Abstract
To better understand the structure and variability of the 45S rDNA cistron and its evolutionary dynamics in grasshoppers, we performed a detailed analysis combining classical and molecular cytogenetic data with whole-genome sequencing in Abracris flavolienata, which shows extraordinary variability in the chromosomal distribution for this element. We found astonishing variability in the number and size of rDNA clusters at intra- and inter-population levels. Interestingly, FISH using distinct parts of 45S rDNA cistron (18S rDNA, 28S rDNA, and ITS1) as probes revealed a distinct number of clusters, suggesting independent mobility and amplification of the 45S rDNA components. This hypothesis is consistent with the higher genomic coverage of almost the entire cistron of 45S rDNA observed in A. flavolineata compared to other grasshoppers, besides coverage variability along the 45S rDNA cistron in the species. In addition, these differences in coverage for distinct components of the 45S rDNA cistron indicate emergence of pseudogenes evidenced by existence of truncated sequences, demonstrating the rDNA dynamics in the species. Although the chromosomal distribution of 18S rDNA was highly variable, the chromosomes 1, 3, 6, and 9 harbored rDNA clusters in all individuals with the occurrence of NOR activity in pair 9, suggesting ancestry or selective pressures to prevent pseudogenization of rDNA sequences in this chromosome pair. Additionally, small NORs and cryptic rDNA loci were observed. Finally, there was no evidence of enrichment and association of transposable elements, at least, inside or nearby rDNA cistron. These findings broaden our knowledge of rDNA dynamics, revealing an independent movement and amplification of segments of 45S rDNA cistron, which in A. flavolineata could be attributed to ectopic recombination.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The 45S rDNA (major rDNA) is a primarily head-to-tail tandemly repeated genomic element that is arranged at one or multiple sites, variable in size, which can be located in one or several chromosomes. The rDNA cistron consists of three genes for 18S, 5.8S, and 28S rRNAs, separated by internal transcribed spacers (ITS1 and ITS2) besides external transcribed spacers and intergenic spacer (Nei and Rooney 2005; Eickbush and Eickbush 2007). Usually, eukaryotes have hundreds to millions of major rDNA copies, which represent one of the most abundant repetitive DNAs. These copies are required for a massive number of ribosomes during a period of fast growth (Eickbush and Eickbush 2007). The copies are clustered in the nucleolar organizer regions (NORs) that generate nucleoli as a result of 45S rDNA transcription during ribosome synthesis.
From the chromosomal point of view, the 45S rDNA is the most widely studied element in eukaryotes by means of fluorescence in situ hybridization (FISH) using distinct parts of the cistron as probes, mostly 18S and 28S/26S. Variability between species in the number and location of rDNA loci is well documented (Garcia et al. 2012; Sochorová et al. 2018). To a lesser extent, intraspecific polymorphisms were also observed, for example, in fish (Castro et al. 2001; Mantovani et al. 2005), grasshoppers (Cabrero et al. 2003; Veltsos et al. 2009), amphibians (Bi et al. 2009; Schmid et al. 2017), and plants (Shishido et al. 2000; Pedrosa-Harand et al. 2006). Chromosomal rearrangements, like translocations and fusions, could be responsible for rDNA movement in distinct chromosomes, causing part of the extensive variability documented. Moreover, the variability of the number and position of major rDNA loci could be caused by transposition mediated by transposable elements (TEs) and ectopic recombination (Schubert 1984; Shishido et al. 2000; Cai et al. 2006; Datson and Murray 2006; Schubert and Wobus 1985; Reed and Phillips 2000; Raskina et al. 2004; Schmid et al. 2017). The dispersion of rDNA sites could also be favored by the reinsertion of circular intermediates (Reed and Phillips 2000; Gornung 2013).
In grasshoppers as in other eukaryotes, the 45S rDNA is the most frequently mapped repetitive DNA on chromosomes, revealing extensive variability, especially in Acrididae (Cabrero and Camacho 2008; Veltsos et al. 2009; Bueno et al. 2013). Recently, a particularly high number of rDNA clusters and high intraspecific variability were observed in the grasshopper Abracris flavolineata (Bueno et al. 2013), in individuals from the Rio Claro/SP (São Paulo), Brazil, population. Using FISH with the 18S rDNA probe, five to nine chromosomes carrying rDNA clusters were observed per haploid genome. The 18S rDNA clusters were variable in size and showed heteromorphisms (monosomicity), i.e., the presence restricted to only one chromosome of a homologous pair. The high variation of rDNA in A. flavolineata is puzzling, and this species could be a good model for understanding the chromosomal evolution of 45S rDNA at the intraspecific level. Therefore, we performed a detailed analysis of the 45S rDNA cistron in A. flavolineata, in a interdisciplinar framework using data of classical and molecular cytogenetics as well as next-generation sequencing data. Our study revealed an intriguing dynamics of the 45S rDNA in A. flavolineata, including rDNA amplification and independent movement of cistron components (genes and spacers) with conserved functionality in a specific chromosome.
Materials and methods
Animal sampling and chromosomal analysis
A total of 48 A. flavolineata adult males were used in this study. They were collected in five regions of Brazil: Rio Claro/SP (São Paulo), 22°24′45″ S, 47°31′28″ W, 10 individuals; Colina/SP, 20°70′ S, 48°52′ W, six individuals; Murici/AL (Alagoas), 9°13′ S, 35°50′ W, four individuals; Cabo/PE (Pernambuco), 8°17′15″ S, 35°2′7″ W, nine individuals; and Sta. Bárbara do Pará/PA (Pará), 1°13′27″ S, 48°17′38″ W, 10 individuals, and also in Posadas/Missiones, Argentina, 27°25′ S, 55°56′ W, nine individuals (Supplementary Material 1). The animals were anesthetized and testis follicles were dissected and fixed in modified Carnoy’s solution (3:1, 100% ethanol/glacial acetic acid) for meiotic chromosomes obtaining. Mitotic chromosomes were obtained from embryo neuroblasts using standard procedures described elsewhere (Webb et al. 1978). Whole animals were stored in 100% ethanol for DNA extraction.
Slides were prepared by maceration of testis or embryo in a drop of 50% acetic acid followed by spreading on a hot plate at 40–45 °C. For conventional analyses, chromosome preparations were stained with 5% Giemsa. Selected unstained slides were used for FISH. Silver nitrate staining was done according to the protocol described by Rufas et al. (1987).
Genome sequencing and computational analysis
DNA of male individuals of A. flavolineata (one without B chromosome, 0B and one with one B chromosome, +1B) was extracted from the saltatorial legs using the Qiagen DNeasy kit (Qiagen Inc., CA, USA) following the manufacturer’s protocol. The extracted DNA was sonicated to obtain fragments of ~ 500 bp and used to prepare paired-end libraries as recommended by Illumina (Illumina Inc., San Diego, CA, USA). The libraries were sequenced with Illumina MiSeq yielding paired-end reads up to 300 nt. Quality trimming of the reads was performed using the Trimmomatic software (Bolger et al. 2014) with minimum quality of Q20 and minimum length of 150 nt. The A. flavolineata 45S rDNA partial sequence was assembled with MITObim (Hahn et al. 2013) using a random selection of 1 million read pairs, 500,000 pairs from each library. As a reference, we used a partial rDNA sequence of a Gomphocerinae species obtained by Mallatt and Giribet (2006) with the accession number AY859546.
In order to compare the genomic coverage in the 45S rDNA cluster of A. flavolineata, we used gDNA paired-end Illumina HiSeq2000 sequencing (2 × 101-nt reads) from other four species: two Acrididae species, Locusta migratoria (SRR2911427, Ruiz-Ruano et al. 2016) and Parascopas sanguineus, one Pamphagidae species Eumigus monticola (SRR3000673, Ruiz-Ruano et al. 2017), and one Pyrgomorphidae species Pyrgomorpha conica (SRR3953136, Ruiz-Ruano et al. 2018). For these four species, we also performed quality trimming using the Trimmomatic software and assembled a partial 45S rDNA sequence as described above, using the previously assembled 45S rDNA sequence of A. flavolineata as a reference.
We estimated the coverage along the 45S rDNA cluster separately in the six Illumina libraries performing a previous selection of homologous reads to the 45S rDNA of its own species using the BLAT aligner (Kent 2002) integrated into a custom script (https://github.com/fjruizruano/ngs-protocols/blob/master/mapping_blat_gs_saver.py). The selected reads were mapped using Bowtie2 (Langmead and Salzberg 2012) with default options, and the coverage was obtained by each position with the software pysamstats (https://github.com/alimanfoo/pysamstats). We compared coverage values between species estimating the genome proportion as the coverage in number of mapped reads divided by the library size.
In addition, we tried to identify external elements associated with the rDNA of A. flavolineata that could be responsible for rDNA mobility. For this purpose, we used the previously selected reads using BLAT and joined the read paired using the “fastq-join” software of the FASTX-Toolkit suit (Gordon and Hannon 2010) with default options. Then, we used the 45S rDNA sequence of A. flavolineata to mask homologous nucleotides using RepeatMasker (Smit et al. 2017). The masked reads were assembled with CAP3 (Huang and Madan 1999), and the resulting contigs were used for comparisons with TEs deposited in Repbase and NCBI, and a database for A. flavolineata genome generated by RepeatExplorer software containing 53 satDNA families and TEs (Milani et al. in preparation). Finally, we estimated the abundance of these contigs in the reads homologous to the rDNA and the complete library using RepeatMasker (Smit et al. 2017). The database for satDNAs and TEs from A. flavolineata genome was also used for an internal BLAST using as query distinct parts of 45S rDNA cistron (18S, ITS1, 5.8S, ITS2, 28S) to check similarity.
Probes and FISH experiments
We determined the chromosomal location along the 45S rDNA in A. flavolineata using a set of probes covering different regions of the element. Firstly, we amplified by PCR a region of the 18S rDNA gene using A. flavolineata genomic DNA with the combination of primers described by Cabral-de-Mello et al. (2010), Sca18S_F and Sca18S_R. This PCR product was used as a probe in FISH experiments in all populations and all 48 individuals studied. It largely corresponds to the region of low genome coverage in comparison to other regions of 45S rDNA revealed by genomic analysis (see “Results”; Fig. 1). FISH using another probe containing a small part of 18S, ITS1+5.8S+ITS2, and a small part of 28S (named ITS1–2 probe) was performed in one individual of each population. Unlike the first probe for the 18S rDNA, this probe corresponds to a region with high coverage for 45S rDNA indicated by genomic analysis (see “Results”; Fig. 1). The probe was obtained using the primers 18S 5′ GCTTTTGTACACACCGCCCGTCGC and ITS4 5′ ATATGCTTAAATTCAGCGGG. Additionally, we performed FISH in one individual of Rio Claro/SP using two probes composed separately of the ITS1 and 28S rDNA cistron regions that showed high coverage according to genomic data compared to 18S rDNA probe (see “Results”; Fig. 1). For the latter FISH experiments, the same individual (one embryo) was used and these probes were each combined with the 18S rDNA probe. The primers for these two regions were designed using the consensus 45S rDNA cistron assembled from the A. flavolineata genome and the Primer3 software (Untergasser et al. 2012), Af_28S_F 5′ CTGGCACTGTGAGGTGACAT and Af_28S_R 5′ TTAATCCCACGGATGGTAGC, and Af_ITS1_F 5′ CTAGCTGTACGGCAGCAATG and Af_ITS1_R 5′ GCCCCTGTTCCAGTTACAAA. The PCR fragments were sequenced using the Macrogen service to check the amplification of the desired sequences.
Genomic coverage for 45S rDNA cistron in five grasshopper species, Abracris flavolineata, Locusta migratoria, Pyrgomorpha conica, Parascopas sanguineus, and Eumigus monticola. Note the higher coverage for A. flavolineata and the variable coverage along 45S rDNA in this species that are in contrast to other species. The coverage for ITS1 and ITS2 is shown only for A. flavolineata. Primers used for amplification of distinct regions of the 45S rDNA cistron from A. flavolineata are also indicated
The probes were labeled with digoxigenin-11-dUTP and biotin-14-dATP by PCR and nick translation, respectively. FISH was done following the Pinkel et al. (1986) protocol with modifications proposed by Cabral-de-Mello et al. (2010). The probes were detected using anti-digoxigenin-Rhodamine (Roche) or streptavidin-Alexa Fluor 488 (Invitrogen). After FISH, preparations were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) and mounted in VECTASHIELD (Vector, Burlingame, CA, USA). Chromosomes and signals were observed using an Olympus microscope BX61 equipped with fluorescence lamp and appropriate filters. Photographs were recorded with a DP70 cooled digital camera. Images were merged and optimized for brightness and contrast with Adobe Photoshop CS6.
Results
rDNA cistron and associated sequences in A. flavolineata genome
The genomic analysis of 45S rRNA cistron of Abracris flavolineata and other four grasshopper species revealed a high variation of coverage (Fig. 1). Eumigus monticola, P. sanguineus, P. conica, and L. migratoria revealed similar coverage along the entire cistron, with a higher copy number for L. migratoria. However, the number of copies in A. flavolineata was higher than in other species, and the copy number varied along the whole cistron. The lowest coverage was found around the position 1000 nt of the 18S rRNA gene, with a similar number of copies observed in the other species (Fig. 1).
The assembly of masked reads to get the elements associated with the rDNA cistron yielded 60 contigs, but we could only annotate 16 of them. Five contigs were homologous with transposable element R2, two contigs with R1, Helitron and Polinton, and one contig with LINE/I, LOA, RTE, Mariner, and the IGS of the 45S rDNA. A higher proportion of reads homologous with 45S rDNA was similar to the IGS, i.e., 2.02%, and the next in abundance was an unannotated contig with about 0.15% (Supplementary Material 2). Comparison of distinct parts of 45S rDNA cistron (18S, ITS1, 5.8S, ITS2, and 28S) with the database for satDNAs and TEs from A. flavolineata did not reveal any coincidence.
Chromosomal location of rDNA clusters
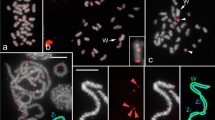
The karyotypes of all individuals studied were the same to previous analysis (Cella and Ferreira 1991; Bueno et al. 2013). Physical mapping by FISH with the 18S rDNA probe performed in 48 individuals from the six populations analyzed revealed extensive variation in four aspects: (i) the number of chromosome pairs carrying rDNA clusters, (ii) the specific chromosomes carrying rDNA, (iii) the size of these clusters that were classified as large (L) and small (S) (Supplementary Material 3), and (iv) heteromorphism (monosomicity) in the presence/absence of rDNA in one homolog of some chromosome pairs (Figs. 2 and 3; Supplementary Material 4). Although the rDNA clusters were invariably located near the pericentromeric regions, in most cases, they were stretched to short arms, forming large blocks (Fig. 2; Supplementary Material 3). The number of rDNA-carrying chromosome pairs ranged from 6 (observed in individuals from Cabo/PE) to 11 (Colina/SP), with frequent occurrence of 8 or 9 (mean = 8.35, SE = 0.16). Significant differences were found among the six populations analyzed, with Colina/SP population showing the highest average number (10), followed by Murici/AL (9), Rio Claro (8.3), Posadas/Missiones and Sta. Bárbara (8.3), and Cabo/PE (7.4) (Supplementary Material 4).
Examples of chromosomal distribution of 18S rDNA in meiotic chromosomes (metaphase I) of Abracris flavolineata revealed by FISH in six different populations (a–f). One individual per population is shown; autosome bivalents are arranged in decreasing order of size, and the centromeres are oriented up and down. Note the variability in the number of rDNA clusters, their size, and location. Bar = 5 μm
The autosomes 1, 3, 6, and 9, in order of decreasing size, carried rDNA in all individuals and populations, whereas autosomes 7 and 11 rarely showed hybridization signals of the 18S rDNA probe, specifically only in 20.8% and 4.2% of cases, respectively (Fig. 3; Supplementary Material 4). The remaining chromosomes carried rDNA at intermediate frequencies: X chromosome (91.7%), autosome 2 (75%), autosome 10 (64.6%), autosome 4 (62.5%), and autosomes 5 and 8 (58.3%). The size of the rDNA clusters also showed variation, with autosomes 1 and 9 always showing large clusters, whereas autosomes 3 and 6 showing large or small clusters. Chromosomes 2, 10, and 11 showed large signals or absence of clusters; chromosome 8 presented small signal or absence of cluster; large or small signals or absence of clusters were observed in chromosomes 4, 5, 7, and X. These patterns varied depending on the population (Fig. 3; Supplementary Material 4).
Remarkably, when FISH experiments were performed using the probe containing ITS1+5.8S+ITS2 and part of 18S and 28S rDNA regions (ITS1–2 probe), we found additional FISH signals in the pericentromeric regions of chromosomes which failed to hybridize with the 18S rDNA probe (Supplementary Material 5). For instance, Supplementary Material 5 shows the presence of rDNA sites on 10 chromosomes in the Cabo/PE population, including the presence of additional terminal sites on four autosome bivalents and interstitial ones on two autosome bivalents and the X univalent. This greatly contrasts with the finding of only pericentromeric sites on a maximum of nine bivalents when the 18S rDNA probe was used (see Fig. 2; Supplementary Material 4). These additional pericentromeric, interstitial, and/or terminal rDNA sites were also observed in the five remaining populations when the ITS1–2 probe was used (Supplementary Material 5). No signals using this probe (ITS1–2) were observed in the B chromosome of individuals from Rio Claro/SP population (Supplementary Material 5), similar to data obtained using only the low genome coverage region of 18S rDNA gene (Bueno et al. 2013).
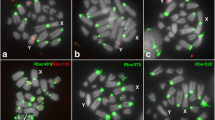
FISH with ITS1 and 28S rDNA probes also revealed distinct chromosomal distribution in comparison with the 18S rDNA probe. Pericentromeric regions of all chromosomes, in some cases also adjacent regions of short arms, were labeled with both probes, and additional terminal signals were observed in most chromosomes. The X chromosome showed one interstitial signal. The pericentromeric signals partially overlapped with 18S rDNA signals (Fig. 4).
FISH mapping of distinct regions of the 45S rDNA cistron in mitotic cells of one embryo from the Rio Claro/SP population, (a, d) 18S rDNA (red), (b) ITS1 (green), (e) 28S rDNA (green), and (c,f) merged signals (green and red) with chromosomes stained with DAPI (gray). Note the distinct chromosomal distribution for 18S rDNA and the other rDNA cistron regions. The X chromosome is indicated. Bar = 5 μm
Nucleolar organizer regions
The analysis of one individual from each population, randomly selected, using silver nitrate impregnation revealed active NORs in the chromosome 9 for all individuals. The chromosome 10 also often exhibited silver nitrate impregnation, except for the individual from the population of Sta. Bárbara do Pará/PA. In contrast, chromosomes 1, 8, and 11 did not show silver nitrate impregnation (Fig. 5). Besides large NORs in some chromosomes, sites with faint silver nitrate staining were observed, indicating the occurrence of small NORs (Fig. 5). Cryptic loci that were stained by silver nitrate but without FISH signals for 18S rDNA were observed in chromosomes 4, 5, and 10 in one individual from Cabo/PE (Fig. 5a) and chromosome 10 in individuals from Murici/AL (Fig. 5c) and Posadas/Misiones, Argentina (Fig. 5d).
Active nucleolar organizer regions (NORs) in diplotene revealed by silver nitrate staining in individuals of Abracris flavolineata from six populations (a–f). Chromosomes carrying NORs are numbered, and black arrowheads indicate cryptic rDNA sites (without FISH signals of 18S rDNA). Blue arrowheads show small NORs. (a, b, d, e) Chromosomes with small NORs are depicted in the insets. Bar = 5 μm; bar in insets = 2.5 μm
Discussion
Variability in the number of major rDNA chromosomal clusters is commonly found at the interspecific level. In animals, it ranges from 1 to 54 sites, although the median number of sites is close to 2 (Cabrero and Camacho 2008; Gornung 2013; Sochorová et al. 2018) and in plants, this range is from 1 to 45 sites with median of four clusters (Roa and Guerra 2012; Garcia et al. 2017). At the intraspecific level, variability in the number and chromosomal distribution of 45S rDNA clusters was documented to a lesser extent (Castro et al. 2001; Cabrero et al. 2003; Mantovani et al. 2005; Bi et al. 2009; Veltsos et al. 2009; Garcia et al. 2017; Schmid et al. 2017). In the present study, we observed remarkable intraspecific variability in six populations of A. flavolineata, and we also observed an unusual pattern, a high rDNA variation within these populations. The high variability was manifested by the occurrence of different numbers of rDNA clusters in individuals from the same population, clusters with distinct sizes and heteromorphisms. For example, in 10 individuals from Rio Claro/SP, six distinct patterns in the rDNA distribution were observed, demonstrating high dynamics of this repetitive DNA family. As proposed for other insect groups, this high dynamics could be caused by different molecular mechanisms leading to the origin and spreading of repetitive DNAs in eukaryote genomes (Dover 1986; Montgomery et al. 1991; Charlesworth et al. 1994; Petrov et al. 2003; Nguyen et al. 2010). In A. flavolineata, the difference in cluster size between distinct chromosomes and the occurrence of heteromorphisms supports the idea of rDNA movement with the emergence of new clusters and cluster deletion (Dubcovsky and Dvorák 1995).
In other orthopterans, an extraordinary variation of the major rDNA, like documented in A. flavolineata (from 10 to 18 per diploid genome), was also reported in Podisma pedestris (Veltsos et al. 2009) and in Eyprepocnemis plorans (Cabrero et al. 2003). In P. pedestris, the number of rDNA sites varied from three to 13 in diploid genomes, and in E. plorans from Spain populations, it varied from four to eight in the haploid complement. In comparison with other grasshoppers, the chromosomal distribution of major rDNA in A. flavolineata is quite exceptional because (i) it is much more variable in the number of sites, (ii) it shows a high incidence of heteromorphisms, (iii) clusters of distinct size occur even in the same chromosome pair, and (iv) it shows a high intrapopulation variation. The occurrence of such extreme variability in geographically distant populations suggests ancestral mobile capacity of rDNA in A. flavolineata chromosomes, before geographical dispersion of the species.
Interestingly, mapping of other 45S rDNA cistron regions revealed distinct patterns compared to 18S rDNA. This finding suggests the mobility of distinct regions of the 45S rDNA cistron in the genome of A. flavolineata independently of each other. The occurrence of chromosomal clusters for distinct regions of 45S rDNA was rarely observed in other organisms. One of the few examples is Drosophila simulans, in which the Y chromosome carries an amplified rDNA spacer without rRNA genes (Lohe and Roberts 1990) and fish from Coregonus genus (Symonová et al. 2013). Unlike A. flavolineata, the use of distinct rDNA probes in another grasshopper species P. pedestris, namely the pTa71 clone from wheat (Gerlach and Bedbrook 1979) and the 18S rDNA, revealed the same chromosomal labeling profile. However, in most species, the mobility of distinct rDNA regions was not studied because the entire 45S or 35S rDNA units were used as probes, and the independent mobility of distinct regions of rDNA could thus be underestimated.
We should also take into account that the multiplication of hybridization signals using the entire rDNA unit as a probe could in fact be generated due to the similarity of distinct parts of rDNA, including the gene spacers, with other repetitive DNAs such as satellite DNAs (satDNAs). Sharing sequence similarity between IGS of rDNA and satDNAs was observed, for example in tomato, potato, and Vicia faba (Maggini et al. 1991; Stupar et al. 2002; Jo et al. 2009). In A. flavolineata, none of the 45S rDNA region used as a probe was similar to satDNAs or transposable elements annotated in the species (Milani et al. in preparation), giving support that certainly the signals observed for distinct rDNA probes used here are only generated by the presence of rDNA sequences. This data also supports that the 45S rDNA in A. flavolineata did not give origin to a satDNA sequence, as reported in other few species (Garrido-Ramos 2017).
Besides the independent chromosomal movement of rDNA segments, our analysis of genome coverage for the 45S rDNA cistron demonstrated the independent multiplication of its distinct regions. This was evidenced by the difference in the number of copies along the entire sequence and also by differences in the number of chromosomal clusters recorded. The multiplication of rDNA clusters in A. flavolineata is evident from a comparison with other species from different grasshopper families having a lower number of 45S rDNA copies as revealed by genome analysis and a lower number of chromosomal clusters per haploid genome, such as P. sanguineus with one cluster (D. C. Cabral-de-Mello, unpublished data), L. migratoria with three clusters (Teruel et al. 2010), and E. monticola and P. conica with two clusters (Ruiz-Ruano et al. 2017, 2018). However, the number of entire copies of 45S rDNA clusters observed in A. flavolineata was similar to other grasshoppers. The data obtained in A. flavolineata thus support the idea of requiring of a lower number of functional 45S rDNA clusters in relation to the total number observed in the genome, some of which are pseudogenized (i.e., truncated), similarly as found in other multi-copy sequences, such as those from multigene families (Eirín-López et al. 2012). The increase in pseudogene quantity could reflect the multiplication of various parts of the 45S rDNA cistron observed here and the independent movement of 45S rDNA fragments. However, we cannot rule out the possibility of moving of the entire rDNA cistron followed by its pseudogenization through elimination of cistron components. Intensive multiplication, movement, and pseudogenization of 45S rDNA in A. flavolineata may have occurred because the original rDNA clusters are not affected and persist as functional sites, which perform their role in ribosome formation. The resulting changes in rDNA distribution are certainly not deleterious because they involve non-functional DNAs. Comparison with other four grasshopper species belonging to three families that showed similar coverage along the entire 45S rDNA cistron but lower than that in A. flavolineata suggests that the intensive multiplication of various rDNA regions and their pseudogenization is not a common feature in grasshoppers.
The intriguing multiplication and spreading of rDNA in A. flavolineata could be caused by various mechanisms, such as chromosomal rearrangements, transposition events associated with TEs, or ectopic recombination, besides the occurrence of extrachromosomal circular DNA (eccDNAs). Considering that the karyotype of A. flavolineata is the ancestral for Acrididae grasshoppers (2n = 23,X0), we suppose that large chromosomal rearrangements are not involved in rDNA movement. Similarly, since the rDNA does not have independent movement capacity and it was only sporadically associated with TEs in A. flavolineata, we suppose that its movement is not mediated by transposition. Moreover, some of the TEs that were found associated with the 45S rDNA cistron like R1 and R2 are not capable of carrying sequences (Han 2010). However, we cannot rule out the possibility that other TEs, not associated with 45S rDNA and not found here, contributed to rDNA mobility. The TE-based movement of ribosomal genes was suggested in other species, such as the frog Craugastor fitzingeri (Schmid et al. 2017) and the plant Aegilops speltoides (Raskina et al. 2004). The spreading of rDNA in A. flavolineata could be attributed to ectopic recombination acting independently on different segments of 45S rDNA, with dispersion of minor rDNA loci (containing few rRNA genes), followed by an increase in copy number and potential elimination of original loci similar to the hypothesis proposed by Dubcovisky and Dvořák (1995). This could be supported by the presence of sites with variable size, including small ones, besides the observation of cryptic and small NORs, like observed in other grasshopper species (Cabrero and Camacho 2008).
Despite the high variability in the chromosomal distribution of rDNA, there were found some chromosomes (1, 3, 6, and 9) consistently showing hybridization signals of the 18S rDNA probe in all populations. This finding suggests that these chromosomes could be ancestral elements carrying rDNA clusters in this species. Of these chromosomes, only the pair 9 showed active NOR after silver staining in all individuals analyzed from the six populations studied. This strongly suggests that in this bivalent, the pseudogenization of copies are prevented, keeping its transcriptional activity. Moreover, it could be the ancient NOR chromosome for the species. The presence of active rDNA sites in the chromosome 9 was reported in 17 of 30 Acrididae grasshopper species studied by Cabrero and Camacho (2008), all with 2n = 23. It was also observed in Orthoscapheus rufipes (Rocha et al. 2011) that belongs to the same tribe (Abracrini) as A. flavolineata, giving further support for the ancestry of NOR in chromosome 9.
These findings broaden our knowledge regarding rDNA dynamics in eukaryotes, revealing high plasticity of major rDNA in A. flavolineata genome in comparison to other grasshoppers (Cabrero and Camacho 2008). This plasticity is reflected in number, position, and cluster size variability of the distinct regions of 45S rDNA. Although this variability is present in the A. flavolineata genome, the activity of the NOR-bearing chromosome is preserved. Finally, our data support an independent movement and amplification of segments of 45S rDNA cistron, which could be attributed to ectopic recombination between non-homologous chromosomes.
References
Bi K, Bogart JP, Fu J (2009) A Populational survey of 45S rDNA polymosphism in the Jefferson salamander Ambystoma jeffersonianum revealed by fluorescence in situ hybridzation (FISH). Curr Zool 55:145–149
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. https://doi.org/10.1093/bioinformatics/btu170
Bueno D, Palacios-Gimenez OM, Cabral-De-Mello DC (2013) Chromosomal mapping of repetitive DNAs in the grasshopper Abracris flavolineata reveal possible ancestry of the B chromosome and H3 histone spreading. PLoS One 8:e66532. https://doi.org/10.1371/journal.pone.0066532
Cabral-de-Mello DC, Moura RC, Martins C (2010) Chromosomal mapping of repetitive DNAs in the beetle Dichotomius geminatus provides the first evidence for an association of 5S rRNA and histone H3 genes in insects, and repetitive DNA similarity between the B chromosome and a complement. Heredity 104:393–400. https://doi.org/10.1038/hdy.2009.126
Cabrero J, Camacho JPM (2008) Location and expression of ribosomal RNA genes in grasshoppers: abundance of silent and cryptic loci. Chromosom Res 16:595–607. https://doi.org/10.1007/s10577-008-1214-x
Cabrero J, Perfectti F, Gómez R, Camacho JPM, Lopéz-Leon MD (2003) Population variation in the a chromosome distribution of satellite DNA and ribosomal DNA in the grasshopper Eyprepocnemis plorans. Chromosom Res 11:375–381. https://doi.org/10.1023/A:1024127525756
Cai Q, Zhang DM, Liu ZL, Wang XR (2006) Chromosomal localization of 5S and 18S rDNA in five species of subgenus Strobus and their implications for genome evolution of Pinus. Ann Bot 97:715–722. https://doi.org/10.1093/aob/mcl030
Castro J, Rodriâguez S, Pardo BG, Sánchez L, Martínez P (2001) Population analysis of an unusual NOR-site polymorphism in brown trout (Salmo trutta L.). Heredity 86:291–302
Cella DM, Ferreira A (1991) The cytogenetics of Abracris flavolineata (Orthoptera, Caelifera, Ommatolampinae, Abracrini). Rev Bras Genet 14:315–329
Charlesworth B, Sniegowski P, Stephan W (1994) The evolutionary dynamics of repetitive DNA in eukaryotes. Nature 371:215–220. https://doi.org/10.1038/371215a0
Datson PM, Murray BG (2006) Ribosomal DNA locus evolution in Nemesia: transposition rather than structural rearrangement as the key mechanism? Chromosom Res 14:846–857. https://doi.org/10.1007/s10577-006-1092-z
Dover GA (1986) Molecular drive in multigene families: how biological novelties arise, spread and are assimilated. Trends Genet 2:159–165. https://doi.org/10.1016/0168-9525(86)90211-8
Dubcovsky J, Dvorák J (1995) Ribosomal RNA multigene loci - nomads of the Triticeae genomes. Genetics 140:1367–1377
Eickbush TH, Eickbush DG (2007) Finely orchestrated movements: evolution of the ribosomal RNA genes. Genetics 175:477–485. https://doi.org/10.1534/genetics.107.071399
Eirín-López JM, Rebordinos L, Rooney AP, Rozas J (2012) The birth-and-death evolution of multigene families revisited. Genome Dyn 7:170–196. https://doi.org/10.1159/000337119
Garcia S, Garnatje T, Kovařík A (2012) Plant rDNA database: ribosomal DNA loci information goes online. Chromosoma 121:389–394. https://doi.org/10.1007/s00412-012-0368-7
Garcia S, Kovařík A, Leitch AR, Garnatie T (2017) Cytogenetic features of rRNA genes across land plants: analysis of the plant rDNA database. Plant J 89:1020–1030. https://doi.org/10.1111/tpj.13442
Garrido-Ramos MA (2017) Satellite DNA: an evolving topic. Genes 8:230. https://doi.org/10.3390/genes8090230
Gerlach WL, Bedbrook JR (1979) Cloning and characterization of ribosomal RNA genes from wheat and barley. Nucleic Acids Res 7:1869–1885
Gordon A, Hannon GJ (2010) Fastx-toolkit. FASTQ/a short-reads preprocessing tools. Available at http://hannonlab.cshl.edu/fastx_toolkit
Gornung E (2013) Twenty years of physical mapping of major ribosomal RNA genes across the Teleosts: a review of research. Cytogenet Genome Res 141:90–102. https://doi.org/10.1159/000354832
Hahn C, Bachmann L, Chevreux B (2013) Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads - a baiting and iterative mapping approach. Nucleic Acids Res 41:129–129. https://doi.org/10.1093/nar/gkt371
Han JS (2010) Non-long terminal repeat (non-LTR) retrotransposons: mechanisms, recent developments, and unanswered questions. Mob DNA 1:15. https://doi.org/10.1186/1759-8753-1-15
Huang X, Madan A (1999) CAP3: a DNA sequence assembly program. Genome Res 9:868–877. https://doi.org/10.1101/gr.9.9.868
Jo S-H, Koo D-H, Kim JF, Hur C-G, Lee S, Yang T-J, Know S-Y, Choi D (2009) Evolution of ribosomal DNA-derived satellite repeat in tomato genome. BMC Plant Biol 9:42. https://doi.org/10.1186/1471-2229-9-42
Kent WJ (2002) BLAT-the BLAST-like alignment tool. Genome Res 12:656–664. https://doi.org/10.1101/gr.229202
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with bowtie 2. Nat Methods 9:357–359. https://doi.org/10.1038/nmeth.1923
Lohe AR, Roberts PA (1990) An unusual Y chromosome of Drosophila simulans carrying amplified rDNA spacer without rRNA genes. Genetics 125:399–406
Maggini F, Cremonini R, Zolfino C, Tucci GF, D'Ovidio R, Delre V, DePace D, Scarascia Mugnozza GT, Cionini PG (1991) Structure and chromosomal localization of DNA sequences related to ribosomal subrepeats in Vicia faba. Chromosoma 100:229–234. https://doi.org/10.1007/BF00344156
Mallatt J, Giribet G (2006) Further use of nearly complete 28S and 18S rRNA genes to classify Ecdysozoa: 37 more arthropods and a kinorhynchn. Mol Phylogenet Evol 40:772–794. https://doi.org/10.1016/j.ympev.2006.04.021
Mantovani M, dos Santos LD, Moreira-Filho O (2005) Conserved 5S and variable 45S rDNA chromosomal localisation revealed by FISH in Astyanax scabripinnis (Pisces, Characidae). Genetica 123:221–216. https://doi.org/10.1007/s10709-004-2281-3
Montgomery EA, Huang SM, Langley CH, Judd BH (1991) Chromosome rearrangement by ectopic recombination in Drosophila melanogaster: genome structure and evolution. Genetics 129:1085–1098
Nei M, Rooney AP (2005) Concerted and birth-and-death evolution of multigene families. Annu Rev Genet 39:121–152. https://doi.org/10.1146/annurev.genet.39.073003.112240
Nguyen P, Sahara K, Yoshido A, Marec F (2010) Evolutionary dynamics of rDNA clusters on chromosomes of moths and butterflies (Lepidoptera). Genetica 138:343–354. https://doi.org/10.1007/s10709-009-9424-5
Pedrosa-Harand A, de Almeida CC, Mosiolek M, Blair MW, Schweizer D, Guerra M (2006) Extensive ribosomal DNA amplification during Andean common bean (Phaseolus vulgaris L.) evolution. Theor Appl Genet 112:924–933. https://doi.org/10.1007/s00122-005-0196-8
Petrov DA, Aminetzach YT, Davis JC, Bensasson D, Hirsh AE (2003) Size matters: non-LTR retrotransposable elements and ectopic recombination in Drosophila. Mol Biol Evol 20:880–892. https://doi.org/10.1093/molbev/msg102
Pinkel D, Straume T, Gray JW (1986) Cytogenetic analysis using quantitative, high sensitivity, fluorescence hybridization. Proc Natl Acad Sci U S A 83:2934–2938. https://doi.org/10.1073/pnas.83.9.2934
Raskina O, Belyayev A, Nevo E (2004) Activity of the En/Spm-like transposons in meiosis as a base for chromosome repatterning in a small, isolated, peripheral population of Aegilops speltoides Tausch. Chromosom Res 12:153–161. https://doi.org/10.1023/B:CHRO.0000013168.61359.43
Reed KM, Phillips RB (2000) Structure and organization of the rDNA intergenic spacer in lake trout (Salvelinus namaycush). Chromosom Res 8:5–16. https://doi.org/10.1023/A:1009214800251
Roa F, Guerra M (2012) Distribution of 45S rDNA sites in chromosomes of plants: structural and evolutionary implications. BMC Evol Biol 12:225. https://doi.org/10.1186/1471-2148-12-225
Rocha MF, Melo NF, Souza MJ (2011) Comparative cytogenetic analysis of two grasshopper species of the tribe Abracrini (Ommatolampinae, Acrididae). Gent Mol Biol 34:214–219. https://doi.org/10.1590/S1415-47572011000200008
Rufas JS, Gimenez-Abian J, Suja JA, Garcia De La Vega C (1987) Chromosome organization in meiosis revealed by light microscope analysis of silver-stained cores. Genome 29:706–712. https://doi.org/10.1139/g87-121
Ruiz-Ruano FJ, López-León MD, Cabrero J, Camacho JPM (2016) High-throughput analysis of the satellitome illuminates satellite DNA evolution. Sci Rep 6:28333. https://doi.org/10.1038/srep28333
Ruiz-Ruano FJ, Cabrero J, López-León MD, Camacho JPM (2017) Satellite DNA content illuminates the ancestry of a supernumerary (B) chromosome. Chromosoma 126:487–500. https://doi.org/10.1007/s00412-016-0611-8
Ruiz-Ruano FJ, Castillo-Martínez J, Cabrero J, Gómez R, Camacho JPM, López-León MD (2018) High-throughput analysis of satellite DNA in the grasshopper Pyrgomorpha conica reveals abundance of homologous and heterologous higher-order repeats. Chromosoma 127:323–340. https://doi.org/10.1007/s00412-018-0666-9
Schmid M, Steinlein C, Feichtinger W, Nanda I (2017) Chromosome banding in amphibia. XXXV. Highly mobile nucleolus organizing regions in Craugastor fitzingeri (Anura, Craugastoridae). Cytogenet Genome Res 152:180–193. https://doi.org/10.1159/000481554
Schubert I (1984) Mobile nucleolus organizing regions (NORs) in Allium (Liliaceae s. lat.) - inferences from the specifity of silver staining. Plant Syst Evol 144:291–305. https://doi.org/10.1007/BF00984139
Schubert I, Wobus U (1985) In situ hybridization confirms jumping nucleolus organizing regions in Allium. Chromosoma 92:143–148. https://doi.org/10.1007/BF00328466
Shishido R, Sano Y, Fukui K (2000) Ribosomal DNAs: an exception to the conservation of gene order in rice genomes. Mol Gen Genet 263:586–591
Smit AFA, Hubley R, Green P (2017) RepeatMasker Open-4.0. Available at http://www.repeatmasker.org
Sochorová J, Garcia S, Gálvez F, Symonová R, Kovarik A (2018) Evolutionary trends in animal ribosomal DNA loci: introduction to a new online database. Chromosoma 127:141–150. https://doi.org/10.1007/s00412-017-0651-8
Stupar RM, Song J, Tek AL, Cheng Z, Dong F, Jiang J (2002) Highly condensed potato pericentromeric heterochromatin contains rDNA-related tandem repeats. Genetics 162:1435–1444
Symonová R, Majtánová Z, Sember A, Staaks GBO, Bohlen J, Freyhof J, Rábová M, Ráb P (2013) Genome differentiation in a species pair of coregonine fishes: an extremely rapid speciation driven by stress-activated retrotransposons mediating extensive ribosomal DNA multiplications. BMC Evol Biol 13:42. https://doi.org/10.1186/1471-2148-13-42
Teruel M, Cabrero J, Perfectti F, Camacho JMP (2010) B chromosome ancestry revealed by histone genes in the migratory locust. Chromosoma 119:217–225. https://doi.org/10.1007/s00412-009-0251-3
Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG (2012) Primer3—new capabilities and interfaces. Nucleic Acids Res 40:e115. https://doi.org/10.1093/nar/gks596
Veltsos P, Keller I, Nichols RA (2009) Geographically localised bursts of ribosomal DNA mobility in the grasshopper Podisma pedestris. Heredity 103:54–61. https://doi.org/10.1038/hdy.2009.32
Webb GC, White MJD, Contreras N, Cheney J (1978) Cytogenetics of the parthenogenetic grasshopper Warramaba (formely Moraba) virgo and its bisexual relatives. IV. Chromosome banding studies. Chromosoma 67:309–339. https://doi.org/10.1007/BF00285964
Acknowledgements
The authors are grateful to Dr. Frantisek Marec for critical reading of an earlier version of the manuscript, to the anonymous reviewers for valuable suggestions and to Dr. Edison Zefa for providing samples from Murici/AL.
Funding
This study was partially supported by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) (process number 2015/16661-1 and 2014/11763-8), Coordenadoria de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq). DCCM received a research productivity fellowship from the Conselho Nacional de Desenvolvimento Científico e Tecnológico-CNPq (process number 305300/2017-2), and Ana Beatriz Stein Machado Ferretti received a scientific initiation scholarship from PIBIC-UNESP, CNPq.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary Material 1
Map showing the geographical location for each population studied. (JPG 245 kb)
Supplementary Material 2
Contigs containing sequences associated with the 45S rDNA, annotation of associated sequence, and frequency of association. (DOCX 70 kb)
Supplementary Material 3
Difference in size for classification of large (L) and small (S) sites of 18S rDNA in chromosomes of Abracris flavolineata, (a) meiotic chromosomes (metaphase I) and (b) mitotic chromosomes from embryo. The large signals (L) occurred in the pericentromeric region extending to the short chromosomal arm, while the small signals (S) appeared as small dots in the short arms near to the cetromeric region. (JPG 657 kb)
Supplementary Material 4
Occurrence of large (L) and small (S) 18S rDNA clusters and their frequency in individual chromosomes of Abracris flavolineata for the six populations studied. Asterisks indicate heteromorphism (monosomicity). (DOCX 43 kb)
Supplementary Material 5
FISH mapping using the probe ITS1–2 in individuals from six populations of Abracris flavolineata studied. (JPG 2147 kb)
Rights and permissions
About this article
Cite this article
Ferretti, A.B.S.M., Ruiz-Ruano, F.J., Milani, D. et al. How dynamic could be the 45S rDNA cistron? An intriguing variability in a grasshopper species revealed by integration of chromosomal and genomic data. Chromosoma 128, 165–175 (2019). https://doi.org/10.1007/s00412-019-00706-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00412-019-00706-8