Abstract
The aim of this study was to determine the responses of nitrifiers and denitrifiers to understand microbial pathways of nitrous oxide (N2O) emissions in grassland soils that received inputs of sheep excreta. Sheep dung and synthetic sheep urine were applied at three different rates, simulating a single, double, or triple overlapping of urine or dung depositions in the field. Quantitative PCR and high-throughput sequencing were combined with process-based modeling to understand effects of sheep excreta on microbial populations and on pathways for N2O production. Results showed that emissions of N2O from urine were significantly higher than from dung, ranging from 0.12 to 0.78 kg N2O-N ha−1 during the 3 months. The N2O emissions were significantly related to the bacterial amoA (r = 0.373, P < 0.001) and nirK (r = 0.614, P < 0.001) gene abundances. It was autotrophic nitrification that dominated N2O production in the low urine-N rate soils, whereas it was denitrification (including nitrifier denitrification and heterotrophic denitrification) that dominated N2O production in the high urine-N rate soils. Nitrifier denitrification was responsible for most of the N2O emissions in the dung-treated soils. This study suggests that nitrifier denitrification is indeed an important pathway for N2O emissions in these low fertility and dry grazed grassland ecosystems.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Grassland, as the largest terrestrial ecosystem in the world, accounts for more than 40% of China’s total land area (Nan 2005). To meet the food demands of a growing population in China, livestock stock densities in these grasslands are increasing. Heavy grazing by higher stocking rates can result in grassland degradation. At the same time, higher stocking rates also create excrement “hot spots,” which are well-known sources of nitrous oxide (N2O) emissions (Bol et al. 2004; Saggar et al. 2004; Cardenas et al. 2007).
Nitrous oxide is a potent greenhouse gas, contributing up to 6% of global warming and is also an ozone depletion gas (Loick et al. 2017). Its concentration has steadily increased at a rate of 0.73 ± 0.03 ppb year−1 over the last three decades (IPCC 2014). N2O is traditionally thought to be mainly produced by autotrophic nitrification and heterotrophic denitrification in the soil (Zhong et al. 2014). The conversion of ammonia (NH3) to nitrite (NO2−), which is the first and rate-limiting step of nitrification, is catalyzed by the ammonia monooxygenase (AMO) enzyme, which is encoded by the amoA genes of ammonia oxidizing bacteria (AOB) and ammonia oxidizing archaea (AOA) (Venter et al. 2004; Francis et al. 2005; Könneke et al. 2005; Wuchter et al. 2006; Prosser and Nicol 2008). Denitrification consists of the sequential reduction of NO3− to N2 via the metalloenzymes nitrate reductase, nitrite reductase, nitric oxide reductase, and nitrous oxide reductase, with N2O as an intermediate product (Zumft 1997; Philippot and Hallin 2005). Another important biotransformation process for N2O production is nitrifier denitrification, and this process may play an important part in the emissions of N2O under some conditions, e.g., soil moisture conditions that are sub-optimal for denitrification (Webster and Hopkins 1996; Kool et al. 2011). Nitrifier denitrification is the oxidation of ammonia (NH3) to NO2− followed by the reduction of NO2− to N2O via NO (Wrage et al. 2001; Stein 2011). As sole reductases in the nitrifier denitrification, a diverse group of nirK and norB genes have been detected in numerous AOB species, which are thus far the only group of microorganisms performing nitrifier denitrification (Stein 2011).
Urea-N is the main form of N in sheep urine returned to soil (Di and Cameron 2002; Shand et al. 2002), and excessive N fertilization had the potential to promote N2O emissions via inhibiting the activity of nosZ genes (Qin et al. 2017). AOA dominated nitrification in lightly grazed soils while AOB dominated nitrification in heavily grazed systems (Pan et al. 2018). AOB contributed more than AOA to N2O emissions from NH4+-N applied arable soils (Wang et al. 2016). Urine application dramatically increased the abundance of AOB amoA, nirK, and nosZ genes, stimulated N2O emissions, and decreased the abundance of AOA in grassland systems (Di et al. 2010, 2014). Fresh dung treatments significantly increased N2O emissions (Ma et al. 2006; Lin et al. 2009) and enhanced soil N retention (Cheng et al. 2016). Wrage et al. (2004) found that application of artificial sheep urine altered the microbial pathways for N2O emissions, and urine significantly promoted N2O production by nitrifier denitrification in an incubation experiment. Additionally, sheep urine and dung tended to overlap in the areas where livestock frequently congregate (such as around drinking water troughs, animal tracks) or in pastures with high stocking rates. The effects of overlapping urine or dung patches on N2O emissions with specific N-cycling pathways in the field are poorly understood.
Therefore, a field experiment that lasted from summer to autumn was conducted in an Inner Mongolia typical grassland. The rainfall in this area is low and falls mainly during July–September which coincides with the plant growing season, and grazing management usually happens during this period. The aim of this study was to improve our understanding of the contribution of the three processes (nitrifier denitrification, autotrophic nitrification and heterotrophic denitrification) to N2O emissions by a combination of molecular biology techniques and processed-based model analysis. We hypothesized that (1) nitrifier denitrification was an important pathway for the production of N2O in grazed grassland soils; (2) N2O emissions from urine and dung patches would be much higher than those without sheep excreta; and (3) AOA would play an important role in these low fertility and dry grassland environments.
Materials and methods
Site description
The field experiment was conducted in Xilingol region Chaokewula Sumu (44° 15″ 24.43′–44° 15″ 40.66′ N, 116° 32″ 8.16′–116° 32″ 28.32′ E) in Inner Mongolia, China, from August 5 to November 4, 2014. The region is located at an attitude of 1111–1121 m above sea level and has a typical continental monsoon climate. This region is very cold in winter and cool in summer. The mean annual temperature is − 0.1 °C, with the minimum and maximum monthly mean temperatures ranging from − 22 °C in January to 18.3 °C in July. The average annual precipitation is 350–450 mm, distributed unevenly among seasons, falling mainly during July–September. The annual potential evapotranspiration ranges from 1600 to 1800 mm. The experiment was set in a fenced area of the typical steppe, which is dominated by Leymus chinensis and some Stipa krylovii, Stipa grandis, and Cleistogenes squarrosa. The soil type is dark chestnut (calcic Chernozem according to ISSS Working Group RB, 1998) (Bai et al. 2010). The initial properties of the surface soil (Chestnut soil) (0–10 cm) were as follows: soil pH (soil:H2O ratio 1:2.5) 7.57; total C (TC) and N (TN) contents 14.699 and 0.950 g kg−1, respectively; soil organic carbon (SOC) 15.84 g kg−1; available phosphorus (Olsen P) 1.158 mg kg−1; available potassium (AK) 156.75 mg kg−1; soil cation exchange capacity (CEC) 15.84 cmol kg−1; and soil bulk density 1.32 g cm−3.
Experiment design and treatment application
Fresh sheep dung was collected every morning (about 4–5 a.m.) for 5 days and kept in a − 20 °C freezer until used. Synthetic sheep urine was prepared according to Shand et al. (2002) before application. The average composition of the fresh dung was 438.037 g total C kg−1 and 15.540 g total N kg−1.
Forty static chambers (50 cm diameter) divided into two rows of rings were installed into the soil to a depth of 10 cm. Another 40 plots of the same size next to the gas rings were used for soil sampling. Therefore, 80 50-cm-diameter plots were set up as described above. A 1-m-wide buffer strip was established between every two plots to avoid interactions and to allow for sampling the plots.
Ten treatments, each with four replicates, were allocated to the plots in a completely randomized design. According to Ma et al. (2006), one adult sheep usually urinates about 70 mL and excretes 60 g at a time and forms a urine patch about 320 cm2 and a dung patch about 400cm2 at a time, respectively, based on the field survey. The treatments included the following: control (CK, nothing was applied); U1, 0.4375 L synthetic sheep urine; W1, 0.4375 L water, to check the effect of water without N, consistent with the U1 treatment; U2, 0.875 L synthetic sheep urine; W2, 0.875 L water, consistent with the U2 treatment; U3, 1.3125 L synthetic sheep urine; W3, 1.3125 L water, consistent with the U3 treatment; D1, 300 g fresh dung; D2, 600 g fresh dung; and D3, 900 g fresh dung. The urine and dung application rates corresponded to about 218, 436, 654, 233, 465, and 698 kg N ha−1, respectively. The urine was applied with a watering can at 80 cm above the soil surface to resemble a real urination of adult sheep. The fresh dung was applied by evenly spreading it over the surface. The higher rates of urine and dung were applied to simulate situations where urine or dung patches were overlapping on top of each other two or three times.
Gas samples and soil samples were collected 12 times during the 91-day field study (0, 1, 3, 7, 14, 21, 28, 42, 56, 61, 70, 91 days, respectively). Gas samples (40 mL) were collected 0 and 40 min after chamber closure by a 20-mL syringe and injected into pre-evacuated 20-mL glass bottles and were collected during 10:00 a.m.–12:00 noon. A gas chromatograph (Shimadzu GC-2010 Plus, Japan) equipped with an electron capture detector (ECD) was used to analyze the concentrations of N2O. The hourly N2O emissions were calculated using the equation detailed in the Supplementary material.
Soil samples were collected from the upper 10 cm layer from three random locations within each corresponding plot. The three samples from each plot were bulked into a single composite sample, packed with ice packs, and transported to the laboratory.
Extraction of total DNA from soils and quantitative PCR of functional genes
DNA was extracted from 0.5 g (fresh weight) of soil with a FastDNA Spin Kit for Soil (MP Biomedicals, LLC., Solon, OH, USA), in accordance with the manufacturer’s protocol. The DNA size and integrity were checked by electrophoresis on a 0.7% agarose gel; the quantity and purity were estimated using a Nanodrop®ND-2000 UV-Vis Spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). The extracted DNA was stored at − 20 °C until analysis.
Quantitative PCR assays targeting amoA gene of AOA and AOB, bacterial nirS, nirK and nosZ genes were carried out in triplicate on a LightCycler 480 (Roche Applied Science) machine. The standard curves were developed as described previously by Di et al. (2009). Each gene fragment was PCR-amplified with primers and PCR conditions detailed in Table S1. Triplicate PCR amplicons were subsequently pooled and purified with a PCR cleanup kit (TransGen Biotech, Beijing, China) before being cloned into the pGEM-T Easy Vector (TransGen Biotech, Beijing, China). The resulting ligation mix was transformed in Escherichia coli JM109 competent cells (TransGen Biotech, Beijing, China) according to the manufacturer’s instructions. The correct inserts were checked by re-amplification of each target gene and positive ones were sequenced with an ABI PRISM® 3730 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA), after cycle sequencing reactions using a BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems). The concentrations of plasmid DNA were measured using a Nanodrop®ND-2000 UV-Vis Spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). The known copy number plasmid DNA was 10-fold serial diluted to generate the standard curve.
Each 20 μL reaction mixture contained 10 μL SYBR Premix Ex Taq (TaKaRa, Dalian, China), 400–750 nM of each primer, 1 μL of 10-fold diluted DNA template (1–10 ng), and 8.4–8.68 μL Milli-Q water. Melting curve analysis was performed at the end of each quantitative PCR run to confirm PCR product specificity by measuring fluorescence continuously with the temperature increasing from 50 to 99 °C. High efficiencies of 87.7–101.1% were obtained for the functional genes amplification, with the R2 values ranging between 0.965 and 0.998.
Cloning, MiSeq sequencing, and phylogenetic analysis
The cloning procedure was similar to that described above for generating standard curves for qPCR analyses. The difference was that positive inserts of each target gene (each containing 25 clones) were sequenced. The sequences were clustered into operational taxonomic unit (OTU) at 97% cutoff using the mothur software (Schloss et al. 2009).
High-throughput sequencing of 16S rRNA genes encompassing the V4 region was conducted with a universal 515F-907R primer of the soil DNA using the Illumina® MiSeq sequencer (Illumina, San Diego, CA, USA). In this study, only sequences > 200 bp in length, with an average quality score > 25, without ambiguous base calls, match the primer and present with at least an 80% match to a previously determined 16S rRNA gene sequences were included in the subsequent analyses. The trimmed and unique sequences were clustered into operational taxonomic units (OTUs) based on 97% pairwise identity.
A representative sequence was then used from each OTU for phylogenetic analysis. The phylogenetic analyses of archaeal and bacterial 16S rRNA as well as nirK gene were then conducted by Molecular Evolutionary Genetic Analysis software (MEGA6.06) with 1000-fold bootstrap support. The DNA sequences used for phylogenetic tree construction were deposited in the DNA Data Bank of Japan (DDBJ) under the accession number LC217919-LC217983 (nirK). All the raw sequence data of 16S rRNA has been deposited to NCBI’s Sequence Read Archive (SRA) under accession number SRP101303.
Process-based model description
In order to evaluate the contributions of the three predominant biotransformation processes (nitrifier denitrification, autotrophic nitrification and heterotrophic denitrification) to N2O production, N2O in soil (N2O soil) was divided into two pools (N2O soil-1 and N2O soil-2) due to independent microbe activity (Baggs 2011). The other N pools were exchangeable NH4+-N, NO3−-N, N2 and N2O air.
As shown in Fig. S1, the oxidation of NH4+ to N2O soil-2 represented the N2O emissions from autotrophic nitrification and nitrifier denitrification, which are important biotransformation processes for N2O production in soils (Kool et al. 2010). Heterotrophic denitrification comprised the reduction from NO3− to N2O soil-1 by nirK and nirS, and reduction from N2O soil-1 to N2 by nosZ. Ammonia volatilization was considered in the model, as the hot, dry, and windy conditions are optimal for ammonia volatilization. In addition, NH3 emissions can increase exponentially with increasing N additions (Jiang et al. 2017). Generally, plants exhibited a preference for NH4+ when the soil N pool was strongly dominated by NH4+ or together NH4+ and NO3− in a natural situation (Nordin et al. 2001). This was the reason why the NH4+-N taken up by plants was also considered in the model.
The N transformations among the pools are described by the following equations:
where C denotes the concentration of each pool mentioned above (also reported in Fig. S1), k is the first-order kinetic parameter of each pathway, and Rainfall is daily rainfall amount during the experiment (Fig. 1a). fhd(WFP, T), fan(WFP, T), fnd(WFP, T), and fav(WFP, T) represent the effects of soil water and temperature on four N processes (heterotrophic denitrification, autotrophic nitrification, nitrifier denitrification, and ammonium volatilization), which can be expressed as the product of the response functions of soil water and soil temperature:
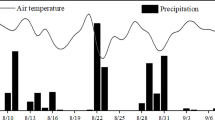
a Daily average air temperature (°C) and rainfall (mm). b Water-filled pore space (WFPS) of soils after treatments throughout the study period. CK, control, nothing was applied; U1, 0.4375 L synthetic sheep urine; U2, 0.875 L synthetic sheep urine; U3, 1.3125 L synthetic sheep urine; W1, 0.4375 L water; W2, 0.875 L water; W3, 1.3125 L water; D1, 300 g fresh dung; D2, 600 g fresh dung; D3, 900 g fresh dung. The urine and dung application rates corresponded to about 218, 436, 654, 233, 465, and 698 kg N ha−1, respectively
The influence of soil temperature on each transformation process was modeled by Ratkowsky (Jansson 2001).
where the values of tmin and tmax are unknown and to be estimated together with other kinetic parameters. The response of different transformation processes to soil water was introduced by piecewise linear equations (Fig. S2) (Müller 1999):
As listed in Table S2, all the unknown parameters were simultaneously estimated by Markov Chain Monte Carlo (MCMC), which has been shown to be more effective than traditional least-square fitting methods (Müller et al. 2007). To generate samples from posterior distributions efficiently, we used the recently developed DiffeRential Evolution Adaptive Metropolis algorithm (DREAM(zs)). The details of DREAM(zs) can be found in Vrugt (2016). To guarantee the accuracy, Gelman-Rubin \( \widehat{R} \) scale reduction factor was applied to judge the convergence of DREAM(zs) (Gelman and Rubin 1992). The ODE solver (ode15s) was employed to build the model structure.
Statistical analysis
All statistical analyses were performed using SPSS version 20, and one-way analysis of variance (ANOVA) followed by Duncan’s multiple range test was used to check for the differences of soil mineral N contents, N2O flux, and functional gene abundance between treatments. Relationships among dynamics of abundances of functional genes, N2O emissions, soil mineral N concentrations, and soil WFPS were determined using Pearson’s correlation coefficients (Hall 2015). It was considered significant for P < 0.05. All the figures were prepared by Origin (Origin Pro 9.0 for Windows).
Results
Environmental conditions
Daily average air temperature was high at the start of the experiment in August and gradually decreased with time, varying from a high of 25 °C to a low of − 8 °C during the experimental period (Fig. 1a). This temperature dynamics corresponded to the mean annual temperature decline in autumn in this region. The rainfall was 75.2 mm during the experimental period, which was slightly lower than the annual average rainfall during this period for the region (100 mm). The soil moisture content (water-filled pore space, WFPS; Fig. 1b) fluctuated with time as a result of rainfall events and varied from a low of 3.10% to a high of 62.97% during the experiment period. The WFPS was below 50% for most of the experimental period.
Total N2O emissions
Results from our study showed that total N2O emissions (Fig. 2) were very low under the dung and water treatments, ranging from 0.005 to 0.02 kg ha−1, and there were no significant differences between the different water or dung rates. The total N2O emissions in the urine treatments were significantly higher than those in the dung and water treatments, ranging from 0.12 to 0.78 kg ha−1 and increased significantly with increasing urine application rates. The percentages of N applied that was emitted as N2O-N under the U1, U2, and U3 treatments were 0.05, 0.07, and 0.1%, respectively, and ranged from 0.0002 to 0.001% under the dung treatments, respectively.
Total N2O emissions as affected by treatments. The vertical bars indicate the standard error of the mean (S.E.M.). CK, control, nothing was applied; U1, 0.4375 L synthetic sheep urine; U2, 0.875 L synthetic sheep urine; U3, 1.3125 L synthetic sheep urine; W1, 0.4375 L water; W2, 0.875 L water; W3, 1.3125 L water; D1, 300 g fresh dung; D2, 600 g fresh dung; D3, 900 g fresh dung. The urine and dung application rates corresponded to about 218, 436, 654, 233, 465, and 698 kg N ha−1, respectively
The contribution of each pathway to N2O emissions by a N transformation model
To quantify the complex N transformations in the grazed grassland soil, we used a complex process-based model (Fig. S1) that considered six pools and ten N transformation processes based on the microbial pathways for N2O formation in soil (Saggar et al. 2004; Cardenas et al. 2007; Loick et al. 2017). Three predominant biotransformation processes, including autotrophic nitrification, heterotrophic denitrification, and nitrifier denitrification for N2O production in the soils, were considered. As mineral nitrogen (NH4+-N and NO3−-N) contents and N2O emissions were slightly affected by water and dung treatments compared with the CK treatment, mathematical models were applied under the three urine treatments (U1, U2, and U3) and the high rates of water (W3) and dung (D3), plus CK. To evaluate the goodness of model-fitting, we performed the evolution of Ȓ statistic (Fig. S5), quantile-quantile (QQ) plots (Fig. S6), and compared observed and simulated data (Fig. S7) in U1, U2, U3, W3, D3, and CK treatments. The evolution of the Gelman-Rubin statistic was used to monitor the convergence of the sampled Markov chains. Convergence was reached for all the chains, i.e., the Ȓ statistic went below the convergence threshold (the dotted black line) (Fig. S5). The normality assumption of the model residual used in the Bayesian framework was assessed via QQ plots (Fig. S6). Although some outlier values were evident in the tails of the distribution, especially in Fig. S6a, the model residuals were overall modeled appropriately. After the convergence, the maximum-a-posterior (MAP) estimations of parameters, which corresponded to the minimal RMSE (root-mean-square error), were chosen to obtain the simulated data. The observed and simulated data for six treatments are shown in Fig. S7. In general, the simulated values fit the observed data well. Although some data of hourly N2O-N flux were underestimated by the model, the trends of simulated data were consistent with those of observed data. This confirmed the validity of the proposed model. The estimated model parameters showed that the three urine treatments led to similar results, and all the parameters in the present model were within a reasonable range (Table S2; Figs. S8 and S9), according to Müller et al. (2004).
The water-treated soils shared similar N transformations with the control soils (Fig. 3). Autotrophic nitrification dominated the generation of N2O emissions but its fraction decreased gradually while nitrifier denitrification played a more important role in N2O emissions with time in the U1 and U2 samples (Fig. 3). In the U3 soils, heterotrophic denitrification accounted for a significant share in generation of N2O at the early stage, while nitrifier denitrification was the dominant pathway to generate N2O at the later stage (Fig. 3). The D3 samples showed similar N transformation with the U3 samples, with less heterotrophic denitrification (Fig. 3).
Fractions of pathway specific N2O emissions over time as affected by U1, U2, U3, W3, D3 and CK treatments. The parameters of the model were estimated by DiffeRential Evolution Adaptive Metropolis algorithm (DREAM(zs)), which was a tool for Bayesian parameter inference. The rate of each transformation was obtained by maximizing the likelihood. CK, control, nothing was applied; U1, 0.4375 L synthetic sheep urine; U2, 0.875 L synthetic sheep urine; U3, 1.3125 L synthetic sheep urine. W1, 0.4375 L water; W2, 0.875 L water; W3, 1.3125 L water; D1, 300 g fresh dung; D2, 600 g fresh dung; D3, 900 g fresh dung. The urine and dung application rates corresponded to about 218, 436, 654, 233, 465, and 698 kg N ha-1, respectively
Abundance of nitrifiers’ and denitrifiers’ functional genes
The abundance of nitrifiers and denitrifiers was evaluated by quantitative PCR of bacterial and archaeal amoA genes, nirS, nirK, and nosZ genes in soil samples for each treatment along the 91-day field study, respectively (Figs. 4 and 5). The abundances of the AOA amoA gene in the soil were generally higher (3.66 × 109–2.18 × 1010 copies g−1 soil) than those of AOB (2.25 × 108–5.22 × 109 copies g−1) (Fig. 4). The application of urine-N or water alone or sheep dung did not lead to major changes in AOA amoA gene copy numbers (Fig. 4a–c). In stark contrast, the AOB amoA gene abundance increased significantly following the application of the sheep excreta and water (P < 0.05) compared with the CK treatment (Fig. 4d–f).
AOA amoA gene copy numbers in different soils as affected by a urine treatments, b water treatments, and c dung treatments. AOB amoA gene copy numbers in different soils as affected by d urine treatments, e water treatment, and f dung treatment. The vertical bars indicate the standard error of the mean (S.E.M.). CK, control, nothing was applied; U1, 0.4375 L synthetic sheep urine; U2, 0.875 L synthetic sheep urine; U3, 1.3125 L synthetic sheep urine; W1, 0.4375 L water; W2, 0.875 L water; W3, 1.3125 L water; D1, 300 g fresh dung; D2, 600 g fresh dung; D3, 900 g fresh dung. The urine and dung application rates corresponded to about 218, 436, 654, 233, 465, and 698 kg N ha−1, respectively
NirS gene copy numbers in different soils as affected by a urine treatments, b water treatments, and c dung treatments. NirK gene copy numbers in different soils as affected by d urine treatments, e water treatments, and f dung treatments. NosZ gene copy numbers in different soils as affected by g urine treatments, h water treatments, and i dung treatments. The vertical bars indicate the standard error of the mean (S.E.M.). CK, control, nothing was applied; U1, 0.4375 L synthetic sheep urine; U2, 0.875 L synthetic sheep urine; U3, 1.3125 L synthetic sheep urine; W1, 0.4375 L water; W2, 0.875 L water; W3, 1.3125 L water; D1, 300 g fresh dung; D2, 600 g fresh dung; D3, 900 g fresh dung. The urine and dung application rates corresponded to about 218, 436, 654, 233, 465, and 698 kg N ha−1, respectively
There was a stronger linear relationship between the NO3−-N concentration in the soil and the AOB amoA gene abundance (r = 0.617, P < 0.001) than with the AOA amoA gene abundance (r = 0.335, P < 0.05) (Table 1). In addition, a significant relationship was found between the N2O flux and the AOB abundance (r = 0.373, P < 0.001), but not to that of AOA (Table 1).
Of all the denitrifying functional genes studied here, nirK was the only gene whose abundance increased significantly following the urine-N application at the high rate (triple urine application) (Fig. 5d). The abundance of all other denitrifying functional genes, including nirS and nosZ, remained stable following the urine, dung or water treatments (Fig. 5). In addition, the abundance of the nirK gene was significantly related to the NO3−-N concentration in the soil (r = 0.380, P = 0.007) and to the N2O emissions (r = 0.614, P < 0.001) (Table 1).
Composition of soil microbial communities
As the abundances of archaeal and bacterial amoA genes and nirK-type denitrifying genes were significantly related to soil NO3−-N content and N2O emissions (Table 1), we conducted high-throughput sequencing of 16S rRNA genes using the universal primers 515F-907R for amoA genes and cloning analysis for nirK genes. The primers and PCR conditions are detailed in Table S1. The samples used to carry out community analysis were sampled on September 2, 2014, under the three urine treatments (U1, U2, and U3) and the high rates of water (W3) and dung (D3), plus control (CK). Significant changes were revealed in the nitrifier and denitrifier functional gene abundances and in the dynamics of soil mineral N contents and hourly N2O fluxes at the sampling dates during the 3-month field experiment under these treatments (Figs. 4 and 5; Figs. S3 and S4).
Approximately 670,269 high-quality sequence reads were obtained (Table S3). The relative abundance of bacterial 16S rRNA genes ranged from 93.5 to 99.8%, and this was much higher than that of archaeal 16S rRNA genes, which ranged from 0.2 to 6.5%. The 16S rRNA genes affiliated with AOA and AOB were selected for phylogenetic analysis from the total MiSeq reads (Fig. S10a and S10b). All the archaeal 16S rRNA genes were affiliated with the soil group І.1b lineage, containing six distinct OTUs within five clusters: 29i4 cluster, 29i4 associated cluster, 54d9 cluster, N. viennensis associated cluster, and N. viennensis cluster (Fig. S10a). With respect to AOB, the bacterial 16S rRNA genes were classified into five OTUs, which fell into three clusters: Nitrosomonas oligotropha, Nitrosospira cluster 3, and Nitrosococcus (Fig. S10b). Clone library construction demonstrated that the nirK genes were clustered into 58 OTUs, three of which were unclassified (Fig. S10c). The other OTUs were phylogenetically related to Bradyrhizobium, Paracoccus, Devosia, Mesorhizobium, Pseudomonas, and Rhodobacter.
In order to understand the dynamics of community distribution of functional genes, we also analyzed the proportional changes of these genes (Fig. 6). The application of different urine-N rates significantly changed the proportions of AOA, AOB, and nirK phylotypes. The N. viennensis associated cluster dominated the AOA community in the typical steppe grassland soils studied here (Fig. 6a). The U3 treatment resulted in the absence of the 54d9 cluster. The majority of AOB, up to 42.9%, was affiliated with Nm. Oligotropha and another 42.9% was affiliated with Nitrosococcus. The proportion of cluster 3 increased, while that of Nm. oligotropha decreased with increasing urine-N rates. The W3 treatment led to a significant increase in Nm. oligotropha and a decrease of cluster 3. The D3 treatment, however, increased the proportion of Nitrosococcus and cluster 3 and decreased that of Nm. oligotropha. As for nirK (Fig. 6c), the relative abundance of the order Rhizobiales significantly increased with increasing urine-N rates, while that of Rhodobacterales decreased. The U3 treatment led to the absent of the order Rhodobacterales. The order Pseudomonadales was exclusively found under the D3 treatment.
Proportional changes of a AOA, b AOB, and c nirK phylotypes in response to excreta treatments. The changes in AOA and AOB composition were analyzed by target 16S rRNA genes in each treatment. The changes in nirK composition were analyzed by cloning analysis. CK, control, nothing was applied; U1, 0.4375 L synthetic sheep urine; U2, 0.875 L synthetic sheep urine; U3, 1.3125 L synthetic sheep urine; W1, 0.4375 L water; W2, 0.875 L water; W3, 1.3125 L water; D1, 300 g fresh dung; D2, 600 g fresh dung; D3, 900 g fresh dung. The urine and dung application rates corresponded to about 218, 436, 654, 233, 465, and 698 kg N ha−1, respectively
Discussion
The percentage of N applied emitted as N2O-N recorded, when a complete emissions envelope was obtained, was equivalent to the emission factor (EF) in the U1 treatment (Fig. S4a). Therefore, the 0.05% EF in the U1 treatment recorded in this study for a single sheep urine deposition was much lower than the international default value of 2% (IPCC 2000) and was much lower than those reported in a number of other studies for pasture soils (Saggar et al. 2007; Barneze et al. 2015) or sheep patch soils (Oenema et al. 1997; de Klein et al. 2003; van Groenigen et al. 2005; Selbie et al. 2014). Likewise, the 0.0013 to 0.002% values measured here for sheep dung were also much lower than those reported in other countries (Oenema et al. 1997; Cardenas et al. 2016). The low N2O emissions from the urine and dung in this study were probably due mainly to the very dry soil conditions (Dobbie and Smith 2003; Liu et al. 2017; Wang et al. 2017). The soil WFPS was mostly between 10 and 30% for large parts of the experimental period and rarely exceeded 50% (Fig. 1b). Such conditions are unfavorable for heterotrophic denitrification to occur to produce N2O from the soil. This is supported from results of process-based modeling analysis (Fig. 3) and the lack of growth of most of the denitrifiers that were measured in this study (Fig. 5). Denitrification can also be driven by microbial oxygen consumption and may occur at low WFPS levels in high C pasture soils (Petersen et al. 2013; Balaine et al. 2016; Friedl et al. 2016). However, the soil organic C content was also very low (15.84 g kg−1) in the present study. Moreover, the significant relationship between N2O emissions and nirK/nosZ ratios (r = 0.572, P < 0.001) further suggests that N2O consumption was a possible explanation for the low N2O emissions (Table 1). Another interesting result revealed by the present study was the little impact of fresh dung on the dynamics of N2O emissions while previous studies have reported markedly increased N2O emissions following dung application (Ma et al. 2006; Lin et al. 2009). This discrepancy may have been caused by the very dry soil conditions of this study where the dung was not decomposed significantly. Further research is required to understand effects of environmental conditions on sheep dung decomposition and effects on N2O emissions in low fertility and dry grassland soils.
Although N2O emissions from both the urine and dung treatments were extremely low, the process-based model indicated that the contribution of each pathway to N2O emissions varied with urine and dung treatments along the sampling time (Fig. 3). The importance of autotrophic nitrification under the U1 and U2 treatments at the early stage was ascribed to the greater advantages of the nitrifiers than denitrifiers after urine addition (Lu et al. 2012; Lu and Jia 2013), as the AOB populations flourished following high N inputs (Fig. 4d) (Di et al. 2009). In contrast, heterotrophic denitrification had a significant share in the generation of N2O in the U3 samples at the early stage (Fig. 3). This phenomenon might be ascribed to the rapid growth of heterotrophic microorganisms, which was stimulated by the large addition of urea, leading to anoxic conditions because of consumption of soil oxygen (Bodelier et al. 1996), although the soil WFPS was fluctuating around 30% (Fig. 1b). The evident growth of nirK-type denitrifier in the U3 samples at the early stage further indicated the importance of heterotrophic denitrification for the generation of N2O (Fig. 5d). Although nitrifier denitrification could be neglected in the W3 and CK soils, it increased with time in the U1 and U2 treated soils, gradually dominating the N2O emissions in the U3 and D3 treated soils (Fig. 3), which corresponded well with the increases in the abundance of bacterial amoA genes (Hink et al. 2017, 2018), as shown by quantitative PCR (Fig. 4d, f). The results agreed with the findings from a study conducted in a dairy farm in the Netherlands, which showed a larger contribution of nitrifier denitrification to the total N2O production after the application of artificial urine (Wrage et al. 2004). Wrage et al. (2004) and Zhu et al. (2013) suggested that larger NO2−-N concentrations following urine application would stimulate nitrifier denitrification. Di et al. (2014) previously reported that the dynamics of nirK gene abundance resembled that of AOB amoA gene abundance. Some AOB populations contain the nirK gene, and therefore, they were able to grow under both aerobic and anaerobic soil conditions performing nitrifier denitrification. Stein (2011) also proposed that AOB, nirK, and norB were the sole nitrifiers and reductases, respectively, in the nitrifier denitrification pathway. It is therefore not surprising that N2O emissions were positively correlated to the abundance of both AOB (r = 0.373, P < 0.001) and nirK genes (r = 0.614, P < 0.001), respectively (Table 1). Given the above, we speculated that nitrifier denitrification dominated N2O production in this grazed dry and low fertility grassland soil. Nitrifier denitrification in natural ecosystems has not received much attention (Butterbach-Bahl et al. 2013; Shcherbak et al. 2014; Stieglmeier et al. 2014; Shi et al. 2017). Our results suggested that nitrifier denitrification, a previously overlooked process, was indeed an important pathway for N2O emissions in this typical steppe grassland soil. The validity of the mathematical model was confirmed by a good fit to the experimental data, as shown by the observed and simulated data of NH4+-N contents, NO3−-N contents, and N2O fluxes (Fig. S7) in the U1, U2, U3, W3, D3, and CK treatments. The Ȓ statistic (Fig. S5) and quantile-quantile (QQ) plots (Fig. S6) confirmed the convergence and effectiveness of the estimated results. In addition, the abundance dynamics of N cycling-related functional genes (Figs. 4 and 5) were also evaluated in the present experiment to further validate the used model. Although the presence of genes does not necessarily mean active expression, the dynamic changes of the functional genes under different treatments along with time might reveal functional activity. It should be pointed out that, although DREAM(zs) is a powerful tool to simultaneously estimate multiple rates from limited data, uncertainties always exist in the estimated results. It is well recognized that using isotopes or inhibitors helps to distinguish the relative contributions of microbial pathways to N2O yields (Shi et al. 2017). From a point view of Bayesian parameter inference, the use of isotopes or inhibitors leads to less uncertainties (i.e., more confidence) in the estimation of results. Further research combining isotopes or inhibitors with process-based modeling introduced in the present study would be useful.
Previous studies of high fertility dairy pasture soils have shown that ammonia oxidation is mainly performed by AOB in the high N status urine patch soil, and AOA population abundance is sometimes inhibited by the high N inputs in the form of animal urine-N (e.g., Di et al. 2009, 2010, 2014). In addition, AOA played a predominated role in acid soil nitrification while AOB tended to dominate nitrification in alkaline soils (Shi Gubry-Rangin et al. 2010; Zhang et al. 2012; Hu et al. 2014). However, results from this study showed that both AOB and AOA were positively correlated to the NO3−-N concentrations in the soil (Table 1), which suggested that both AOA and AOB played a role in ammonia oxidation in this low fertility and slightly alkaline sheep grassland soil. In addition, the community compositions of AOB and AOA found in this low fertility sheep pasture soil were also different from those reported in the high fertility pasture soils. For example, the dominant AOA in this sheep pasture soil after urine-N application was N. viennensis and associated cluster, whereas those in the grassland soils established at Hillsborough, UK, were dominantly by fosmid 29i4-like AOA (Zhou et al. 2015). This discrepancy might be because that N. viennensis cluster was urease-positive and would grow optimally in slightly alkaline typical steppe grassland soils (Tourna et al. 2011). Similarly, the dominant AOB found in this sheep pasture soil were Nitrosococcus and Nitrosospira, whereas those found in the UK grassland soils were Nitrosospira cluster 3-like AOB (Zhou et al. 2015). In fact, both Nitrosococcus and Nitrosospira species of AOB were shown to contain nirK and norB genes and could carry out nitrifier denitrification (Norton et al. 2008; Stein 2011; Klotz et al. 2006). These results further suggested the occurrence of nitrifier denitrification in the typical steppe grassland soils. Based on the nirK clone library analysis (Fig. 6c; Fig. S10c), most of nirK-harboring denitrifiers were related to the nirK from Rhizobiales and Rhodobacterales, similar to findings from previous studies (Bremer et al. 2007; Saito et al. 2008; Pan et al. 2016). It was also worth mentioning that the relative abundance of the order Rhizobiales increased with increasing urine-N rates, while that of Rhodobacterales changed in opposite direction, decreasing with the high rate of urine (U3) treatment (Fig. 6c) but occupied high proportions in the dung and water treatments. The changing trend of the relative abundance of the order Rhizobiales was similar with that of nitrifier denitrification pathway in the urine patches. The predomination of nitrifier denitrification for the N2O emissions in the U3 indicated that nirK-type denitrifiers of Rhizobiales were probably involved in nitrifier denitrification.
Conclusions
Results from this study showed that in this dry and low fertility sheep grassland soil, the N2O emissions from urine patches were significantly higher than those from dung; both were much lower than previously predicted. The microbial pathways of N2O production varied widely with sheep excreta. Nitrifier denitrification, a traditionally overlooked process, was indeed a process producing significant N2O occurred in soils following inputs of urea. Nitrification dominated N2O emissions in environments high in oxygen while heterotrophic denitrification in micro-aerobic conditions. These results help to understand the complex biotransformation processes of N in the dry and low-fertility steppe grassland and are important in developing future management practices for the mitigation of N2O emissions in grazed grasslands.
References
Baggs EM (2011) Soil microbial sources of nitrous oxide: recent advances in knowledge, emerging challenges and future direction. Curr Opin Env Sust 3:321–327
Bai Y, Wu J, Clark CM, Naeem S, Pan Q, Huang J, Zhang L, Han X (2010) Tradeoffs and thresholds in the effects of nitrogen addition on biodiversity and ecosystem functioning: evidence from inner Mongolia grasslands. Glob Chang Biol 16:358–372
Balaine N, Clough TJ, Beare MH, Thomas SM, Meenken ED (2016) Soil gas diffusivity controls N2O and N2 emissions and their ratio. Soil Sci Soc Am J 80:529–540
Barneze AS, Minet EP, Cerri CC, Misselbrook T (2015) The effect of nitrification inhibitors on nitrous oxide emissions from cattle urine depositions to grassland under summer conditions in the UK. Chemosphere 119:122–129
Bodelier P, Libochant JA, Blom CWPM, Laanbroek HJ (1996) Dynamics of nitrification and denitrification in root-oxygenated sediments and adaptation of ammonia-oxidizing bacteria to low-oxygen or anoxic habitats. Appl Environ Microb 62:4100–4107
Bol R, Petersen SO, Christofides C, Dittert K, Hansen MN (2004) Short-term N2O, CO2, NH3 fluxes, and N/C transfers in a Danish grass-clover pasture after simulated urine deposition in autumn. J Plant Nutr Soil Sc 167:568–576
Bremer C, Braker G, Matthies D, Reuter A, Engels C, Conrad R (2007) Impact of plant functional group, plant species, and sampling time on the composition of nirK-type denitrifier communities in soil. Appl Environ Microb 73:6876–6884
Butterbach-Bahl K, Baggs EM, Dannenmann M, Kiese R, Zechmeister-Boltenstern S (2013) Nitrous oxide emissions from soils: how well do we understand the processes and their controls? Phil Trans R Soc B 368(1621):20130122
Cardenas LM, Chadwick D, Scholefield D, Fychan R, Marley CL, Jones R, Bol R, Well R, Vallejo A (2007) The effect of diet manipulation on nitrous oxide and methane emissions from manure application to incubated grassland soils. Atmos Environ 41:7096–7107
Cardenas LM, Misselbrook TM, Hodgson C, Donovan N, Gilhespy S, Smith KA, Dhanoa MS, Chadwick D (2016) Effect of the application of cattle urine with or without the nitrification inhibitor DCD, and dung on greenhouse gas emissions from a UK grassland soil. Agric Ecosyst Environ 235:229–241
Cheng Y, Cai Y, Wang SQ (2016) Yak and Tibetan sheep dung return enhance soil N supply and retention in two alpine grasslands in the Qinghai-Tibetan Plateau. Biol Fert Soils 52:413–422
de Klein CA, Barton L, Sherlock RR, Li Z, Littlejohn RP (2003) Estimating a nitrous oxide emission factor for animal urine from some New Zealand pastoral soils. Soil Res 41:381–399
Di HJ, Cameron KC, Podolyan A, Robinson A (2014) Effect of soil moisture status and a nitrification inhibitor, dicyandiamide, on ammonia oxidizer and denitrifier growth and nitrous oxide emissions in a grassland soil. Soil Biol Biochem 73:59–68
Di HJ, Cameron KC, Shen JP, Winefield CS, O’Callaghan M, Bowatte S, He JZ (2009) Nitrification driven by bacteria and not archaea in nitrogen-rich grassland soils. Nat Geosci 2:621–624
Di HJ, Cameron KC, Shen JP, Winefield CS, O'Callaghan M, Bowatte S, He JZ (2010) Ammonia-oxidizing bacteria and archaea grow under contrasting soil nitrogen conditions. FEMS Microbiol Ecol 72:386–394
Di HJ, Cameron KC (2002) Nitrate leaching in temperate agroecosystems sources, factors and mitigating strategies. Nutr Cycl Agroecosys 64:237–256
Dobbie KE, Smith KA (2003) Nitrous oxide emission factors for agricultural soils in Great Britain: the impact of soil water-filled pore space and other controlling variables. Glob Chang Biol 9:204–218
Francis CA, Roberts KJ, Beman JM, Santoro AE, Oakley BB (2005) Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. P Natl Acad Sci USA 102:14683–14688
Friedl J, Scheer C, Rowlings DW, McIntosh HV, Strazzabosco A, Warner DI, Grace PR (2016) Denitrification losses from an intensively managed sub-tropical pasture—impact of soil moisture on the partitioning of N2 and N2O emissions. Soil Biol Biochem 92:58–66
Gelman A, Rubin DB (1992) Inference from iterative simulation using multiple sequences. Stat Sci 7:457–472
Hall G (2015) Pearson’s correlation coefficient words, 1(9):2015
Hink L, Nicol GW, Prosser JI (2017) Archaea produce lower yields of N2O than bacteria during aerobic ammonia oxidation in soil. Environ Microbiol 19:4829–4837
Hink L, Gubry-Rangin C, Nicol GW, Prosser JI (2018) The consequences of niche and physiological differentiation of archaeal and bacterial ammonia oxidisers for nitrous oxide emissions. ISME J 12:1084
Hu HW, Xu ZH, He JZ (2014) Ammonia-oxidizing archaea play a predominant role in acid soil nitrification. Adv Agron 125:261–302
IPCC (Intergovernmental Panel on Climate Change). Penman J, Kruger D, Galbally I, Hiraishi T, Nyenzi B, Emmanuel S, Buendia L, Hoppaus R, Martinsen T, Meijer J, Miwa K, Tanabe Ked (2000) Good practice guidance and uncertainty management in National Greenhouse Gas Inventories. Hayama, IPCC/OECD/IEA/IGES
IPCC. van Ypersele de Strihou JP. 2014. Climate change 2014—synthesis report
Jansson Per-Erik (2001) Coupled heat and mass transfer model for soil-plant-atmosphere systems
Jiang Y, Deng A, Bloszies S, Huang S, Zhang W (2017) Nonlinear response of soil ammonia emissions to fertilizer nitrogen. Biol Fert Soils 53:269–274
Klotz MG, Arp DJ, Chain PSG, El-Sheikh AF, Hauser L, Hommes NG, Larimer FW, Malfatti SA, Norton JM, Poret-Peterson AT, Vergez LM, Ward BB (2006) Complete genome sequence of the marine, nitrifying purple sulfur bacterium, Nitrosococcus oceani ATCC 19707. Appl Environ Microb 72:6299–6315
Könneke M, Bernhard AE, de la Torre JR, Walker CB, Waterbury JB, Stahl DA (2005) Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437:543–546
Kool DM, Dolfing J, Wrage N, Van Groenigen JW (2011) Nitrifier denitrification as a distinct and significant source of nitrous oxide from soil. Soil Biol Biochem 43:174–178
Kool DM, Wrage N, Zechmeister-Boltenstern S, Pfeffer M, Brus D, Oenema O, Van Groenigen JW (2010) Nitrifier denitrification can be a source of N2O from soil: a revised approach to the dual-isotope labelling method. Eur J Soil Sci 61:759–772
Lin XW, Wang SP, Ma XZ, Xu GP, Luo CY, Li YN, Jiang GM, Xie ZB (2009) Fluxes of CO2, CH4, and N2O in an alpine meadow affected by yak excreta during summer grazing periods on the Qinghai-Tibetan plateau. Soil Biol Biochem 41:718–725
Liu R, Hayden HL, Suter H, Hu H, Lam SK, He J, Mele PM, Chen D (2017) The effect of temperature and moisture on the source of N2O and contributions from ammonia oxidizers in an agricultural soil. Biol Fert Soils 53:141–152
Loick N, Dixon ER, Repullo Ruibérriz de Torres MA, Ciganda V, Lopez-Aizpun MA, Matthews GP, Müller Christoph, Cardenas LM (2017) Using the triple labelling technique to apportion N2O emissions to nitrification and denitrification from different nitrogen sources at different water-filled-pore-spaces. In EGU general assembly conference abstracts Vol. 19, p. 17534
Lu L, Han W, Zhang J, Wu Y, Wang B, Lin X, Zhu J, Cai Z, Jia Z (2012) Nitrification of archaeal ammonia oxidizers in acid soils is supported by hydrolysis of urea. ISME J 6:1978–1984
Lu L, Jia Z (2013) Urease gene-containing archaea dominate autotrophic ammonia oxidation in two acid soils. Environ Microbiol 15:1795–1809
Ma X, Wang S, Wang Y, Jiang G, Nyren P (2006) Short-term effects of sheep excrement on carbon dioxide, nitrous oxide and methane fluxes in typical grassland of Inner Mongolia. New Zeal J Agr Res 49:285–297
Müller C (1999) Modelling soil-biosphere interactions. Oxford University Press
Müller C, Stevens RJ, Laughlin RJ (2004) A 15N tracing model to analyse N transformations in old grassland soil. Soil Biol Biochem 36:619–632
Müller C, Rütting T, Kattge J, Laughlin RJ, Stevens RJ (2007) Estimation of parameters in complex 15 N tracing models by Monte Carlo sampling. Soil Biol Biochem 39:715–726
Nan Z (2005) The grassland farming system and sustainable agricultural development in China. Grassl Sci 51:15–19
Nordin A, Högberg P, Näsholm T (2001) Soil nitrogen form and plant nitrogen uptake along a boreal forest productivity gradient. Oecologia 129:125–132
Norton JM, Klotz MG, Stein LY, Arp DJ, Bottomley PJ, Chain PS, Hauser LJ, Land ML, Larimer FW, Shin MW, Starkenburg SR (2008) Complete genome sequence of Nitrosospira multiformis, an ammonia-oxidizing bacterium from the soil environment. Appl Environ Microb 74:3559–3572
Oenema O, Velthof GL, Yamulki S, Jarvis SC (1997) Nitrous oxide emissions from grazed grassland. Soil Use Manage 13:288–295
Pan H, Li Y, Guan X, Li J, Xu X, Liu J, Zhang Q, Xu J, Di H (2016) Management practices have a major impact on nitrifier and denitrifier communities in a semiarid grassland ecosystem. J Soils Sediments 16:896–908
Pan H, Xie K, Zhang Q, Jia Z, Xu J, Di H, Li Y (2018) Archaea and bacteria respectively dominate nitrification in lightly and heavily grazed soil in a grassland system. Biol Fert Soils 54:41–54
Petersen SO, Ambus P, Elsgaard L, Schjønning P, Olesen JE (2013) Long-term effects of cropping system on N2O emission potential. Soil Biol Biochem 57:706–712
Philippot L, Hallin S (2005) Finding the missing link between diversity and activity using denitrifying bacteria as a model functional community. Curr Opin Microbiol 8:234–239
Prosser JI, Nicol GW (2008) Relative contributions of archaea and bacteria to aerobic ammonia oxidation in the environment. Environ Microbiol 10:2931–2941
Qin S, Ding K, Clough TJ, Hu C, Luo J (2017) Temporal in situ dynamics of N2O reductase activity as affected by nitrogen fertilization and implications for the N2O/(N2O+ N2) product ratio and N2O mitigation. Biol Fert Soils 53:723–727
Saggar S, Bolan NS, Bhandral R, Hedley CB, Luo J (2004) A review of emissions of methane, ammonia, and nitrous oxide from animal excreta deposition and farm effluent application in grazed pastures. New Zeal J Agr Res 47:513–544
Saggar S, Hedley CB, Giltrap DL, Lambie SM (2007) Measured and modelled estimates of nitrous oxide emission and methane consumption from a sheep-grazed pasture. Agric Ecosyst Environ 122:357–365
Saito T, Ishii S, Otsuka S, Nishiyama M, Senoo K (2008) Identification of novel Betaproteobacteria in a succinate-assimilating population in denitrifying rice paddy soil by using stable isotope probing. Microbes Environ 23:192–200
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF (2009) Introducing mothur open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microb 75:7537–7541
Selbie DR, Cameron KC, Di HJ, Moir JL, Lanigan GJ, Richards KG (2014) The effect of urinary nitrogen loading rate and a nitrification inhibitor on nitrous oxide emissions from a temperate grassland soil. J Agr Sci-Cambridge 152:159–171
Shand CA, Williams BL, Dawson LA, Smith S, Young ME (2002) Sheep urine affects soil solution nutrient composition and roots differences between field and sward box soils and the effects of synthetic and natural sheep urine. Soil Biol Biochem 34:163–171
Shcherbak I, Millar N, Robertson GP (2014) Global metaanalysis of the nonlinear response of soil nitrous oxide (N2O) emissions to fertilizer nitrogen. P Natl Acad Sci 111:9199–9204
Shi Gubry-Rangin C, Nicol GW, Prosser JI (2010) Archaea rather than bacteria control nitrification in two agricultural acidic soils. FEMS Microbiol Ecol 74:566–574
Shi X, Hu HW, Zhu-Barker X, Hayden H, Wang J, Suter H, Chen D, He JZ (2017) Nitrifier induced denitrification is an important source of soil nitrous oxide and can be inhibited by a nitrification inhibitor 3, 4-dimethylpyrazole phosphate (DMPP). Environ Microbiol 19:4851–4865
Stein LY (2011) Heterotrophic nitrification and nitrifier denitrification. Ward BB, Arp DJ, Klotz MG 95–116
Stieglmeier M, Mooshammer M, Kitzler B, Wanek W, Zechmeister-Boltenstern S, Richter A, Schleper C (2014) Aerobic nitrous oxide production through N-nitrosating hybrid formation in ammonia-oxidizing archaea. ISME J 8:1135–1146
Tourna M, Stieglmeier M, Spang A, Könneke M, Schintlmeister A, Urich T, Engel M, Schloter M, Wagner M, Richter A, Schleper C (2011) Nitrososphaera viennensis, an ammonia oxidizing archaeon from soil. P Natl Acad Sci 108:8420–8425
van Groenigen JW, Velthof GL, van der Bolt FJ, Vos A, Kuikman PJ (2005) Seasonal variation in N2O emissions from urine patches: effects of urine concentration, soil compaction and dung. Plant Soil 273:15–27
Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu D, Paulsen I, Nelson KE, Nelson W, Fouts DE, Levy S, Knap AH, Lomas MW, Nealson K, White O, Peterson J, Hoffman J, Parsons R, Baden-Tillson H, Pfannkoch C, Rogers Y, Smith HO (2004) Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66–74
Vrugt JA (2016) Markov chain Monte Carlo simulation using the DREAM software package: theory, concepts, and MATLAB implementation. Environ Model Softw 75:273–316
Wang Q, Zhang LM, Shen JP, Du S, Han LL, He JZ (2016) Nitrogen fertiliser-induced changes in N2O emissions are attributed more to ammonia-oxidising bacteria rather than archaea as revealed using 1-octyne and acetylene inhibitors in two arable soils. Biol Fert Soils 52:1163–1171
Wang Q, Liu YR, Zhang CJ, Zhang LM, Han LL, Shen JP, He JZ (2017) Responses of soil nitrous oxide production and abundances and composition of associated microbial communities to nitrogen and water amendment. Biol Fert Soils 53:601–611
Webster FA, Hopkins DW (1996) Contributions from different microbial processes to N2O emission from soil under different moisture regimes. Biol Fert Soils 22:331–335
Wrage N, Velthof GL, Laanbroek HJ, Oenema O (2004) Nitrous oxide production in grassland soils: assessing the contribution of nitrifier denitrification. Soil Biol Biochem 36:229–236
Wrage N, Velthof GL, Van Beusichem ML, Oenema O (2001) Role of nitrifier denitrification in the production of nitrous oxide. Soil Biol Biochem 33:1723–1732
Wuchter C, Abbas B, Coolen MJL, Herfort L, Van Bleijswijk J, Timmers P, Strous M, Teira E, Herndl GJ, Middelburg JJ, Schouten S, Sinninghe Damsté JS (2006) Archaeal nitrification in the ocean. P Natl Acad Sci 103:12317–12322
Zhang LM, Hu HW, Shen JP, He JZ (2012) Ammonia-oxidizing archaea have more important role than ammonia-oxidizing bacteria in ammonia oxidation of strongly acidic soils. ISME J 6:1032–1045
Zhou X, Fornara D, Wasson EA, Wang D, Ren G, Christie P, Jia Z (2015) Effects of 44 years of chronic nitrogen fertilization on the soil nitrifying community of permanent grassland. Soil Biol Biochem 91:76–83
Zhong L, Du R, Ding K, Kang X, Li F, Bowatte S, Hoogendoorn C, Wang Y, Rui Y, Jiang LL, Wang S (2014) Effects of grazing on N2O production potential and abundance of nitrifying and denitrifying microbial communities in meadow-steppe grassland in northern China. Soil Biol Biochem 69:1–10
Zhu X, Burger M, Doane TA, Horwath WR (2013) Ammonia oxidation pathways and nitrifier denitrification are significant sources of N2O and NO under low oxygen availability. P Natl Acad Sci 110:6328–6333
Zumft WG (1997) Cell biology and molecular basis of denitrification. Microbiol Mol Biol R 61:533–616
Funding
This work was financially supported by the National Key Basic Research Program of China (2014CB138801) and Natural Science Foundation of China (41671249).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
ESM 1
(DOCX 1991 kb)
Rights and permissions
About this article
Cite this article
Pan, H., Ying, S., Liu, H. et al. Microbial pathways for nitrous oxide emissions from sheep urine and dung in a typical steppe grassland. Biol Fertil Soils 54, 717–730 (2018). https://doi.org/10.1007/s00374-018-1297-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00374-018-1297-2