Abstract
Purpose
Nitrification and denitrification, two of the key nitrogen (N) transformation processes in the soil, are carried out by a diverse range of microorganisms and catalyzed by a series of enzymes. Different management practices, such as continuous grazing, mowing, and periodic fencing off from grazing, dramatically influenced grassland ecosystems. This study aimed to examine the effects of management practices on the abundance and community structure of nitrifier and denitrifier communities in grassland ecosystems.
Materials and methods
Soil samples were collected from a semiarid grassland ecosystem in Xilingol region, Inner Mongolia, where long-term management practices including free-grazing, different periods of enclosure from grazing, and different frequencies of mowing were conducted. Real-time quantitative polymerase chain reaction (Q-PCR), denaturing gradient gel electrophoresis (DGGE), sequencing, and phylogenetic analysis were applied to estimate the abundance and composition of amoA, nirS, nirK, and nosZ genes.
Results and discussion
The ammonia-oxidizing archaea (AOA) amoA copies were in the range 5.99 × 108 to 8.60 × 108, while those of ammonia-oxidizing bacteria (AOB) varied from 3.02 × 107 to 4.61 × 107. The abundance of AOA was substantially higher in the light grazing treatment (LG) than in the mowing treatments. The quantity and intensity of DGGE bands of AOA varied with pasture management. In stark contrast, AOB population abundance and community structure remained largely unchanged in all the soils irrespective of the management practices. All these results suggested that ammonia oxidizers were dominated by AOA. The higher gene abundance and greater intensity of DGGE bands of nirS and nosZ under the enclosure treatments would suggest greater stimulated denitrification. The ratio of nosZ/(nirS + nirK) was higher in mowing treatments than in the free-grazing and enclosure treatments, possibly leading to more complete denitrification. Correlation analysis indicated that soil moisture and inorganic nitrogen content were the two main soil environmental variables that influence the community structure of nitrifiers and denitrifiers.
Conclusions
In this semiarid neutral to alkaline grassland ecosystem under low temperature conditions, AOA mainly affiliated with Nitrososphaera dominated nitrification. These results clearly demonstrate that grassland management practices can have a major impact on nitrifier and denitrifier communities in this semiarid grassland ecosystem, under low temperature conditions.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Nitrous oxide (N2O), which is both a potent greenhouse gas and an ozone-depletion gas, is produced as a by-product of autotrophic nitrification and as an intermediate product of incomplete biological denitrification (Conrad 1996; Di et al. 2014). In nitrification, ammonium (NH4 +) is converted to nitrite (NO2 −) and then to nitrate (NO3 −), while in denitrification NO3 − is gradually reduced to NO2 − and to nitrogen gases (NO, N2O, and N2). Both nitrification and denitrification in the soil are performed by microorganisms. The first and rate-limiting step of nitrification, the conversation of ammonium to hydroxylamine, is mainly performed by ammonia-oxidizing bacteria (AOB) and ammonia-oxidizing archaea (AOA) which carry the ammonia monooxygenase (amoA) gene (Venter et al. 2004; Francis et al. 2005; Prosser and Nicol 2008). Recent studies have shown that the role of AOB and AOA communities in ammonia oxidation is affected by soil and environmental condition (Chen et al. 2008; Zhang et al. 2012). For example, in some high-fertility or high-nitrogen environments, AOB communities were found to play a dominant role in ammonia oxidation (Di et al. 2009), whereas in strongly acidic soils, AOA may play a dominate role in ammonia oxidation (Zhang et al. 2012). However, our understanding of the community abundance of AOB and AOA and the relative importance of the two in ammonia oxidation in many different ecosystems is poor and urgently requires further research to bridge this knowledge gap.
In the denitrification pathway, the reactions are catalyzed by nitrate reductase, nitrite reductase, nitric oxide reductase, and nitrous oxide reductase (Zumft 1997; Philippot and Hallin 2005). Of the denitrification steps, the reduction of NO2 − to NO involves two structurally different but functionally equivalent genes for nitrite reductase, the nirS gene containing cytochrome cd1 and the nirK gene containing copper (Hallin and Lindgren 1999; Braker et al. 2000). The reduction of N2O to N2 is another critically important step which converts N2O to the inert N2 gas. This last step is catalyzed by the N2O reductase encoded by the nosZ gene (Scala and Kerkhof 2000; Throbäck et al. 2004). If more N2O is reduced to N2, this would reduce the greenhouse gas N2O emissions. However, our understanding on the impact of management on these critically important processes and related N2O emissions is still limited and this limits our ability to manage these processes to reduce greenhouse gas emissions and increase the N use efficiency by plants.
Grassland is one of the largest terrestrial ecosystems in the world. In China, it accounts for more than 40 % of its total land area (Nan 2005). Grassland ecosystem is not only important for food production to feed the growing world population but also plays a critical role in global climate change. The world’s grasslands are an important source of greenhouse gases and will also be impacted by climate change.
Different management practices, such as continuous grazing, mowing, or periodic fencing off from grazing, can significantly affect the functioning of grassland ecosystems (Li et al. 2000; Zhao et al. 2007). For example, mowing of grassland was reported to increase species richness and community stability of herbaceous plants (Collins et al. 1998; Yang et al. 2012), decrease the amount of available N in soil, and affect the abundance and community structure of AOA and AOB in a temperate grassland ecosystem (Chen et al. 2014). Grazing can affect soil physical conditions by animal trampling, which causes soil compaction, and affect nutrient cycling (particularly N) through dung and urine returns to grassland during grazing (Oenema et al. 2007; Houlbrooke et al. 2008; Zhong et al. 2014). In grazed grasslands, about 70–90 % of the N ingested by the grazing animal is returned to the grassland soils in the excreta, particularly in the urine (Di and Cameron 2002). Animal treading was also reported to stimulate denitrification to some extent in grass-clover pastures (Menneer et al. 2005). Cattle grazing led to a significant increase in AOB abundance but not that of AOA, narG, or nosZ genes in a meadow grassland (Zhong et al. 2014). Free livestock grazing was found to have increased the population abundance of nitrification genes and decreased that of denitrification genes (Le Roux et al. 2008; Xie et al. 2014). Another study reported that grazing resulted in an increase in AOB abundance in the topsoil (0–10 cm) but had no effect at 10–20 cm depths (Liu et al. 2011).
However, these studies mainly described the effect of grazing on soil microbial communities and most of them only focused on nitrifiers. The effects of different management practices on nitrification, denitrification processes, and related microbes remain elusive. In particular, there is very little information available on the responses of nitrifying and denitrifying communities in extensively managed semiarid grasslands to mowing or periodic fencing off from grazing. Enclosure is used as a restoration practice for grassland to avoid over grazing, but its impact on key soil microbial communities and functions is poorly understood. An improved understanding of the relationships between different grassland management practices and key N cycling microbial communities is critically important for developing sustainable grassland management systems in the long term.
The aim of this research was to study the impact of different grassland management practices, including free-grazing, enclosure, and mowing, on the abundance and community structure of microbial functional genes related to nitrification and denitrification in the soil in a semiarid grassland under low temperature in Inner Mongolia in northern China. It is hypothesized that animal trampling in the free-grazing treatment would compact the soil and reduce soil aeration, leading to more anaerobic conditions, and this would affect both the nitrifier and denitrifier communities compared with those in the enclosure and mowing treatments. Similarly, the removal of herbage in the mowing treatments would also affect the carbon (C) and nitrogen (N) cycling and this would also affect the microbial communities.
2 Materials and methods
2.1 Site description and soil sampling
This study was conducted in Xilingol region (43°26′–44°39′N, 115°32′–117°12′E) in Inner Mongolia, China. The region has a continental, semiarid climate, with around 102–136 frost-free days. This region is very cold in winter and cool in summer. The mean annual temperature is −0.4 °C, with the minimum and maximum monthly mean temperatures ranging from −19.5 °C in January to 20.8 °C in July (Kawamura et al. 2005). The average annual precipitation is 350 mm, distributed unevenly among seasons, falling mainly during June–August which coincides with the plant-growing season. The annual potential evaportranspiration ranges from 1600 to 1800 mm. The dominant plant species in the study area was Leymus chinensis with some Stipa grandis and Cleistogenes squarrosa. The soil type is dark chestnut (calcic Chernozem according to ISSS Working Group RB, 1998) (Bai et al. 2010).
There were six long-term treatments at the research site, each with three replicates, and these are randomly allocated in the field. The treatments were detailed in Table 1. Soil samples were collected from the upper 10-cm layer from five random locations within each plot using a 5-cm diameter soil auger in October 2013. The five samples from each plot were bulked into a single composite sample, packed with ice packs, and transported to the laboratory. Soil samples were then passed through a 2-mm sieve, stored at 4 °C for soil analysis, and a subsample taken from each soil was stored at −80 °C for nucleic acids extraction.
Soil pH was measured with a soil-to-water ratio of 1:2.5, and soil organic carbon (SOC) was determined by dichromate digestion (Kalembasa and Jenkinson 1973). Gravimetric soil moisture content was analyzed by oven drying at 105 ± 2 °C for 24 h. Available potassium (AK) was extracted by 1 M ammonium acetate and determined by flame photometry. Nitrate (NO3 −) and ammonium (NH4 +) were extracted with 1 M KCl and determined by a flow injection analyzer (SAN++, Skalar, Holland). Total nitrogen (TN) content was analyzed using Kjeldahl acid digestion method with an auto-analyzer (Foss Inc., Hillerød, Sweden).
2.2 Extraction of total DNA from soils and Q-PCR of functional genes
DNA was extracted from 0.5 g (fresh weight) of soil with a FastDNA Spin Kit for Soil (MP Biomedicals, LLC., Solon, OH, USA), in accordance with the manufacturer’s protocol. The DNA size and integrity were checked by electrophoresis on a 0.7 % agarose gel, the quantity and purity were estimated using a Nanodrop®ND-2000 UV-vis spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). The extracted DNA was stored at −20 °C until analysis.
Quantitative polymerase chain reaction (Q-PCR) assays targeting bacterial and archaeal amoA genes, bacterial nirS, nirK, and nosZ genes were carried out in triplicate by real-time PCR with LightCycler 480 (Roche Applied Science). Each 20 μL reaction mixture contained 10 μL SYBR Premix Ex Taq (TaKaRa, Dalian, China), 1 mM of each primer and 1.5 μL of tenfold diluted DNA template (1–10 ng) and 7.9 μL milli-Q water. Melting curve analysis was performed at the end of each real-time PCR run to confirm PCR product specificity by measuring fluorescence continuously with the temperature increasing from 50 to 99 °C.
Real-time PCR standard curve was developed as previously described by Di et al. (2009). Briefly, each gene fragment was PCR-amplified. The details of primers and PCR conditions were shown in Table 2. The PCR amplicons were subsequently purified with a PCR cleanup kit (Nucleospin, Macherey-Nagel) and cloned into the pGEM-T Easy Vector (Promega, Madison, WI, USA). The resulting ligation mix was transformed in Escherichia coli JM109 competent cells (Promega, Madison, WI, USA) according to the manufacturer’s instructions. Plasmids were extracted from the correct insert clones of each target gene and sequenced with an ABI PRISM® 3730 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA), after cycle sequencing reactions using a BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems). Tenfold serial dilutions of a known copy number of the plasmid DNA representing 102–109 copies μL−1 were subjected to real-time PCR assay in triplicate to generate an external standard curve. High efficiencies of 92.3–101.2 % were obtained for the functional genes amplification, with the R values ranging between 0.994 and 0.998.
2.3 PCR-DGGE assays of functional genes
Before denaturing gradient gel electrophoresis (DGGE) analysis, PCR amplifications were performed using the primers and thermal cycling conditions of functional genes from nitrifiers and denitrifiers according to Table 2. Amplification was performed in 20 μL reaction mixtures, including 10 μL 2 × Easy Taq® PCR SuperMix (TransGen Biotech, Beijing, China), 8.6 μL milli-Q water, 0.5 μM of each primer and 1 μL DNA template, except that 1 μL of tenfold diluted DNA was used for AOA PCR amplification. About 150 ng of PCR amplicons was subjected to DGGE analysis carried out in a Dcode Universal Mutation Detection System (Bio-Rad Laboratories, Hercules, CA, USA), and the detailed DGGE analysis profile of the functional genes was listed in Table 2. The PCR products of bacterial and archaeal amoA genes were separated on 8 % (w/v) acrylamide gels while those of nirS, nirK, and nosZ genes were separated on 6 % (w/v) acrylamide gels. After electrophoresis, the gels was stained in 1:10,000 (v/v) SYBR green I (Amresco, Solon, OH, USA) for 30 min and scanned using ChemiDoc XRS System (Bio-Rad). The obtained DGGE profiles were analyzed using Quantity One Software, and cluster analysis was also conducted according to the DGGE bands.
2.4 Sequencing and phylogenetic analysis
DGGE bands with different mobility of every treatment were excised for direct sequencing, then the nucleotide sequences were translated to protein sequences by the EMBOSS Transeq program within the European Bioinformatics Institute website. The protein sequences were aligned with BLAST for the closest relatives from the GenBank database. Phylogenetic analysis of archaeal and bacterial amoA genes and bacterial nirS, nirK, and nosZ genes was then conducted by Molecular Evolutionary Genetic Analysis software (MEGA6.06) with 1000-fold bootstrap support. The DNA sequences of bacterial and archaeal amoA genes and bacterial nirS, nirK, and nosZ genes were deposited in the [DNA Data Bank of Japan (DDBJ) LC060262-LC060279, LC060280-LC060285, LC060286-LC060297, LC060298-LC060305, and LC060306-LC060313], respectively.
2.5 Statistical analysis
All statistical analyses were performed using SPSS version 20, and one-way analysis of variance (ANOVA) followed by Duncan’s multiple range test was used to check for the differences of soil physio-chemical properties and functional genes abundance between treatments. P < 0.05 was considered to be statistically significant. The figures of differences of soil physio-chemical properties and functional genes abundance between treatments were prepared by Origin (Origin Pro 9.0 for Windows). The correlation analysis between microbial community structure and soil properties was analyzed by non-metric multidimensional scaling (NMDS) using the Bray-Curtis dissimilarity measurement. The significant differences between soil properties and each community structure were analyzed by ANOSIM. VEGAN of R language was implemented to do the above two analyses.
3 Results
3.1 Soil physio-chemical properties
Table 3 showed that pasture management practices profoundly affected soil basic characteristics. Generally, soil AK, SOC, and TN of the samples from HG were the lowest among the six treatments. AK in the E83 treatment was significantly higher than those in the HG and E96 treatments. Soil moisture in the HG treatment was lower than that in the E96, markedly lower than those in the LG, E83, and mowing treatments.
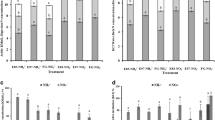
The NH4 +-N concentration in all the treatments was low but was higher in the enclosure treatments (E83 and E97) than HG and mowing (M1 and M1/2) managements (Fig. 1). The NO3 −-N concentration in the LG treatment was higher than those of all the other treatments.
Ammonium-N (▢) concentration and nitrate-N (▨) concentration of samples. The vertical bars indicate the standard errors of triplicate samples (LG light grazing, HG heavy grazing, E96 enclosed since 1996, E83 enclosed since 1983, M1/2 herbage harvested once every second year since 2001, M1 herbage harvested once every year since 2001; similarly hereinafter)
3.2 Abundance of microbial functional genes
3.2.1 Abundance of amoA genes
The archaeal amoA gene copy numbers outnumbered those of AOB in all the soils (Fig. 2). The ratios of archaeal to bacterial amoA gene copy numbers ranged from a high of 24.3 in E83 to a low of 13.1 in M1. The archaeal amoA gene copy numbers varied from 5.99 × 108 to 8.60 × 108 copies per gram dry soil, whereas that of AOB ranged from 3.02 × 107 to 4.61 × 107. The archaeal amoA gene copy numbers were significantly lower in the mowing treatments than in the light grazing treatment (LG). On the other hand, the pasture management did not significantly affect the bacterial amoA gene abundance.
3.2.2 Abundance of denitrification genes nirS, nirK, and nosZ
The copy numbers of nirS gene in the soil samples of M1 were significantly lower than those in the other five treatments, while no significant difference was found between those five treatments (Fig. 3). The nirK gene copy numbers were generally lower than those of the nirS gene except in the M1 treatment, and the effect of grazing management was small on the nirK copy numbers. Furthermore, there was a negative relationship between nirS abundance and TN concentration (R 2 = 0.418, P = 0.004), whereas a positive relationship between nirK abundance and SOC concentration (R 2 = 0.350, P = 0.01). The nosZ gene copy numbers varied from 3.60 × 108 to 6.51 × 108 per gram dry soil, and these were much higher than both the nirS and nirK genes. However, grazing management did not have a significant impact on nosZ gene copy numbers. The nosZ/(nirS + nirK) ratios were 4.15, 5.60, and 7.75 for free-grazing, enclosure, and mowing treatments, respectively.
3.3 DGGE analysis of microbial functional genes
After using the DDBJ to compare the sequences, we found that the DGGE bands with the same mobility in the DGGE gels shared 100 % similarity. Therefore, the bands with the same mobility were marked with the same number. The community structure of archaeal and bacterial amoA, nirS, nirK, and nosZ analyzed by DGGE showed a clear change with management treatments (Fig. 4 and Fig. S1 in the Electronic Supplementary Material). DGGE banding patterns of triplicate soil samples for each treatment showed good reproducibility.
3.3.1 DGGE analysis of amoA genes
In total, 18 bands were detected with different mobility for AOA (Fig. 4a). Bands A3 and A5 were detected only in the LG sample, whereas bands A12, A13, and A14 were unique to HG. Some bands, such as bands A16 and A17 were observed solely in the HG, E96, and E83 treatments. Although some bands (e.g., A6, A8, A9, A10) were present in the profiles of all treatments, their intensity was different. For example, bands A6 and A8 had the highest intensity in the HG treatment, while bands A9 and A10 became stronger in intensity for samples HG, E96, and E83.
The DGGE profile of AOB changed slightly among the management system and six bands with different mobility in total were observed (Fig. S1, Electronic Supplementary Material). Five bands were common with strong intensity in all the samples. Band B6 had the most prominent intensity in the HG treatment but was faint in the LG, E96, and mowing treatments and disappeared altogether in the E83 treatment.
Cluster analysis further confirmed that the different management practices had a strong effect on community structure of AOA (Fig. S2a, Electronic Supplementary Material) but not on that of AOB (Fig. S2b, Electronic Supplementary Material). Moreover, the community structure of AOA was significantly related to soil moisture, pH, TN, and NO3 −-N concentrations (R 2 = 0.580, P = 0.001; R 2 = 0.404, P = 0.027; R 2 = 0.420, P = 0.015 and R 2 = 0.644, P = 0.001, respectively) (Fig. S3a, Electronic Supplementary Material), whereas no significant correlation was found between AOB community and soil characteristics.
3.3.2 DGGE analysis of denitrification genes nirS, nirK, and nosZ
In general, the intensities of DGGE bands of nirS and nosZ were enhanced by the enclosure treatments compared with the grazing and mowing treatments. As for nirS gene (Fig. 4b), band S1 had the highest intensity in the mowing treatments, lower intensity in the LG and enclosure treatments, but disappeared in the HG treatment. Both enclosure treatments showed a more diverse banding pattern with additional bands S3, S4, and S10 compared with the other treatments. However, bands S11 and S12 were unique to the HG treatment, whereas band S6 was only detected in the mowing treatments.
Three common bands of the nirK gene were shown in all the treatments but band K2 was observed in the mowing treatments, and bands K3 and K4 were unique to the enclosure treatments, whereas bands K5 and K7 were only shown in the HG, E96, and E83 treatments (Fig. 4c).
The DGGE band patterns of nosZ (Fig. 4d) showed a clear variation with different pasture management treatments. Band Z2 appeared in all the samples except the mowing treatments, while band Z4 became stronger in intensity for soil samples in the enclosure treatments, and band Z8, although faint, was shown in all but the HG treatments.
Cluster analysis of the band patterns (Fig. S2c–e, Electronic Supplementary Material) showed that there were marked effects of pasture management on community structure of nirS, nirK, and nosZ genes. The period of enclosure also had a major effect on the community structure of nirS (Fig. S2c, Electronic Supplementary Material). Besides, according to the NMDS analysis (Fig. S3b–d, Electronic Supplementary Material), the nirS community structure was significantly related to soil moisture, pH, and NO3 −-N content (R 2 = 0.516, P = 0.007; R 2 = 0.399, P = 0.031; and R 2 = 0.438, P = 0.013, respectively), while changes in the overall nirK community structure were significantly correlated to soil moisture and NH4 +-N concentration (R 2 = 0.557, P = 0.001 and R 2 = 0.399, P = 0.029, respectively). In contrast, the community composition of nosZ was significantly related to soil moisture, SOM, and NO3 −-N concentrations (R 2 = 0.496, P = 0.007; R 2 = 0.481, P = 0.009; and R 2 = 0.490, P = 0.007, respectively).
3.4 Phylogenetic analysis of microbial communities
A neighbor-joining tree was constructed using the translated amino acid sequences of DGGE bands and the related sequences obtained from DDBJ.
3.4.1 Phylogenetic analysis of amoA genes
Phylogenetic analysis of archaeal amoA gene sequences (Fig. 5a) showed that the majority of the AOA sequences were affiliated with cluster Nitrososphaera (previously also designated as soil or І.1b AOA lineage, DeLong 1998), and grouped into three distinct clusters, referred to as Nitrososphaera subclusters 1, 2, and 3. Bands A3 and A5, detected only in the LG treatment, were aligned in cluster 2, while bands A13 and A14 shown exclusively in the HG treatment were affiliated with cluster 1. Almost all the AOB sequences fell into Nitrosospira species, and a high proportion was grouped into cluster 3a, with some in cluster 2 and cluster 3b (Fig. 5b).
3.4.2 Phylogenetic analysis of nirS, nirK, and nosZ genes
The nirS sequences were widespread in the phylogenetic tree, and the nirS tree was divided into four clusters (clusters І–IV) (Fig. 6a). Among them, clusters І and IV were related to the nirS of Rhizobiales and Burkholderiales bacteria, respectively. Cluster III was not related to known denitrifying bacteria, and clusters II showed high similarities to numerous unculturally deduced nirS clones from arable soils. As for nirK (Fig. 6b), each of sequences obtained in this study was branched with known denitrifying bacteria and the sequences were spread throughout the tree. The nirK tree was grouped into three clusters, and they were closely related to nirK from Bradyrhizobium, Mesorhizobium, and Pseudomonadaceae, respectively. The genera Bradyrhizobium and Mesorhizobium are both affiliated to Rhizobiales. In complete contrast to nirK, the phylogenetic tree of nosZ (Fig. 6c) showed that most of the sequences were not related to any known denitrifying bacteria. The nosZ tree was classified into two clusters, and cluster І was related to the nosZ of Rhizobiales, while cluster II was distantly related to the nosZ of known denitrifying bacteria.
4 Discussion
Results from this study clearly demonstrated that pasture management had a major impact on soil fertility with the heavy grazing treatment (HG) having the lowest available K and soil organic matter content. The significant reductions in the nutrient contents would suggest that heavy grazing is not sustainable in the long-term in this grassland ecosystem.
With the microbial communities studied, it is interesting that the abundance of AOA was many times that of AOB in this semiarid grassland. Previous studies have shown that AOA play an important role in nitrification in strongly acid soils (Zhang et al. 2012). In the present study, the soil is weak alkaline but AOA was also the dominant ammonia oxidizer community. This might suggest that soil pH may not be the only factor affecting AOA abundance in different soils. The driving factor for the predominantly higher AOA abundance than AOB abundance is not clear. The very low population abundance of AOB and the lack of a treatment effect on AOB populations might suggest that AOB probably play a lesser role in N cycling compared with their AOA counterparts in this weak alkaline grassland soil under low temperature. Previous studies have shown that AOB are better adapted to higher fertility and higher N environments, whereas AOA are more favored by low fertility and low N environments (Nicol et al. 2008; Jia and Conrad 2009; Di et al. 2009, 2010; Zhang et al. 2012). The soil used in the present study is very low in soil fertility and ammonium content (Fig. 1). This low fertility and dry environment (Table 3) may therefore favored AOA over AOB in this grassland ecosystem. These results confirm the prevalence of AOA in terrestrial ecosystems, particularly in low fertility and stressed environments, such as this semiarid grassland ecosystem.
The only significant pasture management effect on the ammonia oxidizers was the significantly higher AOA abundance in the light grazing (LG) treatment than in the mowing treatments. Previously, Chen et al. (2014) also reported significantly lower AOA abundance in mowing than in non-mowing treatments. It is possible that the recycling of nutrients, particularly N, in the forms of urine and dung in the light grazing treatment might have stimulated a slightly higher AOA population abundance. Wan and Luo (2003) suggested that thaumarchaea could also use organic carbon as an energy source. The soluble organic C in the urine and dung might therefore also played a role. Further research is required to understand possible factors that may control the abundance of AOA in different management systems of grassland.
Although little difference was recorded in the bacterial amoA DGGE patterns among the different treatments, clear differences were detected in the archaeal amoA DGGE patterns between different treatments (Fig. S1, Electronic Supplementary Material and Fig. 4a). The lower numbers of bands and lower intensity of archaeal amoA DGGE bands under the mowing treatments compared with grazing and enclosure treatments would suggest that mowing decreased the diversity of archaeal communities. Even though the HG treatment leads to a more stressed environment with low fertility, the diversity of archaeal amoA communities was still high as demonstrated by the DGGE patterns (Fig. 4a), pointing to the resilience of AOA in this ecosystem. In the semiarid grassland ecosystem, soil moisture, pH, TN, as well as NO3 −-N concentrations were the four main environmental variables that influence AOA community structure (Fig. S3a, Electronic Supplementary Material).
Phylogenetic analysis of the present study showed that most of the AOA sequences were affiliated with the Nitrososphaera cluster (previously also designated as cluster soil (DeLong 1998)), leaving two bands ungrouped. No Nitrosopumilis (previously also designated as marine (DeLong 1998)) origin of AOA in this grassland soils was found (Fig. 5a). This is in line with the findings in other temperate grassland soils (Shen et al. 2011; Chen et al. 2014). All the AOB sequences in this study were grouped within Nitrosospira species (Fig. 5b), similar to those reported in other grassland systems (Di et al. 2009; Chen et al. 2013), upland soils (He et al. 2007), and paddy soils (Bowatte et al. 2006; Chen et al. 2008). The absence of Nitrosomonas in the present study is interesting and requires further investigations to understand the mechanisms behind this finding. Band B6 in this study was grouped to cluster 2, which has previously been associated with acid soils (Stephen et al. 1998), even though the soil in this study is slightly alkaline, suggesting the wider pH adaptability of this group.
These results clearly demonstrate that pasture management practices of grazing, mowing, and enclosure can significantly affect the abundance and community structure of AOA, rather than AOB, confirming the importance of AOA in N cycling in this important grassland ecosystem.
In this study, nirS gene abundance was higher than that of nirK in all the treatments except in the M1 treatment (Fig. 3). This was in contrast with the findings from some previous studies in other ecosystems which showed the dominance of nirK in forest and paddy soils (Wallenstein and Vilgalys 2005; Yoshida et al. 2010; Zhou et al. 2011). It is interesting that the nirS abundance was significantly lower in the M1 treatment, while nirK abundance in the M1 treatment was the highest of all the treatments. Yoshida et al. (2010) and Zhou et al. (2011) found that nirK communities were more sensitive to environmental change than nirS in agricultural soils. These results therefore may partly support the hypothesis that there may be a niche differentiation between nirS and nirK in different ecosystems (Banerjee and Siciliano 2012; Novinscak et al. 2013).
Even though the nosZ gene abundance was the highest of all the denitrifying genes studied in all the treatments, it was not significantly affected by the management practices, including the grazing treatments with animal trampling. This may indicate that the nosZ communities were relatively stable under the different management practices. The soil at the site was dry, and the different treatments did not result in a major difference in soil moisture content. This might be partly responsible for the stable of the nosZ communities. Nevertheless, the DGGE band patterns of nosZ indicated higher intensity for the enclosure treatment (Fig. 4d). Therefore, enclosure might have stimulated greater denitrification. In addition, significantly higher ratios of nosZ/(nirS + nirK) were shown in the mowing treatments, which might suggest that the denitrification reactions might occur to a more complete extent in the mowed grassland. If this were the case, this would have implications on N2O emissions in the mowed grassland treatments. Further research is required to confirm this hypothesis.
Intriguingly, according to our study, the community structure of denitrifiers, irrespective of nirS, nirK, or nosZ, was significantly correlated to soil moisture and inorganic nitrogen content (Fig. S3b–d, Electronic Supplementary Material). These results strongly suggested the impact of environmental variables on diversity of denitrifiers.
Results from the phylogenetic analysis showed that the denitrifiers were comprised of a diverse microbial community. The nirS sequences (clusters І and IV) were closely related to the Rhizobiales and Burkholderiales bacteria in this grassland soil (Fig. 6a). These are different from those reported in paddy soils which were related to Burkholderiales and Rhodocyclales bacteria (Yoshida et al. 2009). Some of the nirS sequences were also unique to this grassland ecosystem (cluster Ш). The nirK sequences, on the other hand, were all grouped to known denitrifiers, mostly relating to Bradyrhizobium sp. and Mesorhizobium sp. (Fig. 6b). Even though Bradyrhizobium contains both nirK-harboring and nirS-harboring denitrifiers, the nirS communities were more diverse than the nirK communities. In contrast, the majority of the nosZ sequences were not grouped to known denitrifying populations, suggesting novel and unique nosZ communities in this grassland soil (Fig. 6c).
5 Conclusions
This field study has shown major impacts of management practices on the community abundance and structure of nitrifiers and denitrifiers in the semiarid grasslands of Inner Mongolia under cold conditions. The results showed that AOA rather than AOB was the predominant ammonia oxidizers in this neutral to alkaline grassland ecosystem. The majority of the AOA species were affiliated with Nitrososphaera cluster whereas the AOB was mainly affiliated to Nitrosospira species. The different management practices only had a significant impact on the abundance and community structure of AOA, but not that of AOB, pointing to the dominant role of AOA in N cycling in this soil system under low temperature conditions. Of the denitrifiers, the nirS communities were greater in number and diversity than the nirK communities, suggesting a greater role by the nirS communities in denitrification. Higher gene abundance and greater DGGE band intensity of nirS and nosZ genes in the enclosure treatments might indicate greater denitrification in this treatment. The higher nosZ/(nirS + nirK) ratios in the mowed grassland treatment might also suggest more complete denitrification in this treatment, possibly reducing N2O emissions in this treatment. Soil moisture and inorganic nitrogen content were found to be the two main soil environmental variables that influence the community structure of nitrifiers and denitrifiers.
References
Bai Y, Wu J, Clark CM, Naeem S, Pan Q, Huang J, Zhang L, Han X (2010) Tradeoffs and thresholds in the effects of nitrogen addition on biodiversity and ecosystem functioning evidence from inner Mongolia grasslands. Glob Chang Biol 16(1):358–372
Banerjee S, Siciliano SD (2012) Spatially tripartite interactions of denitrifiers in arctic ecosystems activities, functional groups and soil resources. Environ Microbiol 14(9):2601–2613
Bowatte S, Jia Z, Ishihara R, Nakajima Y, Asakawa S, Kimura M (2006) Molecular analysis of the ammonia oxidizing bacterial community in the surface soil layer of a Japanese paddy field. Soil Sci Plant Nutr 2(4):427–431
Braker G, Zhou J, Wu L, Devol AH, Tiedje JM (2000) Nitrite reductase genes (nirK andnirS) as functional markers to investigate diversity of denitrifying bacteria in Pacific Northwest marine sediment communities. Appl Environ Microbiol 66(5):2096–2104
Chen XP, Zhu YG, Xia Y, Shen JP, He JZ (2008) Ammonia-oxidizing archaea important players in paddy rhizosphere soil? Environ Microbiol 10(8):1978–1987
Chen Y, Xu Z, Hu H, Hu Y, Hao Z, Jiang Y, Chen B (2013) Responses of ammonia-oxidizing bacteria and archaea to nitrogen fertilization and precipitation increment in a typical temperate steppe in Inner Mongolia. Appl Soil Ecol 68:36–45
Chen YL, Hu HW, Han HY, Du Y, Wan SQ, Xu ZW, Chen BD (2014) Abundance and community structure of ammonia-oxidizing archaea and bacteria in response to fertilization and mowing in a temperate steppe in Inner Mongolia. FEMS Microbiol Ecol 89(1):67–79
Collins SL, Knapp AK, Briggs JM, Blair JM, Steinauer EM (1998) Modulation of diversity by grazing and mowing in native tallgrass prairie. Science 280(5364):745–747
Conrad R (1996) Soil microorganisms as controllers of atmospheric trace gases (H2, CO, CH4, OCS, N2O, and NO). Microbiol Mol Biol Rev 60(4):609–640
DeLong EF (1998) Everything in moderation archaea as ‘non-extremophiles’. Curr Opin Genet Dev 8(6):649–654
Di HJ, Cameron KC (2002) Nitrate leaching in temperate agroecosystems sources, factors and mitigating strategies. Nutr Cycl Agroecosyst 46:237–256
Di HJ, Cameron KC, Shen JP, Winefield CS, O’Callaghan M, Bowatte S, He JZ (2009) Nitrification driven by bacteria and not archaea in nitrogen-rich grassland soils. Nat Geosci 2(9):621–624
Di HJ, Cameron KC, Shen JP, Winefield CS, O’Callaghan M, Bowatte S, He JZ (2010) Ammonia-oxidizing bacteria and archaea grow under contrasting soil nitrogen conditions. FEMS Microbiol Ecol 72(3):386–394
Di HJ, Cameron KC, Podolyan A, Robinson A (2014) Effect of soil moisture status and a nitrification inhibitor, dicyandiamide, on ammonia oxidizer and denitrifier growth and nitrous oxide emissions in a grassland soil. Soil Biol Biochem 73:59–68
Francis CA, Roberts KJ, Beman JM, Santoro AE, Oakley BB (2005) Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc Natl Acad Sci U S A 102(41):14683–14688
Hallin S, Lindgren P (1999) PCR detection of genes encoding nitrite reductase in denitrifying bacteria. Appl Environ Microbiol 65:1652–1657
He JZ, Shen JP, Zhang LM, Zhu YG, Zheng YM, Xu MG, Di H (2007) Quantitative analyses of the abundance and composition of ammonia-oxidizing bacteria and ammonia-oxidizing archaea of a Chinese upland red soil under long-term fertilization practices. Environ Microbiol 9(9):2364–2374
Houlbrooke DJ, Littlejohn RP, Morton JD, Paton RJ (2008) Effect of irrigation and grazing animals on soil quality measurements in the North Otago Rolling Downlands of New Zealand. Soil Use Manag 24(4):416–423
Jia Z, Conrad R (2009) Bacteria rather than archaea dominate microbial ammonia oxidation in an agricultural soil. Environ Microbiol 11(7):1658–1671
Kalembasa SJ, Jenkinson DS (1973) A comparative study of titrimetric and gravimetric methods for the determination of organic carbon in soil. J Sci Food Agric 24(9):1085–1090
Kawamura K, Akiyama T, Yokota HO, Tsutsumi M, Yasuda T, Watanabe O, Wang S (2005) Quantifying grazing intensities using geographic information systems and satellite remote sensing in the Xilingol steppe region, Inner Mongolia, China. Agric Ecosyst Environ 107(1):83–93
Le Roux X, Poly F, Currey P, Commeaux C, Hai B, Nicol GW, Prosser JI, Schloter M, Attard E, Klumpp K (2008) Effects of aboveground grazing on coupling among nitrifier activity, abundance and community structure. ISME J 2(2):221–232
Li SG, Harazono Y, Oikawa T, Zhao HL, He ZY, Chang XL (2000) Grassland desertification by grazing and the resulting micrometeorological changes in Inner Mongolia. Agric For Meteorol 102(2):125–137
Liu T, Nan Z, Hou F (2011) Culturable autotrophic ammonia-oxidizing bacteria population and nitrification potential in a sheep grazing intensity gradient in a grassland on the Loess Plateau of Northwest China. Can J Soil Sci 91(6):925–934
Menneer JC, Ledgard S, McLay C, Silvester W (2005) Animal treading stimulates denitrification in soil under pasture. Soil Biol Biochem 37(9):1625–1629
Nan Z (2005) The grassland farming system and sustainable agricultural development in China. Grassl Sci 51(1):15–19
Nicol GW, Leininger S, Schleper C, Prosser JI (2008) The influence of soil pH on the diversity, abundance and transcriptional activity of ammonia oxidizing archaea and bacteria. Environ Microbiol 10(11):2966–2978
Novinscak A, Goyer C, Dandie CE, Filion M (2013) Abundance, diversity and spatio-temporal dynamics of nirS gene-harbouring denitrifiers in a potato field over the course of a growth season. Syst Appl Microbiol 36(2):112–115
Oenema O, Oudendag D, Velthof GL (2007) Nutrient losses from manure management in the European Union. Livest Sci 112(3):261–272
Philippot L, Hallin S (2005) Finding the missing link between diversity and activity using denitrifying bacteria as a model functional community. Curr Opin Microbiol 8(3):234–239
Prosser JI, Nicol GW (2008) Relative contributions of archaea and bacteria to aerobic ammonia oxidation in the environment. Environ Microbiol 10(11):2931–2941
Scala DJ, Kerkhof LJ (2000) Horizontal heterogeneity of denitrifying bacterial communities in marine sediments by terminal restriction fragment length polymorphism analysis. Appl Environ Microbiol 66:1980–1986
Shen W, Lin X, Gao N, Shi W, Min J, He X (2011) Nitrogen fertilization changes abundance and community composition of ammonia-oxidizing bacteria. Soil Sci Soc Am J 75(6):2198–2205
Stephen JR, Kowalchuk GA, Bruns MAV, McCaig AE, Phillips CJ, Embley TM, Prosser JI (1998) Analysis of β-subgroup proteobacterial ammonia oxidizer populations in soil by denaturing gradient gel electrophoresis analysis and hierarchical phylogenetic probing. Appl Environ Microbiol 64(8):2958–2965
Throbäck IN, Enwall K, Jarvis Å, Hallin S (2004) Reassessing PCR primers targeting nirS, nirK and nosZ genes for community surveys of denitrifying bacteria with DGGE. FEMS Microbiol Ecol 49(3):401–417
Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu DY, Paulsen I, Nelson KE, Nelson W, Fouts DE, Levy S, Knap AH, Lomas MW, Nealson K, White O, Peterson J, Hoffman J, Parsons R, Baden-Tillson H, Pfannkoch C, Rogers YH, Smith HO (2004) Environmental genome shotgun sequencing of the Sargasso Sea. Science 304(5667):66–74
Wallenstein MD, Vilgalys RJ (2005) Quantitative analyses of nitrogen cycling genes in soils. Pedobiologia 49(6):665–672
Wan S, Luo Y (2003) Substrate regulation of soil respiration in a tallgrass prairie results of a clipping and shading experiment. Glob Biogeochem Cycles 17(2):23–31. doi:10.1029/2002GB001971
Watanabe T, Lee CG, Murase J, Asakawa S, Kimura M (2011) Carbon flow into ammonia-oxidizing bacteria and archaea during decomposition of 13C-labeled plant residues in soil. Soil Sci Plant Nutr 57(6):775–785
Xie Z, Le Roux X, Wang C, Gu Z, An M, Nan H, Chen B, Li F, Liu Y, Du G, Feng H, Ma X (2014) Identifying response groups of soil nitrifiers and denitrifiers to grazing and associated soil environmental drivers in Tibetan alpine meadows. Soil Biol Biochem 77:89–99
Yang H, Jiang L, Li L, Li A, Wu M, Wan S (2012) Diversity-dependent stability under mowing and nutrient addition evidence from a 7 years grassland experiment. Ecol Lett 15(6):619–626
Yoshida M, Ishii S, Otsuka S, Senoo K (2009) Temporal shifts in diversity and quantity of nirS and nirK in a rice paddy field soil. Soil Biol Biochem 41(10):2044–2051
Yoshida M, Ishii S, Otsuka S, Senoo K (2010) NirK-harboring denitrifiers are more responsive to denitrification-inducing conditions in rice paddy soil than nirS-harboring bacteria. Microbes Environ 25(1):45–48
Zhang LM, Hu HW, Shen JP, He JZ (2012) Ammonia-oxidizing archaea have more important role than ammonia-oxidizing bacteria in ammonia oxidation of strongly acidic soils. ISME J 6(5):1032–1045
Zhao Y, Peth S, Krümmelbein J, Horn R, Wang Z, Steffens M, Hoffmann C, Peng X (2007) Spatial variability of soil properties affected by grazing intensity in Inner Mongolia grassland. Ecol Model 205(1):241–254
Zhong L, Du R, Ding K, Kang X, Li F, Bowatte S, Hoogendoorn C, Wang Y, Rui Y, Jiang LL, Wang S (2014) Effects of grazing on N2O production potential and abundance of nitrifying and denitrifying microbial communities in meadow-steppe grassland in northern China. Soil Biol Biochem 69:1–10
Zhou ZF, Zheng YM, Shen JP, Zhang LM, He JZ (2011) Response of denitrification genes nirS, nirK, and nosZ to irrigation water quality in a Chinese agricultural soil. Environ Sci Pollut Res 18(9):1644–1652
Zumft WG (1997) Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev 61(4):533–616
Acknowledgments
This work was financially supported by the National Key Basic Research Program of China (2014CB138801) and Natural Science Foundation of China (41271272 and 41230857).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
We declare that all authors report of no conflicts of interest regarding the publication of this paper.
Additional information
Responsible editor: Weijin Wang
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 934 kb)
Rights and permissions
About this article
Cite this article
Pan, H., Li, Y., Guan, X. et al. Management practices have a major impact on nitrifier and denitrifier communities in a semiarid grassland ecosystem. J Soils Sediments 16, 896–908 (2016). https://doi.org/10.1007/s11368-015-1321-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11368-015-1321-1