Abstract
To identify Cryptosporidium species/genotypes in sheep in China and to elucidate the endemic transmission of cryptosporidiosis, a total of 1,701 fecal samples from five farms in four prefectures in Henan Province (central China) were examined. Eighty-two Cryptosporidium-positive samples were analyzed by polymerase chain reaction (PCR)–restriction fragment length polymorphism analysis of the small subunit (SSU) rRNA gene and PCR analysis of the 60 kDa glycoprotein (gp60) gene, and 41 were further analyzed by DNA sequencing of the PCR products. The SSU rRNA-based PCR identified two Cryptosporidium species and one genotype, including the Cryptosporidium cervine genotype (74/82), Cryptosporidium andersoni (4/82), and Cryptosporidium xiaoi (4/82). The cervine genotype was found in all age groups, C. xiaoi in lambs, and C. andersoni in ewes. There were intragenetic differences in the SSU rRNA gene sequences of the Cryptosporidium cervine genotype and C. xiaoi. No Cryptosporidium parvum was detected by both SSU rRNA- and gp60-based PCR assays. These findings suggest that sheep are a potential source for zoonotic infections of the Cryptosporidium cervine genotype.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cryptosporidium species are important zoonotic parasites. It has a wide spectrum of hosts including humans, other mammals, birds, reptiles, amphibians, and fish. It is one of the common causes of diarrhea in humans and domestic animals and exerts significant public health and economic impact (Xiao et al. 2004; Fayer and Xiao 2008). So far, at least 20 Cryptosporidium species are considered valid, and more than 50 host-adapted genotypes with undetermined species status have been described (Fayer 2009; Fayer et al. 2008; Fayer and Santín 2009).
Previous studies have indicated that some animals are important zoonotic reservoirs of Cryptosporidium in humans (Xiao and Fayer 2008). In particular, calves have attracted extensive attention, as they are widely infected with Cryptosporidium parvum, the most important zoonotic Cryptosporidium species (Fayer and Xiao 2008; Xiao and Feng 2008). The role of sheep in transmitting cryptosporidiosis to human is comparatively less studied. Just like cattle, sheep are also commonly raised in many countries including China. The molecular characterizations conducted thus far have identified several species/genotypes in sheep, including C. parvum, Cryptosporidium hominis, Cryptosporidium andersoni, Cryptosporidium suis, Cryptosporidium fayeri, Cryptosporidium bovis-like genotype (recently named as Cryptosporidium xiaoi), Cryptosporidium cervine and sheep genotypes and pig genotype II, with C. parvum, C. xiaoi, and the Cryptosporidium cervine genotype as the major species (Majewska et al. 2000; McLauchlin et al. 2000; Chalmers et al. 2002; Ryan et al. 2005; Leoni et al. 2007; Navarro-i-Martinez et al. 2007; Santín et al. 2007; Santín and Fayer 2007; Soltane et al. 2007; Elwin and Chalmers 2008; Geurden et al. 2008; Giles et al. 2009; Mueller-Doblies et al. 2008; Paoletti et al. 2009; Quílez et al. 2008; Fayer and Santín 2009;Yang et al. 2009). However, there are seemingly some significant differences in the distribution of Cryptosporidium species/genotypes in the different studies. For example, in several studies conducted in Spain, Italy, and UK, C. parvum was much more prevalent than other species in lambs. In contrast, C. xiaoi and Cryptosporidium cervine genotype were the most common ones in studies conducted in Australia and USA (McLauchlin et al. 2000; Ryan et al. 2005; Santín et al. 2007; Santín and Fayer 2007; Geurden et al. 2008; Mueller-Doblies et al. 2008; Quílez et al. 2008; Paoletti et al. 2009; Yang et al. 2009). Results of a recent study conducted in Australia, however, indicate that the use of small subunit rRNA-based polymerase chain reaction (PCR) tools in Cryptosporidium genotyping may underestimate the prevalence of C. parvum (Yang et al. 2009).
In China, the total sheep population was 173.9 million in 2006 (http://www.caaa.cn). Sheep play a critical role in agricultural economy, especially in grassland regions of northwestern China where rearing sheep is a major source of income for local farmers. However, there are no systematic studies of cryptosporidiosis in sheep, and there are very few genetic data on Cryptosporidium species in China. The few studies on the prevalence of Cryptosporidium in sheep were conducted using a microscopy (Wang et al. 2008b). The objectives of this study were to identify the distribution and zoonotic potential of Cryptosporidium species in sheep in China.
Materials and methods
Sample collection and DNA extraction
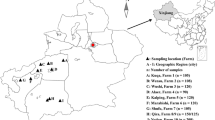
Random fresh fecal samples were collected between July 2006 and July 2007 from four breads of sheep (Small Tail Han, Poll Dorset, Dorset Down, and Rommay) on five sheep farms in four prefectures in Henan Province, China. A total of 1,701 fecal samples were used in the study, including those from preweaned and postweaned lambs, adult sheep, and ewes preparturition and 0–5 weeks after parturition sheep (Table 1). Samples were examined for Cryptosporidium by microscopy of fecal materials concentrated by the Sheather’s sugar flotation technique and stained with the modified acid-fast stain. Cryptosporidium-positive samples were stored in 2.5% potassium dichromate at 4 °C.
DNA extraction
Cryptosporidium oocysts were isolated from the positive fecal samples by the discontinuous density sucrose gradient centrifugation. Genomic DNA was extracted from the purified oocysts using the Mag Extractor-Genome kit (Toyobo Co. Ltd., Osaka, Japan) based on chaotropic extraction followed by absorption of DNA onto silica-coated magnetic beads, using the manufacturer-recommended procedures. The extracted DNA was kept at −20 °C before it was used in molecular analysis.
Cryptosporidium genotyping
Cryptosporidium species and genotypes were determined by nested PCR of the small subunit (SSU) rRNA gene and restriction fragment length polymorphism (RFLP) analysis of the secondary PCR products using restriction enzymes SspI and VspI. Primers and amplification conditions used for PCR-RFLP were adopted from previous publications (Xiao et al. 1999; Xiao et al. 2000; Xiao et al. 2001; Jiang et al. 2005; Feng et al. 2007). The diagnosis of C. andersoni and C. xiaoi were confirmed by DNA sequencing of all positive SSU rRNA PCR products, and the Cryptosporidium cervine genotype by DNA sequencing of PCR products from 33 samples from representative farms and age groups. To exclude the possible presence of light infections of C. parvum or C. hominis, all Cryptosporidium-positive samples were also analyzed by a nested PCR targeting the 60 kDa glycoprotein (gp60) gene, which does not amplify DNA of C. andersoni, C. xiaoi, and the Cryptosporidium cervine genotype. After purification, the secondary PCR products of the SSU rRNA gene were sequenced directly with secondary PCR primers on an ABI PRISMTM 3730 XL DNA Analyzer (Applied Biosystems, USA) by Shanghai Biotechnology Co. Ltd. (Shanghai, China), using the Big Dye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, USA). Sequence accuracy was confirmed by two-directional sequencing and by sequencing a new PCR product if necessary.
Nucleotide sequence accession numbers
The nucleotide sequences of the partial SSU rRNA gene of Cryptosporidium species obtained in this study were deposited in GenBank under accession numbers EU827362 to EU827363, EU827366 to EU827367, EU827369 to EU827370, EU827371 to EU827403, and GU014552 and GU014553.
Results
Prevalence of Cryptosporidium species in sheep
Microscopy analysis of 1,701 fecal samples showed the presence of Cryptosporidium oocysts in 82 (4.8%) samples (Table 1). The percentage of animals shedding oocysts was 10.8%, 4.3%, 2.1%, and 2.5% in preweaned lambs, postweaned lambs, pregnant ewes, and postparturition ewes, respectively (Table 2).
Distribution of Cryptosporidium species/genotype
The SSU rRNA gene of Cryptosporidium species in all 82 microscopy-positive samples was successfully amplified by the nested PCR. RFLP analysis of the SSU rRNA gene products revealed the presence of three Cryptosporidium species/genotypes, including C. xiaoi (4/82), C. andersoni (4/82), and Cryptosporidium cervine genotype (74/82; Tables 1 and 2). DNA sequencing of the SSU rRNA PCR products from four C. andersoni, four C. xiaoi, and 33 Cryptosporidium cervine genotype-positive samples confirmed the identification of these species/genotypes. There was a complete agreement between RFLP and DNA sequencing results. No amplification of the gp60 gene was achieved by a nested PCR analysis of DNA extracted from the 82 microscopy-positive samples, suggesting that C. parvum or C. hominis was not concurrently present among the three Cryptosporidium species/genotypes.
Age patterns of Cryptosporidium species/genotypes
The cervine genotype was the most commonly identified Cryptosporidium, responsible for 90.2% of all Cryptosporidium infections. It was found in all age groups examined in this study, on all five sheep farms and in all four sheep breeds. In contrast, the prevalence of C. xiaoi and C. andersoni was much lower, each responsible for 4.9% of Cryptosporidium infections (Tables 1 and 2). The former was only found in lambs and the latter was only found in ewes (Table 2).
Intragenotypic variations in Cryptosporidium cervine genotype and C. xiaoi
Sequence heterogeneity in the SSU rRNA gene was observed in the Cryptosporidium cervine genotype and C. xiaoi. Four types of nucleotide sequences were obtained from samples positive for the Cryptosporidium cervine genotype and two types for C. xiaoi. The sequences of the Cryptosporidium cervine genotype differed from each other by the presence of an A to G nucleotide substitution (at position 503 of AF442484) and AT deletion (nucleotides 691 and 692 of AF442484) with the majority (27/33) having a nucleotide sequence identical to AF442484 (Table 3; da Silva et al. 2003). The sequences of C. xiaoi differed from each other by the presence of T to C nucleotide substitution (at position 633 of DQ991389). Three of the samples yielded a sequence identical to EU327319 and one sequence identical to EU408317. Both were previously obtained from sheep in UK (Elwin and Chalmers 2008).
Discussion
Cryptosporidiosis was shown to be prevalent in lambs in this study. A 4.8% overall infection rate of Cryptosporidium species was observed. As reported in previous studies conducted in other countries (Xiao et al. 1993; Majewska et al. 2000; Causapé et al. 2002; Sturdee et al. 2003; Santín et al. 2007), the highest infection rate (10.8%) was seen in preweaned lambs. The infection rate decreased significantly in weaned lambs (4.3%) and reached low levels in pregnant and postparturition ewes (2.1% and 2.5%, respectively).
Thus far, nine Cryptosporidium species/genotypes have been identified in sheep, including C. parvum, C. hominis, C. andersoni, C. fayeri, C. suis, the Cryptosporidium cervine genotype, sheep genotype and pig genotype II, and a C. bovis-like genotype (Table 4). The C. bovis-like genotype appears to be a sheep-adapted Cryptosporidium specie and has been recently named as C. xiaoi (Fayer and Santín 2009). Among the species and genotypes identified in sheep, C. parvum, C. xiaoi, and the Cryptosporidium cervine genotype are the three common ones. The remaining ones have each been only detected in less than a handful of animals (Table 4). In this study, only two of the common species/genotypes and C. andersoni were detected in the 82 Cryptosporidium-positive sheep.
Like previously observed in cattle and recently observed in pigs (Santín et al. 2004; Kvác et al. 2009), there seemingly is an age-associated distribution of Cryptosporidium species in sheep. In this study, all age groups of sheep were infected by the Cryptosporidium cervine genotype. In contrast, C. xiaoi was only detected in lambs and C. andersoni in ewes (Table 2). Previously, few studies compared the distribution of Cryptosporidium species in different age groups of sheep. Although an earlier study failed to detect any differences in the distribution of Cryptosporidium cervine genotype and C. xiaoi between preweaned lambs and postparturition ewes (Santín et al. 2007), results of a recent study conducted in UK showed that C. parvum was only found in preweaned lambs and C. xiaoi almost exclusively in postweaned lambs (Mueller-Doblies et al. 2008).
There are also probably geographic differences in the distribution of Cryptosporidium species in sheep (Table 4). An earlier study conducted in Australia suggested that C. parvum was largely absent in sheep. Instead, C. bovis (probably C. xiaoi) and the Cryptosporidium cervine genotype were the two common parasites in sheep (Ryan et al. 2005). This has been confirmed in a subsequent study in USA (Santín et al. 2007) and two small scale studies in Belgium and Tunisia (Soltane et al. 2007; Geurden et al. 2008). In contrast, several studies conducted in UK, Italy, Poland, and Spain showed C. parvum as dominant species in preweaned lambs (Table 4). The Cryptosporidium species distribution in sheep in Henan, China obtained in this study is apparently similar to the one observed in USA and very different from the one seen in recent studies in Europe.
Other factors might have also been attributed to causing the observed differences in the distribution of Cryptosporidium species in published studies. One group of researchers suggested that clinically ill lambs are more likely infected with C. parvum; whereas, healthy lambs are more likely infected with the Cryptosporidium cervine genotype and C. xiaoi (Chalmers et al. 2002; Elwin and Chalmers 2008; Mueller-Doblies et al. 2008). However, C. parvum was frequently detected in apparently normal lambs in other European studies (Majewska et al. 2000; Pritchard et al. 2007; Paoletti et al. 2009), and C. xiaoi was detected in at least one lamb that died of apparent severe cryptosporidiosis (Navarro-i-Martinez et al. 2007). Results of a recent study in Australia have demonstrated that the genotyping technique used would also affect the observed distribution of Cryptosporidium species in sheep. When a genus-specific SSU rRNA-based tool was used, C. xiaoi was identified as the dominant species in preweaned lambs and C. parvum was only identified in two of the 66 positive samples. In contrast, when a PCR technique preferentially detecting C. parvum was used, 63 samples was positive for C. parvum, including ten samples coinfected with C. xiaoi (Yang et al. 2009). A similar strategy was used in the present study, but no C. parvum was detected in any of the microscopy-positive samples.
Among the three Cryptosporidium species/genotypes identified in sheep in Henan, China, only the Cryptosporidium cervine genotype has significant public health importance. It is ranked sixth of the most commonly distributed Cryptosporidium species in humans and has been reported in at least 25 sporadic cases of cryptosporidiosis. All, but one, of the cases were reported in industrialized nations (Wang et al. 2008a; Davies et al. 2009; Pollock et al. 2009). It is also one of the most common Cryptosporidium species found in drinking source water in USA and Canada (Jiang et al. 2005; Ruecker et al. 2007; Karanis et al. 2007; Yang et al. 2008; Jellison et al. 2009). Because of the common occurrence of this parasite in sheep and its absence in cattle, sheep are likely a major source of the Cryptosporidium cervine genotype in humans and source water.
In conclusion, Cryptosporidium cervine genotype was found to be the dominant Cryptosporidium in sheep in China. More studies, especially those conducted in developing countries and/or involving comparison of the distribution of Cryptosporidium species/genotypes in different age groups, are needed for better understanding of the differences in transmission and public health significance of cryptosporidiosis in sheep in various areas.
References
Causapé AC, Quilez J, Sanchez-Acedo C, del Cacho E, Lopez-Bernad F (2002) Prevalence and analysis of potential risk factors for Cryptosporidium parvum infection in lambs in Zaragoza (northeastern Spain). Vet Parasitol 104:287–298
Chalmers RM, Elwin K, Reilly WJ, Irvine H, Thomas AL, Hunter PR (2002) Cryptosporidium in farmed animals: the detection of a novel isolate in sheep. Int J Parasitol 32:21–26
da Silva AJ, Cacciò S, Williams C, Won KY, Nace EK, Whittier C, Pieniazek NJ, Eberhard ML (2003) Molecular and morphologic characterization of a Cryptosporidium genotype identified in lemurs. Vet Parasitol 111:297–307
Davies AP, Campbell B, Evans MR, Bone A, Roche A, Chalmers RM (2009) Asymptomatic carriage of protozoan parasites in children in day care centers in the United Kingdom. Pediatr Infect Dis J 28:838–840
Elwin K, Chalmers RM (2008) Contemporary identification of previously reported novel Cryptosporidium isolates reveals Cryptosporidium bovis and the cervine genotype in sheep (Ovis aries). Parasitol Res 102:1103–1105
Fayer R (2009) Taxonomy and species delimitation in Cryptosporidium. Exp Parasitol (in press)
Fayer R, Xiao L (2008) Cryptosporidium and Cryptosporidiosis. CRC, Boca Raton, FL, USA
Fayer R, Santín M (2009) Cryptosporidium xiaoi n. sp. (Apicomplexa: Cryptosporidiidae) in sheep (Ovis aries). Vet. Parasitol 164:190–200
Fayer R, Santín M, Trout JM (2008) Cryptosporidium ryanae n. sp. (Apicomplexa: Cryptosporidiidae) in cattle (Bos taurus). Vet Parasitol 156:191–198
Feng Y, Alderisio KA, Yang W, Blancero LA, Kuhne WG, Nadareski CA, Reid M, Xiao L (2007) Cryptosporidium genotypes in wildlife from a New York watershed. Appl Environ Microbiol 73:6475–6483
Geurden T, Thomas P, Casaert S, Vercruysse J, Claerebout E (2008) Prevalence and molecular characterisation of Cryptosporidium and Giardia in lambs and goat kids in Belgium. Vet Parasitol 155:142–145
Giles M, Chalmers R, Pritchard G, Elwin K, Mueller-Doblies D, Clifton-Hadley F (2009) Cryptosporidium hominis in a goat and a sheep in the UK. Vet Rec 164:24–25
Jellison KL, Lynch AE, Ziemann JM (2009) Source tracking identifies deer and geese as vectors of human-infectious Cryptosporidium genotypes in an urban/suburban watershed. Environ Sci Technol 43:4267–4272
Jiang J, Alderisio KA, Xiao L (2005) Distribution of Cryptosporidium genotypes in storm event water samples from three watersheds in New York. Appl Environ Microbiol 71:4446–4454
Karanis P, Kourenti C, Smith H (2007) Waterborne transmission of protozoan parasites: a worldwide review of outbreaks and lessons learnt. J Water Health 5:1–38
Kvác M, Hanzlíková D, Sak B, Kvetonová D (2009) Prevalence and age-related infection of Cryptosporidium suis. C. muris and Cryptosporidium pig genotype II in pigs on a farm complex in the Czech Republic. Vet Parasitol 160:319–322
Leoni F, Mallon ME, Smith HV, Tait A, McLauchlin J (2007) Multilocus analysis of Cryptosporidium hominis and Cryptosporidium parvum isolates from sporadic and outbreak-related human cases and C. parvum isolates from sporadic livestock cases in the United Kingdom. J Clin Microbiol 45:3286–3294
Majewska AC, Werner A, Sulima P, Luty T (2000) Prevalence of Cryptosporidium in sheep and goats bred on five farms in west-central region of Poland. Vet Parasitol 89:269–275
McLauchlin J, Amar C, Pedraza-Díaz S, Nichols GL (2000) Molecular epidemiological analysis of Cryptosporidium spp. in the United Kingdom: results of genotyping Cryptosporidium spp. in 1, 705 fecal samples from humans and 105 fecal samples from livestock animals. J Clin Microbiol 38:3984–3990
Mueller-Doblies D, Giles M, Elwin K, Smith RP, Clifton-Hadley FA, Chalmers RM (2008) Distribution of Cryptosporidium species in sheep in the UK. Vet Parasitol 154:214–219
Navarro-i-Martinez L, da Silva AJ, Bornay-Llinares FJ, Moura IN, del Aguila C, Oleaga A, Pieniazek NJ (2007) Detection and molecular characterization of Cryptosporidium bovis-like isolate from a newborn lamb in Spain. J Parasitol 93:1536–1538
Paoletti B, Giangaspero A, Gatti A, Iorio R, Cembalo D, Milillo P, Traversa D (2009) Immunoenzymatic analysis and genetic detection of Cryptosporidium parvum in lambs from Italy. Exp Parasitol 122:349–352
Pollock KG, Ternent HE, Mellor DJ, Chalmers RM, Smith HV, Ramsay CN, Innocent GT (2009) Spatial and temporal epidemiology of sporadic human cryptosporidiosis in Scotland. Zoonoses. Public. Health (in press)
Pritchard GC, Marshall JA, Giles M, Chalmers RM, Marshall RN (2007) Cryptosporidium parvum infection in orphan lambs on a farm open to the public. Vet Rec 161:11–14
Quílez J, Torres E, Chalmers RM, Hadfield SJ, Del Cacho E, Sánchez-Acedo C (2008) Cryptosporidium genotypes and subtypes in lambs and goat kids in Spain. Appl Environ Microbiol 74:6026–6031
Ruecker NJ, Braithwaite SL, Topp E, Edge T, Lapen DR, Wilkes G, Robertson W, Medeiros D, Sensen CW, Neumann NF (2007) Tracking host sources of Cryptosporidium spp. in raw water for improved health risk assessment. Appl Environ Microbiol 73:3945–3957
Ryan UM, Bath C, Robertson I, Read C, Elliot A, Mcinnes L, Traub R, Besier B (2005) Sheep may not be an important zoonotic reservoir for Cryptosporidium and Giardia parasites. Appl Environ Microbiol 71:4992–4997
Santín M, Fayer R (2007) Intragenotypic variations in the Cryptosporidium sp. cervine genotype from sheep with implications for public health. J Parasitol 93:668–672
Santín M, Trout JM, Fayer R (2007) Prevalence and molecular characterization of Cryptosporidium and Giardia species and genotypes in sheep in Maryland. Vet Parasitol 146:17–24
Santín M, Trout JM, Xiao L, Zhou L, Greiner E, Fayer R (2004) Prevalence and age-related variation of Cryptosporidium species and genotypes in dairy calves. Vet Parasitol 122:103–117
Soltane R, Guyot K, Dei-Cas E, Ayadi A (2007) Prevalence of Cryptosporidium spp. (Eucoccidiorida: Cryptosporiidae) in seven species of farm animals in Tunisia. Parasite 14:335–338
Sturdee AP, Bodley-Tickell AT, Archer A, Chalmers RM (2003) Long-term study of Cryptosporidium prevalence on lowland farm in the United Kingdom. Vet Parasitol 116:97–113
Wang R, Wang J, Sun M, Dang H, Feng Y, Ning C, Jian F, Zhang L, Xiao L (2008a) Molecular characterization of the Cryptosporidium cervine genotype from a sika deer (Cervus nippon Temminck) in Zhengzhou, China and literature review. Parasitol Res 103:865–869
Wang YL, Cui B, Jian FC, Zhang LX, Ning CS, Dong HP, Lu QB, Gao GQ (2008b) Epidemiological investigation of ovine cryptosporidiosis in Henan Province. Vet Sci Chin 38:160–164
Xiao L, Fayer R (2008) Molecular characterisation of species and genotypes of Cryptosporidium and Giardia and assessment of zoonotic transmission. Int J Parasitol 38:1239–1255
Xiao L, Feng Y (2008) Zoonotic cryptosporidiosis. FEMS Immunol Med Microbiol 52:309–323
Xiao L, Herd R, Rings D (1993) Diagnosis of Cryptosporidium on a sheep farm with neonatal diarrhea by immunofluorescence assays. Vet Parasitol 47:17–23
Xiao L, Fayer R, Ryan U, Upton SJ (2004) Cryptosporidium taxonomy: recent advances and implications for public health. Clin Microbiol Rev 17:72–97
Xiao L, Alderisio K, Limor J, Royer M, Lal AA (2000) Identification of species and sources of Cryptosporidium oocysts in storm waters with a small-subunit rRNA-based diagnostic and genotyping tool. Appl Environ Microbiol 66:5492–5498
Xiao L, Singh A, Limor J, Graczyk TK, Gradus S, Lal A (2001) Molecular characterization of Cryptosporidium oocysts in samples of raw surface water and wastewater. Appl Environ Microbiol 67:1097–1101
Xiao L, Escalante L, Yang C, Sulaiman I, Escalante AA, Montali RJ, Fayer R, Lal AA (1999) Phylogenetic analysis of Cryptosporidium parasites based on the small-subunit rRNA gene locus. Appl Environ Microbiol 65:1578–1583
Yang R, Jacobson C, Gordon C, Ryan U (2009) Prevalence and molecular characterisation of Cryptosporidium and Giardia species in pre-weaned sheep in Australia. Vet Parasitol 161:19–24
Yang W, Chen P, Villegas EN, Landy RB, Kanetsky C, Cama V, Dearen T, Schultz CL, Orndorff KG, Prelewicz GJ, Brown MH, Young KR, Xiao L (2008) Cryptosporidium source tracking in the Potomac River watershed. Appl Environ Microbiol 74:6495–6504
Acknowledgments
This study was supported in part by the National Natural Science Foundation of China (number 30771881, 30871863, and 30928019), Henan Province Great Special Fund of Public Welfare (Henan Province Financial Administration number 2008-145), and Ministry of Health Special Funds of Public Sector Research (number 200808012).We thank Professors Tengyun Gao, Ming Li, and Feng Lin for their assistance in sample collection. We also thank Heping Dong, Qingbin Lu, Gengqu Gao, Chaofeng Ma, and Ke Shi for technical assistance.
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Wang, Y., Feng, Y., Cui, B. et al. Cervine genotype is the major Cryptosporidium genotype in sheep in China. Parasitol Res 106, 341–347 (2010). https://doi.org/10.1007/s00436-009-1664-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-009-1664-x