Abstract
The extent of 5S and 45S ribosomal DNA (rDNA) variation was investigated in wild and domesticated common beans (Phaseolus vulgaris) chosen to represent the known genetic diversity of the species. 5S and 45S rDNA probes were localized on mitotic chromosomes of 37 accessions by fluorescent in situ hybridization (FISH). The two 5S rDNA loci were largely conserved within the species, whereas a high variation in the number of 45S rDNA loci and changes in position of loci and number of repeats per locus were observed. Domesticated accessions from the Mesoamerican gene pool frequently had three 45S rDNA loci per haploid genome, and rarely four. Domesticated accessions from Andean gene pool, particularly from the race Peru, showed six, seven, eight or nine loci, but seven loci were found in all three races of this gene pool. Between three and eight loci were observed in accessions resulting from crosses between Andean and Mesoamerican genotypes. The presence of two to eight 45S rDNA loci in wild common beans from different geographic locations indicates that the 45S rDNA amplification observed in the Andean lineage took place before domestication. Our data suggest that ectopic recombination between terminal chromosomal regions might be the mechanism responsible for this variation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Common bean (Phaseolus vulgaris, 2n=22) is the world’s most important grain legume for direct food consumption, especially in Latin America and Africa. Although little is known about its genomic organization, the evolution and domestication history of this species have been intensively studied over the last few years. P. vulgaris is thought to have originated in a region encompassing Ecuador and northern Peru (Kami et al. 1995), and dispersed both northwards and southwards establishing the Mesoamerican and Andean gene pools, respectively (reviewed in Gepts 1998). Independent domestication took place in each gene pool (Becerra Velasquez and Gepts 1994). Based on morphological, agroecological and molecular data, the resulting domesticated forms have been grouped into six or seven races (Singh et al. 1991; Beebe et al. 2000).
Molecular cytogenetic analysis in common bean has been restricted to a few genotypes. Using a combination of double FISH with 5S and 45S rDNA probes, chromosome morphology and heterochromatin distribution, ideograms of two European cultivars were established (Moscone et al. 1999). Later, the chromosomal and genetic maps of common bean were integrated by means of direct hybridization of pooled RFLP clones from the University of Florida map (Vallejos et al. 1992) to the mitotic chromosomes of two cultivars of the species (Pedrosa et al. 2003). The cultivars analyzed in these previous studies revealed a high degree of variation in the number and size of the 45S rDNA loci within the species. Based on this small, random sample, it has been suggested that a higher degree of amplification and dispersion of 45S rDNA units in Andean beans has occurred in comparison to Mesoamerican beans (Pedrosa et al. 2003). However, the origin and extent of variation remained unclear.
Variation in the number of 5S and 45S rDNA loci among plant species of the same genus has frequently been reported (Melo and Guerra 2003; Marcon et al. 2005; Vaio et al. 2005). Fewer examples of variation among different accessions of the same species exist (Frello and Heslop-Harrison 2000; Hayasaki et al. 2001; Raskina et al. 2004b). This variation has led to the hypothesis that rDNA clusters are mobile, with different mechanisms believed to be responsible for this mobility (Schubert and Wobus 1985; Dubcovsky and Dvorak 1995). The high degree of variation observed in common bean, along with its well-documented genealogy, make common bean an ideal species for studying this phenomenon.
In the present work, the 5S and 45S rRNA gene loci have been chromosomally assigned by fluorescent in situ hybridization (FISH) in 37 wild and domesticated common bean accessions from different gene pools and of different geographical origins. Amplification in the number of 45S rDNA loci was confirmed in Andean, in contrast to Mesoamerican accessions. The possible mechanisms, as well as the establishment of this variation, are discussed.
Materials and methods
Plant material
All P. vulgaris accessions analyzed in this study are listed in Table 1. The material was chosen to represent the wide range of diversity present in the species, including cultivars from different gene pools and races, and wild forms from different geographical locations. Seeds from all samples were obtained from the germplasm banks of the Embrapa Arroz e Feijão, Brazil, and International Center for Tropical Agriculture (CIAT), Colombia.
Chromosome preparation
Root tips obtained from germinating seeds were pre-treated in 2 mM 8-hydroxyquinoline for 5 h at 16°C or for 20–24 h at 10°C, fixed in ethanol/acetic acid 3:1 (v/v) and stored in fixative at −20°C for up to several weeks. For digestion of the cell wall, root tips were macerated in 2–3% (w/v) cellulase ‘Onozuka R-10’ (Serva) plus 20–30% (v/v) pectinase (Sigma-Aldrich) in 0.01 M citric acid–sodium citrate buffer pH 4.8 for 1–1.5 h at 37°C. Digested material was transferred to a drop of 45% acetic acid and flamed before squashing. One root tip was collected per seed and used per preparation; thus, each preparation corresponded to one individual.
DNA probes
R2, a 6.5-kb fragment of a 18S-5.8S-25S rDNA repeat unit from Arabidopsis thaliana (Wanzenböck et al. 1997), and D2, a 500-bp fragment of a 5S rRNA repeat unit from Lotus japonicus (Pedrosa et al. 2002), were used as probes. Probes were labeled by nick translation (Roche Diagnostics, Life Technologies) with biotin-14-dATP (Life Technologies), biotin-16-dUTP (Roche), digoxigenin-11-dUTP (Roche), Cy3-
dUTP (Amersham Biosciences) or SpectrumGreen-dUTP (Vysis).
Fluorescence in situ hybridization (FISH)
Slides were selected and pre-treated as described in Pedrosa et al. (2001). Chromosome and probe denaturation, post-hybridization washes and detection were performed according to Heslop-Harrison et al. (1991), except for the stringent wash which was performed with 0.1× SSC at 42°C. Hybridization mixes consisted of: 50% (v/v) formamide, 10% (w/v) dextran sulphate, 2× SSC and 2–5 ng/μl probe. The slides were denatured for 5 min at 75°C and hybridized for up to 2 days at 37°C. Biotin-labeled probes were detected using ExtrAvidin-FITC conjugate (Sigma-Aldrich) or mouse anti-biotin in combination with anti-mouse-TRITC (Dako) in 1% (w/v) BSA. Digoxigenin-labeled probes were detected using sheep anti-digoxigenin-FITC (Roche) and amplified with anti-sheep-FITC (Dako), also in 1% (w/v) BSA. All preparations were counter-stained and mounted with 2 μg/ml DAPI in Vectashield (Vector).
Data analysis
Photographs were taken on a Zeiss Axioplan (Carl Zeiss) equipped with a mono cool view CCD camera (Photometrics, Tucson, AZ, USA) and the IPLab spectrum software (IPLab, Fairfax, USA) or on a DMLB Leica equipped with a Cohu CCD video camera and the Leica Qfish software. Digital images were imported into Adobe Photoshop version 8 for final processing.
The number of individuals and cells analyzed per sample is indicated in Table 1. For a subset of accessions, the intensity of the 5S and 45S rDNA signals on chromosomes 6 and 10 were scored as intense (+++), intermediate (++) and weak or very weak (+). Those chromosomes are easily identified by the presence of both rDNA loci at different positions (chromosomes 1 and 8, respectively, in Pedrosa et al. 2003). Chromosomes were renamed according to the linkage group names of the core linkage map of common bean (Freyre et al. 1998).
To test whether the difference in the number of 45S rDNA loci between the Andean and Mesoamerican accessions is significant, the non-parametric Mann–Whitney test was performed (http://www.faculty.vassar.edu/lowry/utest.html). ‘Cargamanto’ G5702 was considered twice (both numbers of 45S rDNA loci). ‘Frijol de Pichon’, ‘Pajarito’ G24404, ‘Pajarito’ G24423, ‘XR-235-1-1’ as well as the introgressed and hybrid accessions were excluded from this calculation.
Results
In order to evaluate the degree of variation in the number and position of the 45S rDNA loci in common bean, this gene family was first localized by FISH in domesticated accessions of different races of each gene pool. The 5S rDNA probe was included to confirm its conserved status in a larger sample and for aiding chromosome identification. The observed number of 45S rDNA loci is summarized in Table 1, where accessions were ordered by gene pool and race and numbered for reference. In the Mesoamerican gene pool, two accessions from the race Durango, three from the race Jalisco and three from the race Mesoamerica, as well as two other cultivars from the Mesoamerican gene pool which originated from crosses of genotypes from different races, showed a low number of 45S rDNA loci, mostly three loci per haploid genome (Figs. 1a, 2; Table 1). A fourth, very weak, 45S rDNA locus was observed in two of those accessions: ‘Apetito’ (number #19 in Table 1) from the race Durango, and ‘Jamapa’ (#20) from the race Mesoamerica. ‘Jamapa’ was previously reported as bearing three 45S rDNA loci (Pedrosa et al. 2003), but an additional locus was observed in most of the analyzed cells in two individuals (Fig. 1b). Four loci were also observed in ‘XR-235-1-1’ (#25), a Mesoamerican breeding line that carries some chromosome segments from P. coccineus (Vallejos et al. 1992). These results suggest that three is the most common number of loci for representatives of the Mesoamerican gene pool, with one additional locus appearing, probably independently, in some accessions (Fig. 2).
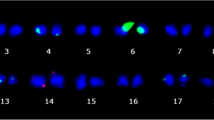
Localization of 5S (red) and 45S (green) rDNA probes on P. vulgaris chromosomes. Chromosomes 6 and 10 were numbered according to similarity to the ‘Calima’ ideogram (see Fig. 3). a ‘Pinto UI 114’. b ‘Jamapa’. c ‘Jalo EEP558’. d ‘Nuña Mani’. e ‘Pajarito’ G24423. Insert shows upper chromosome 10, with a lower contrast, with very weak green signals. f ‘Frijol de Pichon’. g ‘Pajarito’ G24404. Note that only five 45S rDNA signals are present. Arrows indicate weak signals on chromosomes other than 6 and 10. Bar in g =2.5 μm
Distribution of the analyzed accessions according to their number of 45S rDNA loci. Upper panel different gene pools and types, as presented in Table 1. Races are indicated as: C Chile, D Durango, J Jalisco, M Mesoamerica, N Nueva Granada, P Peru. Lower panel hybrids between both gene pools (Andean + Mesoamerica Andean accession with Mesoamerican introgression; Andean × Mesoamerica cross between Andean and Mesoamerican accessions followed by selfing). Accessions that showed inter-individual variation (‘Pajarito’ G24404, ‘Frijol de Pichon’ and ‘Cargamanto’ G5702) have been represented twice
Among the samples from the Andean gene pool, three accessions from the race Nueva Granada, six from the race Peru and two from the race Chile exhibited six, seven, eight or nine 45S rDNA loci, seven being the most common number (Figs. 1c, 2). Eight loci were present in two accessions of race Peru, although only seven loci were observed in a second individual of one of those accessions (‘Cargamanto’ G5702, #7). Nine loci were observed in one nuña or popping bean accession of Peru (‘Nuña Mani’, #10, Fig. 1d) and in one previously analyzed accession of the race Nueva Granada (‘Calima’). Each of these 45S rDNA loci was located on a different chromosome, except for the nine loci observed in ‘Calima’, which were distributed over eight chromosomes (Pedrosa et al. 2003). Six loci were found in a single accession from the race Peru (‘Overitos’, #11). Since little information is available on this accession, it is necessary to consider the possibility that it is not a pure race Peru accession.
Indeed, five accessions we studied from Colombia were previously considered to belong to the Andean gene pool but were recently demonstrated to be Andean accessions with Mesoamerican introgressions based on molecular and morphological attributes (Amirul Islam et al. 2004). These accessions are indicated in Table 1 as Andean + Mesoamerican, as is one of the samples of ‘Cargamanto’ (G4658, #29), which is considered to contain Mesoamerican introgression based on morphological seed characters. It should be noted that ‘Cargamanto’ G4658 only has three 45S rDNA loci, while two other samples of ‘Cargamanto’ (G7231 and G5702, #6 and #7, respectively) have eight loci. The other introgressed Andean + Mesoamerican accessions included in this study revealed three, five or eight 45S rDNA loci, possibly due to different degrees of introgression (Fig. 2). ‘Criollo’ (#30, three loci) and ‘Boyaca 82’ (#28, five loci) have smaller and darker seeds (more common in Mesoamerican beans) than ‘Culateño’, ‘Revoltura’ and ‘X-15943’, which bear eight loci each (#31, #32 and #33, respectively).
To test the possible correlation between the high degree of Mesoamerican introgression and a lower number of 45S rDNA loci, we analyzed recombinant inbred lines from two different crosses between Mesoamerican (three loci) and Andean (seven loci) accessions: ‘BAT93’ (#23) × ‘Jalo EEP558’ (#3) and ‘DOR 364’ (#24) × ‘Chaucha Chuga’ G19833 (#8). For the four lines analyzed, we observed three and four loci in two lines (#34 and #36, respectively) and six loci in two other lines (#35 and #37), numbers that are intermediate between the numbers of loci of the parental accessions (Fig. 2). No general correlation between a lower number of 45S rDNA loci and a higher proportion of Mesoamerican alleles at microsatellite loci (Blair et al. 2003) was observed. Nevertheless, there is apparently no strong bias towards having more or less 45S rDNA loci in the genome.
To find out whether the variation in the number of 45S rDNA loci between Mesoamerican and Andean accessions occurred before or after the domestication process, five wild common bean accessions were also analyzed, one of them (‘Frijol de Pichon’, #27) being considered an ancestral-like accession, because it has the ancestral I phaseolin type (Kami et al. 1995; Chácon et al. 2005). A Mexican wild bean (‘JSG & MAS-262’, #26) had four 45S rDNA loci. One Colombian accession (‘Pajarito’ G24404, #13) had two or three loci, whereas another (‘Pajarito’ G24423, #14) had five, with one weak and two very weak sites (Fig. 1e). The Colombian wild beans used to be placed in the Mesoamerican gene pool (Koenig and Gepts 1989; Becerra Velasquez and Gepts 1994) but they were shown to be genetically distant from the other Mesoamerican wild beans and may represent an independent gene pool (Tohme et al. 1996). An Andean Argentinean accession (‘DGD-629’, #12) showed eight loci, one of them being very weak and not always detectable. In the Peruvian accession (‘Frijol de Pichon’, #27), the putative ancestral-like type belonging to the Northern Andean gene pool, three or four loci were detected (Fig. 1f). It is interesting to note that out of the three wild accessions with two or more individuals analyzed, two accessions showed variation in the number of loci between individuals. Also surprising was the presence of an apparently heterozygous individual in one of the two samples of ‘Pajarito’ (G24404, #13), with five chromosomes bearing 45S rDNA signals in the diploid complement (Fig. 1g). Common bean is considered to be a predominantly self-pollinating species, although gene flow involving wild populations has been reported (Tohme et al. 1996; Papa and Gepts 2003). This heterozygosity strongly indicates that individuals with four and six loci in the diploid complement occur within the same ‘Pajarito’ G24404 wild population. Altogether, this data set indicates a high degree of variation among wild bean forms, supporting the hypothesis of amplification in the number of loci in the Andean lineage prior to domestication.
Besides the difference in the number of 45S rDNA loci among accessions, the intensity or size of the signals also seemed to vary. In order to confirm this observation, chromosomes 6 and 10, which can easily be identified by the additional presence of a 5S rDNA locus, were analyzed in more detail in a subset of the accessions (Fig. 3). Differences in the relative intensity of signals were only indicated when they could be consistently observed. It is possible to state that the same locus may have a different number of repeats in different accessions (compare, for example, the signal intensity for chromosome 10 in Figs. 1a, d, e, 3), or even in different individuals of the same accession, as observed in chromosome 6 of ‘Frijol de Pichon’ (#27, Fig. 3). Accessions with a similar number of loci may also vary in terms of the number of repeats in a locus. The number of 45S rDNA loci in the chromosomes of ‘BAT93’ (#23) and ‘36 Sal Rico de Minas Gerais’ (#21), for example, was always three, but the intensity of the signals in chromosomes 6 and 10 was very distinct (Fig. 3). Variation in the position of a locus along the chromosome was only observed in ‘Jamapa’ (#20), which had a weak interstitial signal on one chromosome pair (Fig. 1b). All other 45S rDNA loci were observed at terminal positions (see Fig. 1).
Signal intensity variability of 5S (grey) and 45S (black) rDNA sites on chromosomes 6 (previously named chromosome 1) and 10 (previously named chromosome 8) of different common bean accessions. Chromosomes were identified by comparison to the ‘Calima’ ideogram (Pedrosa et al. 2003). Except for the standard accession ‘Calima’, accessions were ordered according to Table 1. Number of ‘+’ sign represents relative intensity of signals. (+) indicates very weak signals not visible in all cells and individuals. Intra-accession variation in ‘Frijol de Pichon’ is depicted
In contrast to the 45S rDNA, two 5S rDNA loci at proximal and interstitial positions on chromosomes 6 and 10, respectively, were observed in all accessions analyzed. Variation in the relative intensity of signals was, however, observed in at least five accessions of Andean and Mesoamerican origin, as well as in the ancestor-like wild accession, ‘Frijol de Pichon’ (#27, see Figs. 1f, 3).
Discussion
The localization of the 5S and 45S rRNA genes in 37 accessions of common bean confirmed the high degree of variation observed for the 45S rDNA (from two to nine 45S rDNA-bearing chromosomes) and the conserved status of the 5S rDNA previously detected in a few cultivars of the species (Moscone et al. 1999; Pedrosa et al. 2003). A major, highly-significant (P<0.0001) difference in the number of 45S rDNA loci was observed between the Andean and the Mesoamerican gene pools, with Andean beans having usually seven, sometimes eight, and rarely six or nine loci, and Mesoamerican accessions having three, or rarely four 45S rDNA clusters. Changes in the 45S rDNA independent of similar changes in the 5S rDNA loci have previously been observed in other plant groups, suggesting that each tandemly-repeated sequence has its own evolutionary fate (Hanson et al. 1996; Adams et al. 2000). Although intraspecific variation in the number of 45S rDNA loci seems not to be rare in plants (Schubert and Wobus 1985; Shishido et al. 2000; Hayasaki et al. 2001; Raskina et al. 2004b), we are not aware of any example of more than a fourfold variation in the number of 45S rDNA loci within a species, as we observed in common bean. Moreover, variation in the number of 45S rDNA loci was sometimes observed among individuals of the same accession.
Mechanisms of rDNA variation
The inter- and intraspecific variation in the position of 5S and 45S rDNA loci has been attributed to at least three different mechanisms: (1) amplification of satellite DNA may differentially alter chromosome morphology without changing the order of other sequences (Navratilova et al. 2003); (2) structural chromosomal rearrangements, such as translocations, were demonstrated to apparently change the position of rDNA loci in Lotus japonicus (Hayashi et al. 2001); (3) dispersion of rDNA repeats, amplification of the new minor loci and deletion of original major loci is believed to be the mechanism by which rDNA loci repeatedly changed position during radiation of Triticeae species, without changing the colinearity of other markers (Dubcovsky and Dvorak 1995). How exactly the dispersion step would occur is not clear, but this mechanism could also explain the change in number of rDNA loci and the dispersion of other heterochromatic sequences through a genome. Dubcovsky and Dvorak (1995) seem to favor the hypothesis of intrachromosomal recombination and insertion of a circular intermediate into a new locus, but recent studies in Aegilops suggest that the movement of rDNA may be mediated by En/Spm-like transposons (Raskina et al. 2004a, b).
Although the ‘dispersion-amplification-deletion’ model has been suggested to be the single mechanism for explaining variation in the number of rDNA loci (Dubcovsky and Dvorak 1995), this variation may also be explained by ectopic (interlocus) recombination. Interlocus unequal crossing over has been proposed to play a role in concerted evolution, especially among terminal 45S rDNA loci (Arnheim et al. 1980; Wendel et al. 1995). Furthermore, it has been argued that it could account for changes in sizes of 45S rDNA loci and that species with terminal 45S rDNA sites would show a greater degree of concerted evolution, a higher number of loci, and more variability in locus number and size between and within species than species with interstitial loci (Hanson et al. 1996). Results from Avena agadiriana (Hayasaki et al. 2001), Aegilops speltoides (Raskina et al. 2004b), Aloe (Adams et al. 2000), Paeonia (Zhang and Sang 1999), as well as our present data, agree with these predictions, further supporting the role of interlocus unequal crossing over in terminal rDNA variability.
The terminal position of the rDNA loci is an important condition in this model because it allows frequent rearrangement to occur, without disrupting other gene linkages in the cell (Hanson et al. 1996). Studies in Asparagales have suggested that lack of the Arabidopsis-type telomeric sequence was also an important factor (Pich et al. 1996; Adams et al. 2000), but the extremely high degree of variation observed in common bean, despite the presence of the Arabidopsis-type telomeric sequence (Guerra and Kenton 1996), demonstrate that this is not necessary. Instead, what may be necessary but possibly not sufficient, is the presence of other classes of repetitive sequences, such as satellite DNA, in the chromosome ends involved. The presence of such sequences would enable homologous recombination to take place between these non-allelic sites, in a similar way as proposed for concerted evolution. If this ectopic recombination occurs between a chromosome end that bears a distal rDNA site and another end that does not have one, a new rDNA locus could be generated. Subtelomeric repeats were detected in most common bean chromosome arms (Sonnante et al. 1994; A. Pedrosa-Harand, unpublished results) and several other plant species (Sharma and Raina 2005).
Differential 45S rDNA amplification in common bean
The detailed analysis of the variation in the 45S rDNA locus in common bean has revealed how this variation has developed during the evolutionary history of this species. In agreement with the low number of loci (1–3) observed in the closely-related Phaseolus species (Zheng et al. 1994; Guerra et al. 1996; Moscone et al. 1999), the ancestral-like wild bean accession investigated here showed three or four 45S rDNA sites. It is likely, therefore, that three was the ancestral number of 45S rDNA loci in this species and that these loci were homologous to the three loci observed in P. coccineus, the closest related species to common bean (Delgado-Salinas et al. 1999; Moscone et al. 1999). The synteny of 5S and 45S rDNA loci in two of these three chromosomes (chromosomes 6 and 10) in both species further reinforces these chromosome homeologies. Wild common bean dispersion towards north and south from its putative center of origin in Peru/Ecuador seems to have been accompanied by amplification of 45S rDNA sites mainly in the Andean gene pool.
It is clear, however, that the variation in the number of 45S rDNA loci is a result of various independent amplifications, as indicated by rare occurrence of a higher number of loci in single accessions of different races, both in the Andean and Mesoamerican gene pools (Jalisco, Mesoamerica, Nueva Granada and Peru). Similarly, variation in the number of loci among individuals of the same accession indicates that increase, but possibly also reduction, in the number of loci is still taking place. Reduction in the number of repeats and probably the total elimination of a locus are likely to have occurred, at least for chromosome 10 of ‘Pajarito’ G24404. Nevertheless, the most parsimonious hypothesis still predicts that a major amplification event happened in the Andean lineage after its separation from the Mesoamerican lineage, in agreement with the evolutionary model proposed for the species (Kami et al. 1995; Tohme et al. 1996). High variation among wild bean forms, especially the presence of eight loci in the Andean wild accession ‘DGD-629’, from the southern end of its distribution where domestication has been postulated to have had occurred (Beebe et al. 2001; Chácon et al. 2005), suggests that amplification happened before domestication.
The process of domestication in common bean is still a matter of debate, although it is generally accepted that independent domestications took place in the Andean and Mesoamerican gene pools (Becerra Velasquez and Gepts 1994). Some pieces of evidence support single domestication events, while others support multiple domestications in each gene pool. A recent study comprising a large number of accessions from both gene pools indicates that Mesoamerican landraces are the result of multiple domestications or single domestication followed by hybridization to wild populations, while Andean landraces were derived from a single event (Chácon et al. 2005). The small number of wild bean samples investigated in the present study does not allow us to support any specific scenario. It is interesting to note, however, that the Mexican wild accession investigated has four 45S rDNA loci, while the majority of cultivars of the Mesoamerican gene pool only have three sites. In the Andean gene pool, variation in the number of 45S rDNA loci was observed even within a race, but seven loci were present in accessions of all races, suggesting that this is possibly the number of loci present in the wild common bean that was domesticated in this region. Considering the gradual amplification hypothesis and the high degree of variation in the number of loci observed among wild common bean accessions in the present day, it is possible that the whole range of variation would be detected among Andean wild beans if more samples were to be analyzed. If this were true, an additional indication of the place where Andean domestication took place could be obtained. However, we cannot exclude the possibility that present-day wild beans have not maintained the number of 45S rDNA loci that was present at the time of domestication.
The most intriguing point in the variation observed in common bean, is that it is very pronounced in Andean lineages of the species, but relatively limited in the Mesoamerican gene pool. This does not correlate with a general higher genetic variability in the Andean lineage, since the Andean gene pool is postulated to have a narrower genetic base than the Mesoamerican gene pool (Koenig and Gepts 1989; Beebe et al. 2000; Beebe et al. 2001). If we consider that both elimination and amplification are probably the result of the same mechanism discussed above, the accumulation of rDNA repeats in the Andean lineage may have occurred either due to genetic drift or natural selection, which shifted the equilibrium towards accumulation of repeats exclusively in this group.
References
Adams SP, Leitch IJ, Bennett MD, Chase MW, Leitch AR (2000) Ribosomal DNA evolution and phylogeny in Aloe (Asphodelaceae). Am J Bot 87:1578–1583
Amirul Islam FM, Beebe S, Munoz M, Tohme J, Redden RJ, Basford KE (2004) Using molecular markers to assess the effect of introgression on quantitative attributes of common bean in the Andean gene pool. Theor Appl Genet 108:243–252
Arnheim N, Krystal M, Schmickel R, Wilson G, Ryder O, Zimmer E (1980) Molecular evidence for genetic exchanges among ribosomal genes on non-homologous chromosomes in man and apes. Proc Natl Acad Sci USA 77:7323–7327
Becerra Velasquez VL, Gepts P (1994) RFLP diversity of common bean (Phaseolus vulgaris) in its centres of origin. Genome 37:256–263
Beebe S, Skroch PW, Tohme J, Duque MC, Pedraza F, Nienhuis J (2000) Structure of genetic diversity among common bean landraces of Middle American origin based on correspondence analysis of RAPD. Crop Sci 40:264–273
Beebe S, Rengifo J, Gaitán E, Duque MC, Tohme J (2001) Diversity and origin of Andean landraces of common bean. Crop Sci 41:854–862
Blair MW, Pedraza F, Buendia HF, Gaitán-Solís E, Beebe SE, Gepts P, Tohme J (2003) Development of a genome-wide anchored microsatellite map for common bean (Phaseolus vulgaris L.). Theor Appl Genet 107:1362–1374
Chácon MI, Pickersgill SB, Debouck DG (2005) Domestication patterns in common bean (Phaseolus vulgaris L.) and the origin of the Mesoamerican and Andean cultivated races. Theor Appl Genet 110:432–444
Delgado-Salinas A, Turley T, Richman A, Lavin M (1999) Phylogenetic analysis of the cultivated and wild species of Phaseolus (Fabaceae). Syst Bot 24:438–460
Dubcovsky J, Dvorak J (1995) Ribosomal RNA multigene loci: nomads of the Triticeae genomes. Genetics 140:1367–1377
Frello S, Heslop-Harrison JS (2000) Chromosomal variation in Crocus vernus Hill (Iridaceae) investigated by in situ hybridization of rDNA and tandemly repeated sequence. Ann Bot 86:317–322
Freyre R, Skroch PW, Geffroy V, Adam-Blondon A-F, Shirmohamadali A, Johnson WC, Llaca V, Nodari RO, Pereira PA, Tsai S-M, Tohme J, Dron M, Nienhuis J, Vallejos CE, Gepts P (1998) Towards an integrated linkage map of common bean. 4. Development of a core linkage map and alignment of RFLP maps. Theor Appl Genet 97:847–856
Gepts P (1998) Origin and evolution of common bean: past events and recent trends. HortScience 33:1124–1130
Guerra M, Kenton A (1996) Distribution of telomere DNA in mitotic and polytene nuclei of the anther tapetum of a tetraploid hybrid bean Phaseolus vulgaris × P. acutifolius. Brazil J Genet 19:313–318
Guerra M, Kenton A, Bennett M (1996) rDNA sites in mitotic and polytene chromosomes of Vigna unguiculata (L.) Walp. and Phaseolus coccineus L. revealed by in situ hybridization. Ann Bot 78:157–161
Hanson RE, Islam-Faridi MN, Percival EA, Crane CF, Ji Y, McKnight TD, Stelly DM, Price HJ (1996) Distribution of 5S and 18S-28S rDNA loci in a tetraploid cotton (Gossypium hirsutum L.) and its putative diploid ancestors. Chromosoma 105:55–61
Hayasaki M, Morikawa T, Legget JM (2001) Intraspecific variation of 18S-5.8S-26S rDNA sites revealed by FISH and RFLP in wild oat, Avena agadiriana. Genes Genet Syst 76:9–14
Hayashi M, Miyahara A, Sato S, Kato T, Yoshikawa M, Taketa M, Hayashi M, Pedrosa A, Onda R, Imaizumi-Anraku H, Bachmair A, Sandal N, Stougaard J, Murooka Y, Tabata S, Kawasaki S, Kawaguchi M, Harada K (2001) Construction of a genetic linkage map of the model legume Lotus japonicus using an intraspecific F2 population. DNA Res 8:301–310
Heslop-Harrison JS, Schwarzacher T, Anamthawat-Jónsson K, Leitch AR, Shi M, Leitch IJ (1991) In situ hybridization with automated chromosome denaturation. Technique 3:109–115
Kami J, Becerra Velasquez V, Debouck DG, Gepts P (1995) Identification of presumed ancestral DNA sequences of phaseolin in Phaseolus vulgaris. Proc Natl Acad Sci USA 92:1101–1104
Koenig R, Gepts P (1989) Allozyme diversity in wild Phaseolus vulgaris: further evidence for two major centers of genetic diversity. Theor Appl Genet 78:809–817
Marcon AB, Barros IC, Guerra M (2005) Variation in chromosome numbers, CMA bands and 45S rDNA sites in species of Selaginella (Pteridophyta). Ann Bot 95:271–276
Melo NF, Guerra M (2003) Variability of the 5S and 45S rDNA sites in Passiflora L. species with distinct base chromosome numbers. Ann Bot 92:309–316
Moscone EA, Klein F, Lambrou M, Fuchs J, Schweizer D (1999) Quantitative karyotyping and dual-color FISH mapping of 5S and 18S-25S rDNA probes in the cultivated Phaseolus species (Leguminosae). Genome 42:1224–1233
Navratilova A, Neumann P, Macas J (2003) Karyotype analysis of four Vicia species using in situ hybridization with repetitive sequences. Ann Bot 91:921–926
Papa R, Gepts P (2003) Asymmetry of gene flow and differential geographical structure of molecular diversity in wild and domesticated common bean (Phaseolus vulgaris L.) from Mesoamerica. Theor Appl Genet 106:239–250
Pedrosa A, Jantsch MF, Moscone EA, Ambros PF, Schweizer D (2001) Characterisation of pericentromeric and sticky intercalary heterochromatin in Ornithogalum longibracteatum (Hyacinthaceae). Chromosoma 110:203–213
Pedrosa A, Sandal N, Stougaard J, Schweizer D, Bachmair A (2002) Chromosomal map of the model legume Lotus japonicus. Genetics 161:1661–1672
Pedrosa A, Vallejos CE, Bachmair A, Schweizer D (2003) Integration of common bean (Phaseolus vulgaris L.) linkage and chromosomal maps. Theor Appl Genet 106:205–212
Pich U, Fuchs J, Schubert I (1996) How do Alliaceae stabilize their chromosome ends in the absence of TTTAGGG sequences? Chromosome Res 4:207–213
Raskina O, Belyayev A, Nevo E (2004a) Activity of the En/Spm-like transposons in meiosis as a base for chromosome repatterning in a small, isolated, peripheral population of Aegilops speltoides Tausch. Chromosome Res 12:153–161
Raskina O, Belyayev A, Nevo E (2004b) Quantum speciation in Aegilops: molecular cytogenetic evidence from rDNA cluster variability in natural populations. Proc Natl Acad Sci USA 101:14818–14823
Schubert I, Wobus U (1985) In situ hybridization confirms jumping nucleolus organizing regions in Allium. Chromosoma 92:143–148
Sharma S, Raina SN (2005) Organization and evolution of highly repeated satellite DNA sequences in plant chromosomes. Cytogenet Genome Res 109:15–26
Shishido R, Sano Y, Fukui K (2000) Ribosomal DNAs: an exception to the conservation of gene order in rice genomes. Mol Gen Genet 263:586–591
Singh SP, Gepts P, Debouck DG (1991) Races of common bean (Phaseolus vulgaris, Fabaceae). Econ Bot 45:379–396
Sonnante G, Stockton T, Nodari RO, Becerra Velásquez VL, Gepts P (1994) Evolution of genetic diversity during the domestication of common-bean (Phaseolus vulgaris L.). Theor Appl Genet 89:629–635
Tohme J, González DO, Beebe S, Duque MC (1996) AFLP analysis of gene pools of a wild bean core collection. Crop Sci 36:1375–1384
Vaio M, Speranza P, Valls JF, Guerra M, Mazzella C (2005) Localization of the 5S and 45S rDNA sites and cpDNA sequence analysis in species of the Quadrifaria group of Paspalum (Poaceae, Paniceae). Ann Bot. DOI 10.1093/aob/mci168
Vallejos CE, Sakiyama NS, Chase CD (1992) A molecular marker-based linkage map of Phaseolus vulgaris L. Genetics 131:733–740
Wanzenböck E-M, Schöfer C, Schweizer D, Bachmair A (1997) Ribosomal transcription units integrated via T-DNA transformation associate with the nucleolus and do not require upstream repeat sequences for activity in Arabidopsis thaliana. Plant J 11:1007–1016
Wendel JF, Schnabel A, Seelanan T (1995) Bidirectional interlocus concerted evolution following allopolyploid speciation in cotton (Gossypium). Proc Natl Acad Sci USA 92:280–284
Zhang D, Sang T (1999) Physical mapping of ribosomal RNA genes in the peonies (Paeonia, Paeoniaceae) by fluorescent in situ hybridization: implications for phylogeny and concerted evolution. Am J Bot 86:735–740
Zheng J, Irifune K, Hirai K, Nakata M, Tanaka R, Morikawa H (1994) In situ hybridization to metaphase chromosomes in six species of Phaseolus and Vigna using ribosomal DNA as the probe. J Plant Res 107:365–369
Acknowledgements
A.P-H. was partially supported by a grant from the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Brazil, and by the Hertha-Firnberg program from the Austrian Science Fund (FWF) on behalf of the Federal Ministry for Education, Science and Culture (BMBWK), Austria. C.C.S.A. was supported by a grant from CNPq, Brazil. The work was partially supported by a grant to D.S. from the Gregor Mendel Institute of Molecular Plant Biology (GMI), Austria.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by J. S. Heslop-Harrison
Rights and permissions
About this article
Cite this article
Pedrosa-Harand, A., de Almeida, C.C.S., Mosiolek, M. et al. Extensive ribosomal DNA amplification during Andean common bean (Phaseolus vulgaris L.) evolution. Theor Appl Genet 112, 924–933 (2006). https://doi.org/10.1007/s00122-005-0196-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-005-0196-8