm for 10 min. The precipitates from 42 samples were finally transferred separately to 1.5-mL microtubes.
Staining method
At first, a thin slide was prepared from formalin-ethyl acetate sedimentation. After the slide was dried, methanol was added to fix sedimentation. Subsequently, carbolfuchsin was added to the slide for 5 min and gently washed with 50% alcohol for 3 to 5 s. Next, 1% sulfuric acid was gradually added to the slide for 1 to 2 min until the removal of all colors. Finally, the slide was completely dried in the open air, and its background was stained with methylene blue solution for 1 min. After drying the slide, Cryptosporidium oocysts appeared as a red oval shape with blue background (Mahmoudi et al., 2020).
DNA extraction and PCR
A commercial kit (BIO BASIC, Canada) was employed to extract DNA, which was carried out in accordance with the manufacturer’s instructions. Forward (5′ GACATATCATTCAAGTTTCTGACC 3′) and reverse (5′ CTGAAGGAGTAAGGAACAACC 3′) primers were planned for the multiplication of the SSU rRNA gene of Cryptosporidium spp. To the microtubes, 1 µL of each of reverse primers, 3 µL of extracted DNA, 12.5 µL of master mix, and 7.5 µL of sterile distilled water were added. The final volume of material inside the microtube was 25 µL. All the microtubes were then transferred to a thermocycler, in which different stages of PCR for this parasite were conducted: hot start stage (1 cycle at 94 °C and 120 s), denaturation stage (35 cycles at 94 °C and 120 s), annealing stage (35 cycles at 58 °C and 60 s), extension stage (35 cycles at 68 °C and 120 s), and final extension stage (1 cycle at 72 °C and 420 s). The PCR product was loaded on a 1% agarose gel and electrophoresed for 25 min in a 1% Tris/borate/EDTA solution. Finally, a gel imaging device was used to capture digital images. Bands of 830 bp were found in Cryptosporidium spp., and the PCR product was sent to Kawsar Biotech Company (Iran) for sequencing. The Chromas program was used to visualize the sequences of positive PCR products (version 2.6). The sequences were then compared to those in the GenBank database using BLAST software.
Results
PCR identified 17 (40.47%) positive samples of Cryptosporidium, while the number detected by acid-fast staining method was 14 (33.33%). Phylogenic analyses predicated based on obtained sequence data were conducted by maximum likelihood using MEGAX. All characters were run unordered and equally weighted. Alignment gaps were treated as missing data. Bootstrap analyses were conducted using 1000 replicates. The PCR result of analyzing samples for each river is detailed in Table 1. As illustrated in the table, all 17 isolates were recognized as C. parvum, with the GenBank accession numbers ON515714, ON515715, ON515716, ON515717, ON515718, ON515719, ON515720, ON515721, ON515722, ON515723, ON515724, ON515725, ON515726, ON515727, ON515728, ON515729, and ON515730. Also, according to the phylogenetic tree shown in Fig. 2, all the isolated species belonged to C. parvum. There were several livestock and agricultural centers near the sample collection centers and around the rivers.
Discussion
In this study, Cryptosporidium was identified by PCR in 17 (40.47%) water samples, and based on sequencing, all isolated species were C. parvum. In addition, 14 positive samples (33.33%) were reported using acid-fast staining method. Our results generally demonstrated the high prevalence of the mentioned parasite in the waters of Lake Urmia’s catchment area.
The frequency of Cryptosporidium among children with diarrhea ranges from 3 to 13% in developing countries such as Brazil, Venezuela, Indonesia, Thailand, South Africa, Ghana, India, and Bangladesh. However, in the developed regions of the world such as the UK, the USA, Canada, Australia, and Denmark, 1–4% of these children are infected with this parasite (Vanathy et al., 2017). In a survey, Tamomh et al. evaluated the prevalence of Cryptosporidiosis in children admitted to Kosti Teaching Hospital, Kosti, Sudan. In the study, 150 children aged about 5 years were examined. Of these 150 patients, 70 (46.7%) had diarrhea, but 80 (53.3%) showed no such disorder. The Cryptosporidium prevalence was 27.1% in children with diarrhea and 8.8% in non-diarrheal children. Overall, the frequency of this parasite in hospitalized children was 17.3% (26 out of 150) (Tamomh et al., 2021).
In a general and systematic study, the prevalence of Cryptosporidium in Iran among different animals, including rodents, horses, cattle, sheep, camels, birds, and dogs, were reported to be 20.8, 19.5, 14.4, 9.1, 8.4, 7.5, and 4.9%, respectively (Haghi et al., 2020). Also, the global incidence of this parasite was reported as 29.1% in cattle, 24.4% in sheep, 8.2% in goats, 22.6% in pigs, 4.7% in horses, 26% in buffalo (Hatam-Nahavandi et al., 2019), 8% in dogs (Taghipour et al., 2020), and 6% in cats (Taghipour et al., 2021). In a comprehensive study by Keshavarz et al., Iranian cattle were found to be infected mainly with C. parvum (72.6%), C. andersoni (17.7%), and C. bovis (7.8%) (Keshavarz et al., 2009).
In 2016, Berahmat et al. conducted a systematic review and meta-analysis survey based on studies and articles published from 1990 to 2015. Their findings of phylogenetic analysis using gp60 and 18S rRNA markers exhibited that most infections belonged to C. parvum (especially subgroup IIaA15G2R1), followed by C. hominis in Iran. Moreover, the occurrence of Cryptosporidiosis estimated using the random effect model over a 24-year period was reported as 3.65, 2.94, 1.29, and 4.54%, in children, healthy individuals, gastroenteritis patients, and immunocompromised cases in Iran, respectively. The prevalence of this parasite in humans in different parts of the country was between 0.83 and 24%. Human Cryptosporidium infection, the main causative agent of C. parvum, is the most widespread infection in Khorasan Razavi, Yazd, Bushehr, and Kurdistan provinces of Iran. Species with low prevalence include C. andersoni, C. bovis, and C. meleagridis (Berahmat et al., 2017).
To evaluate the incidence of Cryptosporidium species isolated from HIV-positive patients, Ghaffari and associates performed an investigation in southwestern Iran during 2014–2015. After reviewing the fecal samples of these patients, the incidence of the parasite was 10.8%, and the most frequent species included C. parvum (70.38%) followed by C. hominis (25.92%) and C. meleagridis (3.7%) (Ghafari et al., 2018). In another study, Mahmoudi et al. obtained 221 diarrheal stool samples from children admitted to Motahhari Hospital in Urmia, Northwestern Iran. Based on their obtained results from PCR and acid-fast staining methods, 7 (3.1%) and 4 (1.8%) samples were found to be positive, respectively. After sequencing, the research group explored that all isolated species were C. parvum (Mahmoudi et al., 2020). In another investigation by the same author, the prevalence and detection of Cryptosporidium were evaluated in Iranian waters. During 2009–2010, 49 surface water samples were collected from rivers and surface waters in Gilan and Tehran provinces. In total, 24 water samples (48.97%) collected were found to be positive, and C. parvum was the most common species (11/24; 45.8%) (Mahmoudi et al., 2015). However, other researchers reported the prevalence of this parasite to be 4–63.33% in Iranian waters (Mahmoudi et al., 2013, 2017; Naeini et al., 2011). Mahmoudi et al. performed an analysis to determine the prevalence and detection of Cryptosporidium in Iranian waters. During 2009–2010, 49 surface water samples were obtained from rivers and surface waters in Gilan and Tehran provinces. A total of 24 water samples (48.97%) collected were found to be positive, and C. parvum was the most common species (11/24; 45.8%) found. In a systematic review and meta-analysis study, Daraei and colleagues assessed the occurrence of Cryptosporidium spp. from January 1, 1983 to September 10, 2019 in different waters in the world. According to IFA, PCR, microscopy, and ELISA methods, Cryptosporidium was found in 25.7% of purified water and 40.1% of untreated water. Also, the frequency of this parasite reported was 46.9% in wastewater, 45.3% in surface water, 31.6% in raw water, 25.5% in drinking water, 24.5% in reservoir water, 18.8% in groundwater, 7.5% in swimming pool water, and 0.20% in marine water (Daraei et al., 2021). In another research by Koloren et al., the prevalence of Cryptosporidium spp. in environmental waters in Turkey ranged from 38.3 to 42%. In addition, C. parvum was the most prevalent species in the samples examined (Koloren & Ayaz, 2016). The results of our study are consistent with other investigations showing the high prevalence of Cryptosporidium in water. In most studies, especially on water resources, as in our study, after sequencing, C. parvum was the most dominant and most identified species among other species of Cryptosporidium. One of the potential explanations for the higher Cryptosporidium spp. levels in surface water downstream of cattle farms is exposure to agricultural effluent runoff or field drainage ditches (Ong et al, 1996). Inefficient farm and livestock management in the catchment area can also contribute to the contamination of the water with Cryptosporidium spp. (Ionas et al., 1998). As a result, wastewater, manure, and livestock, as well as domestic and wild animal waste, have been identified as the primary Cryptosporidium-to-water transfer pathways (Edwards et al., 2008; Rostami et al., 2020). The presence of livestock and agricultural centers located near rivers due to the entry of livestock and agricultural fertilizers and even human wastewater into river waters is one of the main reasons for the high prevalence of Cryptosporidium in surface and river waters. Near the rivers we sampled, there were many agricultural and livestock centers, particularly cattle farms.
According to available evidence, parasitic densities are higher in fall and winter seasons because of high daily rainfall levels, and rainfall may promote the entry of parasitic fecal particles into water bodies. In wet seasons and after rainfall runoff, the density of Cryptosporidium oocysts is higher as well. However, the sampling process and location are important factors in parasite detection, and parasite levels are lower in upstream than downstream water bodies. Based on the prevalence of Cryptosporidium spp. in waters, wastewater was ranked first, followed by surface water, raw water, drinking water, water reservoirs samples, groundwater, swimming pool water, and marine water (Daraei et al., 2021). As exhibited in Berahmat et al.’s study, the incidence of Cryptosporidium infection was higher in autumn (10%) and summer (6%), as well as in patients with diarrhea (Berahmat et al., 2017). In our study, we collected our samples from late summer to mid autumn, the time when heavy rainfall occurs. Moreover, Cryptosporidium was found downstream of all rivers, except for Aji Chai, Simineh River, Rouzeh Chai, and Baranduz Chai; in addition to the downstream, the infection was detected in the middle of these rivers. The reason could be the existence of many livestock breeding centers around the middle parts of these rivers. On the other hand, we could not find Cryptosporidium in any part of Azarshahr Chai River, which may be due to the seasonality of this river and the lack of livestock breeding centers around this river.
Morgan and research team compared Cryptosporidium detection methods (PCR vs. microscopic) in feces and applied acid-fast staining method for microscopic analysis. The number of stool samples tested after being referred for screening due to diarrhea was 511, among which 36 were identified by PCR, and 29 positive samples were detected by routine microscopy (Morgan et al., 1998). Kar et al. found out that PCR was the most powerful method for detecting C. parvum oocysts in calf feces, though the results of carbolfuchsin and modified Ziehl–Neelsen staining were comparable (Kar et al., 2011). In 2019, Mahmoudi et al. applied two methods, PCR and acid-fast staining, to identify Cryptosporidium in diarrhea stool samples. The ultimate results showed that seven samples by PCR and four samples by acid-fast staining method were positive for Cryptosporidium (Mahmoudi et al., 2020). In the detection of Cryptosporidium species, microscopy-based methods are inefficient and nonspecific. Furthermore, these techniques necessitate lengthy labor and a high degree of expertise. However, nucleic acid-based methods for detecting C. parvum and other Cryptosporidium species are highly sensitive and specific (Hassan et al., 2021). In the present study, the number of positive cases reported based on PCR method was more than that reported by acid-fast staining method, which was predictable with regard to the findings of other studies, signifying that the detection ability of PCR is higher than acid-fast staining and it is more reliable.
About 155 species of other mammals are known to be in human hosts of C. parvum, indicating that this parasite is adapting and growing to infect many diverse hosts, and this species can be a major problem of common zoonotic diseases (Pumipuntu & Piratae, 2018). As the most dominant species for humans and livestock, C. parvum has an economic burden on livestock centers (Pérez-Cordón et al., 2016). A previous study has highlighted that C. parvum is the predominant water-polluting species in Ireland every year and every season (Mahon & Doyle, 2017). The results of a study in China emphasized that C. parvum is highly associated with gastrointestinal cancers and could be a potential risk factor for such diseases (Zhang et al., 2020). Our results showed that species isolated from positive samples were all C. parvum, which supports the importance of what has been stated about this species. C. parvum is a prevalent zoonotic species; thus, it can infect a wide range of animals and humans. Therefore, a very important issue that needs to be taken into account is gastrointestinal diseases, particularly gastrointestinal cancers, caused by this species both in humans and animals. Another issue that can be significant, considering our results and others, is that C. parvum is adapting and growing and more resistant in nature and different hosts, which likely causes extensive and more severe pathogens that can give rise to many health and economic problems (especially livestock economics) throughout the world. Since the most common route of transmission of this parasite to humans and animals is through water, the hygiene of water resources, especially surface water, which is directly exposed to use by various animals as well as humans, is a major determinant in diseases caused by this parasite, especially the species C. parvum.
Of 42 rivers sampled in the present study, nine were located in West Azarbaijan Province and five rivers in East Azarbaijan. Considering the collection of water samples from three parts of each river, the number of samples collected from the rivers of West and East Azerbaijan was 27 and 15 samples, respectively. Based on the molecular PCR method, the prevalence of Cryptosporidium in the rivers of West Azarbaijan Province was 44.44%, and in the rivers of East Azarbaijan was 33.33%, which suggests the higher incidence of this protozoan in the former than latter province. The reason for this discrepancy could be the large number of livestock and agricultural centers in West Azarbaijan compared to East Azerbaijan Province. Another reason could be the presence of abundant vegetation and grazing areas and limited mechanical and industrial centers of animal husbandry and agriculture in West Azarbaijan Province, compared to its neighboring province, West Azarbaijan, which has caused the feeding and grazing of livestock, such as cattle and sheep, to be conducted traditionally, ultimately facilitating the entry of this protozoan into the environment and thus surface water and rivers.
Conclusions
The present study is the first to assess the species and prevalence of Cryptosporidium in the waters of Lake Urmia. For this study, water samples were collected from late summer to mid autumn, but for further investigation and the achievement of more accurate results, sampling in all seasons is recommended. The prevalence of this parasite was high in the studied catchment area; therefore, to reduce the contamination and transmission of this protozoan, the treatment of livestock centers located around rivers appears to be necessary. Likewise, preventing the entry of sewage or effluents of animal husbandry centers, as well as human wastewater, into rivers can be an effective measure to make the prevalence of this protozoan in the rivers of this catchment area decline.
Availability of data and materials
The authors declare that all data supporting the findings of this study are available within the article.
References
Bartram, J., Thyssen, N., & Gowers, A., Pond, K., & Lack, T. (2002). Water and health in Europe: joint report from the European Environment Agency and the WHO Regional Office for Europe. WHO Regional Office Europe. Availabe at: https://www.google.com/url?sa=t&rct=j&q=&esrc=s&source=web&cd=&cad=rja&uact=8&ved=2ahUKEwjS2Pad7Or9AhVtxgIHHSvRBpgQFnoECBEQAQ&url=https%3A%2F%2Fwww.euro.who.int%2F__data%2Fassets%2Fpdf_file%2F0007%2F98449%2FE76521.pdf&usg=AOvVaw0cmhqeeKWVrADN4to6J0V4
Berahmat, R., Spotin, A., Ahmadpour, E., Mahami-Oskouei, M., Rezamand, A., Aminisani, N., & Mikaeili-Galeh, T. (2017). Human cryptosporidiosis in Iran: a systematic review and meta-analysis. Parasitology Research, 116(4), 1111–1128.
Clode, P. L., Koh, W. H., & Thompson, R. A. (2015). Life without a host cell: what is Cryptosporidium? Trends in Parasitology, 31(12), 614–624.
Cunha, F. S., Peralta, R. H. S., & Peralta, J. M. (2019). New insights into the detection and molecular characterization of Cryptosporidium with emphasis in Brazilian studies: A review. Revista do Instituto de Medicina Tropical de São Paulo, 61, e28.
Daraei, H., Oliveri Conti, G., Sahlabadi, F., Thai, V. N., Gholipour, S., Turki, H., Fakhri, Y., Ferrante, M., Moradi, A., & Mousavi Khaneghah, A. (2021). Prevalence of Cryptosporidium spp. in water: A global systematic review and meta-analysis. Environmental Science and Pollution Research, 28(8), 9498–9507.
Edwards, A. C., Kay, D., McDonald, A. T., Francis, C., Watkins, J., Wilkinson, J., & Wyer, M. D. (2008). Farmyards, an overlooked source for highly contaminated runoff. Journal of Environmental Management, 87(4), 551–559.
Fayer, R. (2004). Cryptosporidium: a water-borne zoonotic parasite. Veterinary Parasitology, 126(1–2), 37–56.
Ghafari, R., Rafiei, A., Tavalla, M., Choghakabodi, P. M., Nashibi, R., & Rafiei, R. (2018). Prevalence of Cryptosporidium species isolated from HIV/AIDS patients in southwest of Iran. Comparative Immunology, Microbiology and Infectious Diseases, 56, 39–44.
Haghi, M. M., Khorshidvand, Z., Khazaei, S., Foroughi-Parvar, F., Sarmadian, H., Barati, N., & Ghasemikhah, R. (2020). Cryptosporidium animal species in Iran: a systematic review and meta-analysis. Tropical Medicine and Health, 48(1), 1–15.
Hassan, E. M., Örmeci, B., DeRosa, M. C., Dixon, B. R., Sattar, S. A., & Iqbal, A. (2021). A review of Cryptosporidium spp. and their detection in water. Water Science and Technology, 83(1), 1–25.
Hatam-Nahavandi, K., Ahmadpour, E., Carmena, D., Spotin, A., Bangoura, B., & Xiao, L. (2019). Cryptosporidium infections in terrestrial ungulates with focus on livestock: a systematic review and meta-analysis. Parasites & Vectors, 12(1), 1–23.
Ionas, G., Learmonth, J., Keys, E., & Brown, T. (1998). Distribution of Giardia and Cryptosporidium in natural water systems in New Zealand—a nationwide survey. Water Science and Technology, 38(12), 57–60.
Kar, S., Gawlowska, S., Daugschies, A., & Bangoura, B. (2011). Quantitative comparison of different purification and detection methods for Cryptosporidium parvum oocysts. Veterinary Parasitology, 177(3–4), 366–370.
Keshavarz, A., Haghighi, A., Athari, A., Kazemi, B., Abadi, A., & Mojarad, E. N. (2009). Prevalence and molecular characterization of bovine Cryptosporidium in Qazvin province, Iran. Veterinary Parasitology, 160(3–4), 316–318.
Koloren, Z., & Ayaz, E. (2016). Genotyping of Cryptosporidium spp. in environmental water in Turkey. Acta parasitologica, 61(4), 671–679.
Mac Kenzie, W. R., Hoxie, N. J., Proctor, M. E., Gradus, M. S., Blair, K. A., Peterson, D. E., & Rose, J. B. (1994). A massive outbreak in Milwaukee of Cryptosporidium infection transmitted through the public water supply. New England Journal of Medicine, 331(3), 161–167.
Mahmoudi, M. R., Bandepour, M., Kazemi, B., & Mirzaei, A. (2017). Detection and enumeration of Cryptosporidium oocysts in environmental water samples by real-time PCR assay. Journal of Basic Research in Medical Sciences, 4(3), 42–47.
Mahmoudi, M. R., Kazemi, B., Mohammadiha, A., Mirzaei, A., & Karanis, P. (2013). Detection of Cryptosporidium and Giardia (oo) cysts by IFA, PCR and LAMP in Surface Water from Rasht, Iran. Transactions of the Royal Society of Tropical Medicine and Hygiene, 107(8), 511–517.
Mahmoudi, M. R., Mojarad, E., Kazemi, B., Haghighi, A., Mirzaei, A., Mohammadiha, A., Jahantab, S., Xiao, L., & Karanis, P. (2015). Cryptosporidium genotypes and subtypes distribution in river water in Iran. Journal of Water and Health, 13(2), 600–606.
Mahmoudi, M., Tappeh, K. H., Abasi, E., & Aminpour, A. (2020). Utility of different diagnostic tools for detection of Cryptosporidium in children with diarrhea. Archives of Pediatric Infectious Diseases, 8(1).
Mahon, M., & Doyle, S. (2017). Waterborne outbreak of cryptosporidiosis in the south east of Ireland: weighing up the evidence. Irish Journal of Medical Science (1971-), 186(4), 989–994.
Morgan, F. U., Pallant, L., Dwyer, B., Forbes, D., Rich, G., & Thompson, R. (1998). Comparison of PCR and microscopy for detection of Cryptosporidium parvum in human fecal specimens: clinical trial. Journal of Clinical Microbiology, 36(4), 995–998.
Naeini, K. M., Asadi, M., & Chaleshtori, M. H. (2011). Detection and molecular characterization of Cryptosporidium species in recreational waters of Chaharmahal va Bakhtiyari province of Iran using nested-PCR-RFLP. Iranian Journal of Parasitology 6(1), 20.
Ong, C., Moorehead, W., Ross, A., & Isaac-Renton, J. (1996). Studies of Giardia spp. and Cryptosporidium spp. in two adjacent watersheds. Applied and Environmental Microbiology, 62(8), 2798–2805.
Pérez-Cordón, G., Robinson, G., Nader, J., & Chalmers, R. M. (2016). Discovery of new variable number tandem repeat loci in multiple Cryptosporidium parvum genomes for the surveillance and investigation of outbreaks of cryptosporidiosis. Experimental Parasitology, 169, 119–128.
Pumipuntu, N., & Piratae, S. (2018). Cryptosporidiosis: a zoonotic disease concern. Veterinary World, 11(5), 681.
Rostami, M., Fasihi-Harandi, M., Shafiei, R., Aspatwar, A., Derakhshan, F. K., & Raeghi, S. (2020). Genetic diversity analysis of blastocystis subtypes and their distribution among the domestic animals and pigeons in northwest of Iran. Infection, Genetics and Evolution, 86, 104591.
Rouhani, S., Raeghi, S., & Spotin, A. (2017). Spermatogenic and phylo-molecular characterizations of isolated Fasciola spp. from cattle, north west Iran. Pakistan journal of biological sciences: PJBS, 20(4), 204–209.
Taghipour, A., Khazaei, S., Ghodsian, S., Shajarizadeh, M., Olfatifar, M., Foroutan, M., & Karanis, P. (2021). Global prevalence of Cryptosporidium spp. in cats: a systematic review and meta-analysis. Research in Veterinary Science.
Taghipour, A., Olfatifar, M., Bahadory, S., Godfrey, S. S., Abdoli, A., Khatami, A., & Shahrivar, F. (2020). The global prevalence of Cryptosporidium infection in dogs: a systematic review and meta-analysis. Veterinary Parasitology, 281, 109093.
Tamomh, A. G., Agena, A. M., Elamin, E., Suliman, M. A., Elmadani, M., Omara, A. B., & Musa, S. A. (2021). Prevalence of cryptosporidiosis among children with diarrhoea under five years admitted to Kosti teaching hospital, Kosti city, Sudan. BMC Infectious Diseases, 21(1), 1–6.
Vanathy, K., Parija, S. C., Mandal, J., Hamide, A., & Krishnamurthy, S. J. T. p. (2017). Cryptosporidiosis: A mini review. Tropical Parasitology, 7(2), 72–80.
Xu, Z., Guo, Y., Roellig, D. M., Feng, Y., & Xiao, L. (2019). Comparative analysis reveals conservation in genome organization among intestinal Cryptosporidium species and sequence divergence in potential secreted pathogenesis determinants among major human-infecting species. BMC Genomics, 20(1), 1–15.
Zhang, N., Yu, X., Zhang, H., Cui, L., Li, X., Zhang, X., & Wang, X. (2020). Prevalence and genotyping of Cryptosporidium parvum in gastrointestinal cancer patients. Journal of Cancer, 11(11), 3334.
Funding
This article is extracted from Rasoul Sharafi MS Thesis of parasitology. This work, with the project number 2799, was financially supported by Urmia University of Medical Sciences, Urmia, Iran.
Author information
Authors and Affiliations
Contributions
Arash Aminpour and Ali Ahmad Aghapour designed the work. Rasoul Sharafi collected the samples and carried out the work under supervision of Arash Aminpour and Ali Ahmad Aghapour. Arash Aminpour and Rasoul Sharafi wrote the manuscript. Arash Aminpour, Ali Ahmad Aghapour, and Rasoul Sharafi read the whole manuscript. Arash Aminpour confirmed the final version of the manuscript.
Corresponding author
Ethics declarations
Ethics approval
The study was approved by the Ethics Committee of Urmia University of Medical Sciences, Urmia, Iran (approval number: IR.UMSU.1399.223).
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sharafi, R., Aghapour, A.A. & Aminpour, A. Prevalence and molecular analysis of Cryptosporidium spp. collected from surface water. Environ Monit Assess 195, 499 (2023). https://doi.org/10.1007/s10661-023-11097-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10661-023-11097-w