Abstract
Key message
Endogenous hormone secretion proteins along with stress and defense proteins play predominant role in banana embryogenesis. This study reveals the underlying molecular mechanism during transition from vegetative to embryogenic state.
Abstract
Banana (Musa spp.) is well known globally as a food fruit crop for millions. The requirement of quality planting material of banana is enormous. Although mass multiplication through tissue culture is in vogue, high-throughput techniques like somatic embryogenesis (SE) as a mass multiplication tool needs to be improved. Apart from clonal propagation, SE has extensive applications in genetic improvement and mutation. SE in banana is completely genome-dependent and most of the commercial cultivars exhibit recalcitrance. Thus, understanding the molecular basis of embryogenesis in Musa will help to develop strategies for mass production of quality planting material. In this study, differentially expressed proteins between embryogenic calli (EC) and non-embryogenic calli (NEC) with respect to the explant, immature male flower buds (IMFB), of cv. Grand Naine (AAA) were determined using two-dimensional gel electrophoresis (2DE). The 2DE results were validated through qRT-PCR. In total, 65 proteins were identified: 42 were highly expressed and 23 were less expressed in EC compared to NEC and IMFB. qRT-PCR analysis of five candidate proteins, upregulated in EC, were well correlated with expression at transcript level. Further analysis of proteins showed that embryogenesis in banana is associated with the control of oxidative stress. The regulation of ROS scavenging system and protection of protein structure occurred in the presence of heat shock proteins. Alongside, high accumulation of stress-related cationic peroxidase and plant growth hormone-related proteins like indole-3-pyruvate monooxygenase and adenylate isopentenyltransferase in EC revealed the association with the induction of SE.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Banana is an important crop having elevated from its status of subsistence crop to a high-value crop (Ravi and Uma 2011). It is widely cultivated as a multifaceted crop providing nutritional and economic security for millions across the globe. Banana being a vegetatively propagated crop has a low multiplication index through suckers (Singh et al. 2011) which emphasized the need for mass production protocols for the supply of quality planting material. Tissue culture technology has revolutionized the banana industry, but it only caters to approximately 8% of the global demand for planting material (Singh et al. 2011). Occurrence of somaclonal variations due to prolonged subcultures by industries has been a bottleneck (Sales and Butrado 2014). Thus, the growing demand for quality planting material has necessitated the need to look for alternate mass production tools. Somatic embryogenesis (SE) has been exploited as an alternative high-throughput technique for mass multiplication for its enhanced multiplication efficiency within a short span of time (Hussein et al. 2006).
SE is a technique in which somatic cells of a plant develop into whole new plants which are similar to zygotic embryo-derived plants (Ji et al. 2011). SE is a promising tool for producing quality planting material in many horticultural crops like Prunus cerasus L., Mangifera indica, Vitis vinifera, Citrus sinensis Osbeck, and Cucumis sativus (Tang et al. 2000; Patena et al. 2002; Marsoni et al. 2008; Pan et al. 2009; Wisniewska et al. 2012) SE in banana through embryogenic cell suspension (ECS) has been reported among various Indian commercial varieties (Akbar 2011). ECS is considered to be the best model system for mass multiplication of interested genotypes and improvements of traits through gene transfer technology (Pan et al. 2009). The improvement of in vitro regeneration system is much needed for processing genetic transformation in banana (Ganapathi et al. 1999). However, success rate of embryogenic callus induction and regeneration of plants through SE are limited to few of the banana genotypes (Sidha et al. 2006; Uma et al. 2012).
The molecular mechanism behind SE can be elucidated both by proteomic and transcriptomic approaches (Marsoni et al. 2008). Many SE-inducing genes like ABA-inducible gene (Yang and Zhang 2010), AGL 15 (Heck et al. 1995), BABY BOOM (Boutilier et al. 2002), Calmodulin (Overvoorde and Grimes 1994), DcAGP (Baldwin et al. 2001), LEC (Lotan et al. 1998), SERK (Nolan et al. 2003) and WUSCHEL (Zuo et al. 2002) were identified through transcriptome approach. Although transcriptomic approach has been established as a superior tool, it does not afford a clear biological representation of an organism at a specific point of time. There has been no evident correlation established between the expression level of mRNA and the abundance level of proteins as the stability and translatability of mRNA and protein stability vary from time to time (Sharifi et al. 2012). Also, mRNA expression profiling does not provide any thorough information on protein levels which include protein structure, function, activity and post-translational modifications. As proteins are the final gene products directly involved in cellular mechanism, it is highly pertinent to identify proteins accumulated with SE through proteomic approach (Pan et al. 2009).
Proteomics approach allows identification of differentially expressed proteins during the growth and development of plants (Canovas et al. 2004). The emergence of advanced high-throughput techniques like two-dimensional gel electrophoresis (2DE) and mass spectrometry (MS) takes proteomic approach to a next level of research (Marsoni et al. 2008). More recently, proteomic studies on SE were carried out in several plant species which includes Coffea arabica, Medicago truncatula, Picea glauca, Vitis vinifera, Citrus sinensis Osbeck, Cyclamen persicum, Boesenbergia rotunda, Crocus sativus, Cucumis sativus and Zea mays. Basic information generated has led to enhancement of SE efficiency in respective crops (Yuffa et al. 1993; Imin et al. 2004; Lippert et al. 2005; Marsoni et al. 2008; Pan et al. 2009; Rode et al. 2011; Tan et al. 2012; Sharifi et al. 2012; Wisniewska et al. 2012; Sun et al. 2013).
SE in banana has been well reported across various genotypes (Schoofs 1997; Ganapathi et al.1999; Akbar 2011). But till date, there is no evidence on molecular mechanism/s underlying the process of SE for any banana genomic groups. Thus, understanding the molecular mechanism/s of SE process in banana is expected to facilitate the development of a universal protocol for inducing embryogenic calli across genotypes. Thus, in the present study, major commercial cultivar Grand Naine, one of the few banana genotypes with successful history of SE is used.
To the best of our knowledge, this is the first study which applies 2DE in addition with matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF–MS) to identify the differentially expressed proteins that are associated with the induction of SE in banana. In this study, we have focused mainly on the identification and classification of differentially expressed proteins that are primarily responsible for embryogenic callus induction. The promising roles of the proteins expressed during the induction of SE are discussed in detail.
Materials and methods
Plant material and sample collection
The male flower buds of cv. Grand Naine (AAA), were collected from commercial plantations in Theni district of Tamil Nadu, India. The collected buds were virus-indexed to confirm the freeness from known banana viruses like banana bunchy top virus, banana bract mosaic virus and banana streak virus. Buds were reduced to the size of 0.8 × 2 cm and surface-sterilized with 70% ethanol for 1 min. The immature whorls from 2nd to 18th position of the male flower buds were isolated with a fine sterile scalpel and used as explant. The explants, immature male flower buds (IMFB) were cultured on callus induction medium (MA1) which was supplemented with 4 mg/L 2, 4-d (2, 4-dichlorophenoxyacetic acid), 1 mg/L 3-IAA, 1 mg/L NAA and 30 g/L sucrose (Akbar 2011). The pH of the medium was adjusted to 5.8 using 0.1 M NaOH. The medium contained 2 g/L CleriGel™ (HiMedia) as solidifying agent. The sterilization of culture medium was carried out by autoclaving at 121 °C for 20 min. The medium was poured on 90-mm Petri plates and allowed to solidify. The cut edges of the IMFB were placed in contact with the surface of the medium and the dishes were sealed with parafilm to avoid contamination and reduce evaporation. The plates were incubated and kept undisturbed in total darkness at 25 ± 2 °C for first 3 months. After 3–6 months of incubation, the plates were observed for developing embryogenic calli (EC) and non-embryogenic calli (NEC).The callus thus formed were identified based on their morphology observed under stereomicroscope, while cell structures were observed under the compound microscope. The callus containing pro-embryogenic mass (PEM) was considered as embryogenic callus which contains more number of starch granules around their nucleus and the rest as non-embryogenic calli. Lugol’s iodine staining (Tsai et al. 2009) and 4′, 6-diamidino-2-phenylindole (DAPI) staining (Wang et al. 2013) were followed to confirm the starch granules and nucleus of embryogenic cells, respectively. The percentage of EC was calculated based on the INIBAP technical guidelines (Strosse et al. 2003). The EC, NEC and IMFB were collected in triplicates, weighed and treated with liquid nitrogen. The frozen samples were stored in deep freezer at −80 °C for proteomic analysis and gene expression studies.
Protein extraction
Total protein extracts were prepared from frozen IMFB, EC and NEC according to the previously described method (Hurkman and Tanaka 1986) with minor modifications. The samples (~100 to 200 mg) in triplicates were ground to a very fine powder in a mortar and pestle using liquid nitrogen. While still frozen, the resulting powder was transferred to a sterile microfuge tube and suspended in 1 mL of ice-cold extraction buffer containing 50 mM Tris buffer (pH 8.5), 5 mM EDTA disodium salt, 100 mM KCl, 30% sucrose, 1% polyvinyl pyrrolidone (PVP), 1% dithiothreitol (DTT) and 40 µL of protease inhibitor cocktail (Cat no. P8340, Sigma-Aldrich, USA). After vortexing for 30 s, 1 mL of Tris-saturated phenol (pH 8.8) was added to each tube. The tubes were incubated for 30 min on ice. The upper phenolic phase was transferred to a new microfuge tube after centrifugation at 11,000×g for 10 min at 4 °C. The collected proteins in the phenolic phase were precipitated by adding five volumes of prechilled 100 mM ammonium acetate in methanol. The mixture was incubated overnight at −20 °C. The protein pellets were collected by centrifugation at the same speed as above, for 20 min at 4 °C. The protein pellets obtained were resuspended in 100 mM ammonium acetate and were again incubated at −20 °C for 30 min. The centrifugation was repeated for 10 min at 4 °C. The final protein pellets thus obtained were washed twice with 80% ice-cold acetone and were air-dried and stored in deep freezer for further analysis. The dried pellets were resuspended in lysis buffer containing 7 M urea, 2 M thiourea, 4% CHAPS, 40 mM DTT and 2% IPG buffer (pH 4–7) for 2D gel electrophoresis. The sample mixture was vortexed for 1 h at RT. The protein concentration was determined using 2-D quantification kit (GE Healthcare Bio-Science Crop, Piscataway, NJ, USA) using BSA standard. The protein samples were further purified using 2-D Clean-Up kit (GE Healthcare Bio-Science Crop, Piscataway, NJ, USA) prior to use.
Two-dimensional gel electrophoresis (2DE)
Total protein (200 µg) extract of IMFB, EC and NEC samples was loaded separately on 13-cm IPG strips, (pH 4–7) (GE Healthcare Bio-Sciences AB, Uppsala, Sweden) by active overnight rehydration. The first-dimensional separation of proteins on gel was performed using IEF 100 unit (Hoefer Inc, San Francisco, USA). The parameters followed for analysis were as follows: 250 V for 2 h, 500 V for 1 h, 3500 V for 3 h, increasing to 4500 V over 1 h and holding at 4500 V for 3 h. Finally, hold step was maintained at 1000 V to obtain sharp bands and for completion of the run that lasted for a total of 31,974 Vh.
Following IEF, the focused strips were equilibrated with two types of SDS-equilibration buffer solutions [6 M urea, 75 mM Tris HCl (pH 8.8), 29.3% glycerol, 2% SDS and 0.002% bromophenol blue] for 15 min. The first buffer contained 130 mM dithiothreitol (DTT) and the second buffer contained 135 mM iodoacetamide (IAA). The alkylated strips were then subjected to two-dimensional separations at the top of 12% SDS-polyacrylamide gel prepared according to Laemmli (1970). The electrophoresis was performed using SE 600 unit (Hoefer Inc, San Francisco, USA) at 50 V for 20 min and then at 150 V for 5 h. Molecular masses of the unknown proteins in the gel were determined based on molecular weight of the pre-stained PAGE RULER (Fermentas, Thermo Fisher Scientific, Mumbai, India) loaded on the left side of the gel. Each protein sample was run in replicates of three.
Gel staining and analysis of images
The gels were stained with silver nitrate for analytical purpose as described by Blum et al. (1987) and with colloidal coomassie brilliant blue stain (CBB) for preparative purpose as described by Neuhoff et al. (1988) with minor modifications. The gels were destained with several changes of distilled water to remove the excess stain. The destained gels were scanned using EPSON scanner (EPSON PERFECTION V 750 PRO, Seiko Epson Corp, Japan) at a resolution of 800 dpi and the images were saved for further analysis. Gel images were analyzed for spot detection and quantification using Hoefer 2D view software (Hoefer Inc, San Francisco, USA). After detecting the spot and subtracting the background, the gel images were aligned, annotated, matched, and the quantitative determination of the spot volumes was performed. The spots which were differentially expressed by more than 1.5-fold were alone selected for subsequent data analysis. The volumes of reproducible spots from three replicate gels were normalized against total spot volume.
Excision of spot and in-gel trypsin digestion
The protein spots of interest exhibiting more than 1.5-fold change in abundance were subjected to further characterization using MS. The spots were manually excised from the preparative colloidal CBB-stained gels by using one-touch spot picker (Gel Company, San Fransisco, USA). The protein spots were then rinsed with proteomic-grade water and, then, with acetonitrile (ACN) destaining solution containing 50 mM ammonium bicarbonate (NH4HCO3) for 15 min. After discarding the aqueous phase, the gel pieces were subjected to ACN wash for two more times and were allowed to dry in vacuum centrifuge for 20 min. Then, the proteins in the gel were reduced by adding freshly prepared 10 mM DTT in 50 mM NH4HCO and incubating at 56 °C for 45 min. After discarding the aqueous phase, proteins in the gel were alkylated immediately by adding 55 mM IAA in 50 mM NH4HCO3. The mixture was incubated for 30 min at room temperature in the dark. The pieces of gel were washed twice with 50 mM NH4HCO3 and ACN (1:1) mixture for 15 min and were allowed to dry under vacuum. Trypsin (Trypsin gold, promega, WI, USA) was prepared by dissolving the enzyme in 50 mM acetic acid and diluting the same with 25 mM NH4HCO3 to give a final working concentration of 100 ng/µL. The freshly prepared enzymatic solution was added to rehydrate the gel for 30 min and enough amount of 25 mM NH4HCO3 (2–3 µL) to keep the gel in wet condition. The reaction mixture was incubated overnight at 37 °C for digestion.
Mass spectrometry analysis
The resulted peptide fragments from the above step were further concentrated and desalted using C18 Zip Tips (Millipore Corporation, Bedford, USA) and directly applied on standard, robust, ground steel target plate. To that mixture, same volume of 4-hydroxy-α-cyano-cinnamic acid (HCCA) in 50% aqueous ACN containing 0.1% trifluoroacetic acid (TFA) was added to perform the MALDI-TOF–MS (MALDI-TOF, Bruker Daltonics, Billerica, USA) analysis of proteins at the Shrimpex Biotech Service Pvt Ltd, Chennai, India. The MS was operated at 19 kV accelerating voltage according to the manufacturer’s instructions, and the spectra of mass acquired between 600 and 4000 m/z range were captured in the positive ion reflection mode.
Annotation and identification of proteins
Proteins were identified using local MASCOT (http://www.matrixscience.com) search engine. The peptide tags were submitted as query sequence to SWISS-PROT database and non-redundant database of National Center for Biotechnology Information (NCBI nr). The search was performed using the following parameters: Trypsin for enzyme, Viridiplantae (green plants) for taxonomy, carbamidomethylation (C) for fixed modification, and methionine oxidation (M) for variable modification. The variation allowed was up to one missed cleavage of trypsin enzyme, 1 Da of molecular mass tolerance for peptides and monoisotopic ions. For positive identification of proteins, a threshold level of three peptide matches and 20% sequence query coverage were considered as requirements (Zhang et al. 2009).
RNA preparation and qRT-PCR
Total RNA was isolated from IMFB, NEC and EC using Spectrum Plant Total RNA kit (Sigma-Aldrich, St. Louis, USA). The concentration of total RNA was determined by measuring the absorbance at 260 and 280 nm using Colibri spectrometer (Berthold Detection Systems GmbH, Pforzhelm, Germany). The mRNA was isolated from total RNA using MRN70, mRNA miniprep kit (Sigma-Aldrich, St. Louis, USA) according to the manufacturer’s instructions. First-strand cDNA was synthesized from mRNA using high-capacity cDNA reverse transcription kit (Applied Biosystems, California, USA) with oligo-dT primers according to manufacturer’s protocol. Synthesized cDNA was diluted (1:10) with nuclease-free water and used as template for gene expression analysis. A total of five genes were selected based on their expression resulting from 2DE analysis of early somatic embryogenesis, and primers were designed using NCBI primer designing tool (Table 1). The qRT-PCR assays were performed using LightCycler 480 instrument (Roche Diagnostics, Germany). Reaction volume of 20 μL was set up in triplicates for each primer set with 5 μM of forward and reverse primers, 2X Syber Green PCR Master mix (Roche, Germany), and final reaction volume was made up with nuclease-free water. RPS2 gene was used as an internal reference gene (Chen et al. 2011). The qRT-PCR cycling programs were set as follows: 95 °C for 10 min (1 cycle); 94 °C for 10 s, 58–62 °C for 15 s, 72 °C for 15 s (50 cycles). Finally, expression level of each gene was relatively quantified based on calculated normalized ratio using advanced relative quantitation of Roche LightCycler software version 1.5.
Results
Induction of somatic embryogenesis
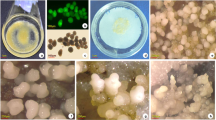
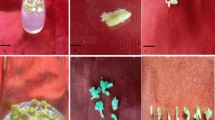
The immature male flower bud whorls ranging from 2nd to 18th position of cv. Grand Naine were cultured on MA1 callus induction medium augmented with 3-IAA, NAA and 2, 4-d as plant growth regulators. IMFB between 8th and 16th positions were more responsive for callus production. One or few somatic embryos appeared on the surface of callus after 3–6 months of incubation under dark. The morphological changes and the cell structure of calli were observed under stereomicroscope and compound microscope, respectively. The EC were found to be white, translucent, thin and friable in nature with many somatic embryos on their surface (Fig. 1c). The regions beneath the somatic embryos, PEM having more number of friable embryogenic cells was considered as an appropriate material for initiation of embryogenic cell suspension (ECS). As reported by Pan et al. (2011), two different types of cells were observed in EC: The first type of cells contained large nucleus with nucleolus (Fig. 2b), while the second type of cells contained more number of starch granules (Fig. 2c). These embryogenic cells were stained with DAPI and Lugol’s stain to confirm the presence of nucleus and starch granules. The DAPI-stained cells of EC contain prominent nucleus in blue color at the centre of the cell (Fig. 2d) and Lugol’s iodine-stained cells of EC show packed starch granules in deep purple color (Fig. 2e). The NEC were found to be tough, compact and often remains watery (Akbar 2011) and lacked embryo like structure on their surface (Fig. 2d). The cells of NEC were amorphous, highly vacuolated and lacked prominent nucleus (Fig. 2e). Though some NEC were morphologically similar to meristematic globules, their cytological features do not signify embryogenic potential (Riberio et al. 2012). Different types of NEC obtained on callus induction medium are given in Supplementary file I.
Induction of somatic embryogenesis in Musa spp. Immature floral hands (2nd to 18th positions) from male flower buds were used as explants and initiated on callus induction medium containing 1 mg/mL IAA, 1 mg/mL NAA and 4 mg/mL 2, 4-d. The panels represents a male flower bud, b initiated immature male flower buds on MA1 medium, c embryogenic calli, d non-embryogenic calli. SE somatic embryo and PEM pro-embryogenic mass
Cell structure of EC (a–e) and NEC (f). a Two types of cells found in EC, b type 1 cells of EC have large nucleus with nucleolus, c type 2 cells of EC have numerous starch granules, d DAPI-stained cells of EC show nucleus in blue color, e Lugol’s iodine-stained cells of EC show packed starch granules in deep purple color, f cells of NEC which are amorphous and highly vacuolated. T1 type 1 cell, T2 type 2 cell, S starch grains, N nucleus, Nu nucleolus, V vacuole. Bar in a represents 80 µm, bars in b–f represents 100 µm (color figure online)
After 6 months of explant initiation, the callogenesis was evaluated based on INIBAP technical guidelines (Strosse et al. 2003), and it was observed that induction frequency of EC (13.3%) was much lower than NEC (86.7%). The heterogeneous, yellow, compact NEC was used for the present study as it records more than 50% of total NEC.
Protein extraction and 2DE analysis
Extraction of quality proteins from banana samples is a major hindrance due to the presence of high phenol content (Carpentier 2007). Thus, various protocols namely (1) trichloroacetic acid–acetone method (Carpentier et al. 2005), (2) phenol/SDS–ammonium acetate method (Rodrigues et al. 2012) and (3) phenol–ammonium acetate method (Hurkman and Tanaka 1986) were tried. Based on high quality and quantity of total protein and more number of resolved protein bands in SDS-PAGE, phenol–ammonium acetate protocol was found to be the best method. Hence, this protocol was followed to extract total soluble proteins from the explants. More than 1500 individual protein spots across three replicate gels of EC, NEC and IMFB could be distinguished, and proteomic profiling of IFMB was observed to be completely different from that of EC and NEC profiling. Many highly abundant proteins of EC and NEC were spotted in the range of 28–32 and 45–55 kDa, respectively, compared with IMFB (Fig. 3). Gel replicates of each sample are given in the supplementary material II.
Differentially expressed protein spots were detected by comparing the proteomic profiling of IMFB, EC and NEC in all possible combinations using Hoefer 2D view software. Statistical analysis of comparison of proteins resulted in the detection of 114 differentially expressed protein spots with significant change in abundance (more than 1.5-fold). The clear spots which could be excised from CBB-stained gel alone were selected for further downstream analysis. A total of 65 excised protein spots were trypsin-digested and then subjected to MALDI-TOF–MS analysis. All proteins were successfully identified through MASCOT search engine software. Functional annotation of the 65 proteins through Blast2GO analysis indicated that most of the identified proteins were involved in defense response (10%) followed by stress response, oxidation–reduction process and protein biosynthesis (each 9%). About 12% of the proteins did not hit with any of the known proteins and they were considered as unknown proteins (Fig. 4b). Among the identified proteins, 29 were uniquely expressed, 13 were overexpressed and 23 were downregulated in EC than NEC and IMFB.
The theoretical molecular weight (MW) of maximum number of spots exactly matched with their experimental MW except for few spots (3, 9, 19, 33, 42 and 56). The lower experimental MW of these spots might be due to possible protein degradation (Sharifi et al. 2012). On the other hand, low correlation was found between the theoretical and experimental isoelectric point (pI) values for few protein spots (1, 4,7,39 and 43) which could be due to the variability in pH gradient across rehydrated gel strips and difference in protein migration during IEF program. Interestingly, the annotation results of differentially expressed proteins showed that spots 18 and 26 were identified as glyceraldehyde-3-phosphate dehydrogenase and, similarly, spots 46 and 47 as cell division proteins. The reason for more than one spot corresponding to the same protein could be attributed to the different isoforms of the same protein and/or their post-translational modifications (Sharifi et al. 2012).
Along with actin (spot 59) and elongation factor (spot 58), three proteins involved in oxidation–reduction process include arogenate dehydrogenase (spot 62), ascorbate peroxidase (spot 64) and FAD-dependent oxidoreductase-like protein (spot 65) which were overexpressed in the IMFB than EC and NEC. During the transitions from somatic to callogenesis state, these proteins produced varied expression which could be attributed to stress-related gene reprogramming. The identified proteins along with their corresponding spot ID number, protein identity, accession number, database, percent of query sequence coverage, theoretical and experimental pI and MW are given in supplementary material III.
Hierarchical clustering of identified proteins based on the expression
The differentially expressed proteins identified were clustered based on their expression patterns displayed across three samples. The expression values of proteins are based on the spot intensity and are expressed using a linear scale in which the larger number represents higher expression levels. The minimum and maximum values of the color scale were set from 6 (minimum positive expression: red) to 8 (maximum positive expression: green) as most of the values fall in this range. Around 65% of differentially expressed protein spots were highly accumulated in EC which are indicated in green color of the map (Fig. 5).
Validation of protein expression through qRT-PCR analysis
Five proteins (MYB transcription factor, glyceraldehydes 3-phosphate dehydrogenase, Nudix hydrolase, indole-3-pyruvate monooxygenase and cationic peroxidase) which are highly upregulated in EC were selected for qRT-PCR analysis. There was a high correlation between the expression of transcripts and their respective proteins levels of all selected genes in EC, whereas no correlation was observed for MYB transcription factor in NEC and indole-3-pyruvate monooxygenase in IMFB (Fig. 6). Similar kind of non-correlation results was also observed between EC and NEC of Vitis vinifera (Zhang et al. 2009) which could be due to the variation in stability of mRNA and protein levels (Sharifi et al. 2012).
Quantitative RT-PCR validation of 2DE protein results. Relative quantification was carried out to measure fold changes in selected gene expression among IMFB, NEC and EC relative to internal reference gene. RPS2 was used as a reference gene. Data (technical replicates of three biological experiments) are reported as mean ± standard error
Discussion
To understand the molecular mechanisms underlying the SE of banana, the differentially expressed proteins between EC and NEC were classified based on their function. It was understood that most of the identified proteins correspond to the enzymes that are involved in various metabolic, defense and stress-related processes, signal transduction, protein synthesis and degradation and hormone responsive. The differentially accumulated proteins based on their functional classification are discussed below.
Carbohydrate and energy metabolism
In our present study, two forms of glyceraldehyde 3-phosphate dehydrogenase (GAPDH, spot 18 and 26) were highly induced only in EC. Large amounts of starch granules present in embryonic cells are required for the cell to attain the embryogenic state (Pan et al. 2011; Xu et al. 2011; Ma et al. 2012; Torres et al. 2015), and one of the GAPDH isoforms has been proven to be necessary for starch metabolism (Backhausen et al. 1998). Thus, the abundant expression of GAPDH in EC suggested that this enzyme may play a crucial role in providing energy for rapid cell differentiation and proliferation of PEM of EC during early SE. Apart from its involvement in glycolysis pathway, GAPDH may also play a vital role in non-metabolic processes like apoptosis and regulation of gene expression (Zaffagnini et al. 2013). Similar to zygotic embryogenesis, somatic embryos develop with the orientation of apical and basal asymmetry resulting in the formation of proper embryo and suspensor, respectively. During development, the suspensor region is completely degraded through apoptosis (Smertenko and Bozhkov 2014), and high accumulation of GAPDH in EC can be related to this programmed cell death of suspensor region of the developed somatic embryos over the PEM of cells.
Another carbohydrate synthesis enzyme glycosyltransferase (GT, spot 32), involved in the synthesis of polysaccharides, was also highly abundant in EC alone. The main process of GT is the synthesis of polysaccharides by forming glycosidic bonds and it also plays a major part in cell wall formation (Scheible and Pauly 2004). Thus, overexpression of GT in EC implied that this enzyme might be involved in the synthesis of polysaccharides aiding the formation of cell wall of the embryogenic cells.
DNA and RNA metabolism associated proteins
Maturase K (MATK) which functions as a post-transcriptional splicing factor of RNA and involved in plant developmental process (Owiti et al. 2011) was overexpressed in EC. The induction of MATK gene is reported to help in overcoming the developmental stress of EC as it acts as a signal for the production of precursor metabolites and energy (Garavaglia et al. 2010). S-Adenosyl-l-methionine-dependent methyltransferase-like protein (SAM MTases, spot 28), which is involved in methylation of protein and nucleic acid was highly expressed in EC. The rate of DNA methylation is high during early embryogenesis which might be due to the exogenous auxins in callus culture medium (Zavattieri 2010). Thus, our study reconfirms that the important role of SAM MTases in EC is enhancement of proliferation of pro-embryogenic cells. Nucleoside diphosphates linked to moiety X (Nudix) hydrolases (spot 5) were found to be accumulated in EC alone. Similar trend was also observed in EC of Vitis vinifera (Zhang et al. 2009). This enzyme is involved in the protection of EC from oxidative stress and it plays a major role in the maintenance of ATP and NAD+ levels by reprocessing the nucleotide from ADP-ribose molecule (Ito 2012).
Defense and stress response protein
Dudits et al. (1991) documented that the cellular stress is playing an important role in the somatic embryo development from differentiated cells. The overexpression of stress- and defense-related proteins in EC was also evidenced in many crops such as Vitis vinifera (Marsoni et al. 2008), Citrus sinensis (Pan et al. 2009) and Crocus sativus (Sharifi et al. 2012). Our results are in conformity with earlier studies exhibiting more number of defense- and stress-responsive proteins which were differentially expressed in EC. Two resistance gene analog proteins (NBS-LRR, spot 9 and RGA3, spot 31) were found upregulated in EC.
Interestingly, in our present study, one small heat shock protein (HSP 25.3, spot 13) and two large heat shock proteins (HSP 70, spot 12 and HSP 90, spot 61) were found to be differentially expressed. HSP 25.3 and HSP 70 were found to be uniquely expressed in EC. The small HSPs are the key components of heat shock response and help in adaptation to different stresses, through refolding of proteins into native state and protect the unfolded state of newly synthesized proteins by avoiding them from aggregating and misfolding (Zhang et al. 2009; Correia et al. 2012). Similarly, high expression of HSPs was also observed in EC of Vitis vinifera (Zhang et al. 2009) and Crocus sativus (Sharifi et al. 2012). Thus, the induction of HSPs during early embryonic stage indicated their importance in SE to overcome the stress/es associated with cell dedifferentiation, i.e., transition from vegetative state to embryonic state.
Many reports evidenced that oxidative stress plays a vital role in embryogenesis through increased levels of reactive oxygen species (ROS) (Feher et al. 2003). The overproduction of ROS results in severe cellular damage (Mittler 2002) but can be well regulated by ROS scavenging mechanism mediated by peroxidase (POX) enzyme. In carrot, cationic peroxidase is known to play a major role in SE process by inhibiting the glycosylation inhibitors (Cordewener et al. 1991). In our study, cationic peroxidase was overaccumulated in EC than NEC. Thus, cationic POX might play an important role as a free radical scavenger and ROS-regulated stress signaling pathway during early embryogenic induction.
Protein synthesis and processing
In this experiment, we identified nine differentially expressed proteins (spot 1, 14, 21, 22, 33, 39, 44, 45 and 54) which are involved in protein synthesis and processing. The possible overaccumulation of ribosomal protein (spot 1 and 39) in EC during early embryonic stages is reported to be due to faster cell division process as this protein is mainly involved in G1 phase of cell cycle (Wisniewska et al. 2012).
Two spots (spot 14 and 22) of 26S protease regulatory subunit which are involved in protein turnover process were identified to be differentially expressed between EC and NEC. Similar results were also observed in developmental stages of SE in Cyclamen persicum (Rode et al. 2011). Papaevgeniou and Chondrogianni (2014) reported the occurrence of proteolysis during regulation of cellular processes like cell cycle and apoptosis to prevent the accumulation of misfolded proteins and the catabolised proteins, consequently delivering amino acids for the synthesis of new proteins (Vierstra 1996). Apart from proteolysis, 26S proteasome is also found to be involved in auxin signaling during developmental process of embryogenesis in Arabidopsis (Brukhin et al. 2005). The high accumulation of eukaryotic translation factor 3 (eIF 3F, spot 21) in EC of banana showed its significant role in cell proliferation and cell differentiation through its involvement in translation of newly synthesized proteins. Sharifi et al. (2012) and Wisniewska et al. (2012) also explained the importance of eIF in Crocus sativus and Cucumis sativus L. during the callus and SE development, respectively.
Transcription, transport and signal transduction proteins
In our present study, around 11% of proteins which are involved in transcription and various signal cascade pathways, were differentially expressed between NEC and EC. Of these, three proteins (spot 23–calcium binding protein, spot 53–Ran binding protein and spot 56–ABC transporter G protein) were highly expressed in NEC, and the rest of the proteins (spot 2, 6, 7, 10, 11, 17, 30 and 38) were highly expressed in EC. Interestingly, high expression of MYB transcription factor (MYB TF) was also reported in cotton (Yang et al. 2012). The important role of MYB TF was also evidenced in the regulation of embryogenesis pathway (Vinodhana and Ganesan 2013) and during the formation of Arabidopsis somatic embryos (Wang et al. 2009). Thus, the unique expression of MYB TF in banana EC also reiterated its possible key role in SE.
Protein phosphates 2C (PP2C, spot 2), a major plant serine/threonine protein phosphates belonging to metal ion-dependent phosphoprotein phosphates (PPM) family were expressed only in EC. Stress-responsive signaling as well as auxin and brassinosteroid signaling are well regulated by PP2C (Singh and Pandey 2012; Uhrig et al. 2013) Thus, the overexpression of PP2C in EC appears to be correlated with the overexpression of hormone-responsive genes. It was also observed that serine/threonine protein kinase (spot 17) was expressed only in EC. Similar expression of this gene was observed during the process of SE in cucumber (Wisniewska et al. 2012). This enzyme is reported to play a major role in signal transduction pathways needed for embryonic development, cell proliferation, cell differentiation and apoptosis (Hu et al. 2010). Similarly, increased accumulation of FBD-associated F-box protein (spot 30) in EC than NEC indicated its possible role in proliferation of embryogenic cells as it is involved in many cellular functions like signal transduction and regulation of the cell cycle transition (Kuroda et al. 2002).
Our results are contradicting with that of sandalwood (Anil and Rao 2000) and sugarcane (Suprasanna et al. 2004) where calcium binding proteins were overexpressed during the induction of SE. But the overexpression of calcium-binding proteins (KIC, spot 24) only in NEC suggested the occurrence of calcium-mediated signaling during the formation of NEC. It was also observed that another transport-related protein—Ran binding protein (spot 53), a small nuclear GTP-binding protein, was highly induced in NEC than EC. Similarly, ATP binding cassette (ABC) transporter G family protein (spot 56) was upregulated in NEC, while in potato, its expression was reported during the transition of PEM to somatic embryos during the late embryogenesis (Sharma et al. 2008). Thus, the over accumulation of KIC, Ran binding protein and ABC transporter protein in NEC leads to speculate that the pathway involved in SE is species-specific. Thus, in-depth study on the role of these enzymes in banana during embryogenesis is expected to throw more light.
Secondary metabolism
A total of nine differentially expressed proteins (spot 23, 25, 35, 40, 18, 3, 4, 15 and 27) involved in the production of various secondary metabolites were obtained in both EC and NEC.
Acyl-acyl-carrier-protein (ACP, spot 35) desaturase which is involved in fatty acid biosynthesis was highly abundant in NEC leading to high fat accumulation. Similar results were observed in NEC of Saccharum sp. (Nieve et al. 2003). Granule-bound starch synthase (GBSS, spot 40), a class of waxy protein, secreting the amylose component of starch was highly expressed in NEC. Thus, it is speculated that the overexpression of ACP and GBSS might be the reasons for the tough, compact and waxy nature of the NEC. Overexpression of Caffeoyl-CoA O-methyltransferase (CCOMT, spot 48), which catalyses the lignin biosynthesis pathway by transferring methylated lignin precursors, has also been reported in NEC of Crocus sativus (Sharifi et al. 2012). Similar trend was observed in our study, which reiterated that accumulation of CCOMT might increase the production of cell wall lignin, eventually leading to the arrest of SE formation.
Pectinesterase, a major cell-wall-associated enzyme involved in mechanical stability of cell wall, was highly abundant in EC than NEC of Cyclamen persicum (Hoenemann et al. 2010). Similarly, Verdeil et al. (2001) suggested that higher content of pectin in the embryogenic cells of Cocos nucifera is one of the reasons for the formation of friable callus. Also, pectin plays a major role in maintaining the structural integrity of embryogenic cell (Xu et al. 2011). Thus, the overexpression of pectinesterase inhibitor (PEI) in NEC of banana suggested that it might inhibit the pectin accumulation in the cell wall of NEC offering hindrance for friable cells formation. Wu et al. (2002) also reported that overexpression of PEI is the main reason for hard masses and lumps in banana fruit tissues. Mitochondrial dicarboxylate carrier protein (DIC) acts as a key player in many metabolic processes like synthesis of primary amino acid, biosynthesis of isoprenoid and gluconeogenesis and fatty acid metabolism (Palmieri et al. 2008). High accumulation of mitochondrial dicarboxylate carrier proteins (spot 4) in EC indicated their possible role during the embryogenic induction in banana as the plant growth regulators like abscisic acid, cytokinin and gibberellic acid are the derivatives of terpenoids.
The enzyme hydroxyphenylpyruvate reductase (HPPR, spot 27) mediates the metabolic biosynthesis of rosmarinic acid (RA), an antioxidant and antibacterial compound (Barberini et al. 2013). Many researchers evidenced that RA secondary metabolite is being induced in callus and suspension cultures in several species like Saccharum officinalis, Anchusa officinalis and Coleus blumei (Kintzios et al. 1999). Thus, overexpression of HPPR in banana EC reiterated its important role in early developmental stage of somatic embryogenesis. The differentially expressed proteins which are engaged in various secondary metabolisms imply a definite role during SE. But their specific role in SE needs detailed investigation.
Cell-division-associated proteins
Early embryogenesis is attained with a morphogenesis process during which the somatic embryos appear on the surface of EC. In the present study, high accumulation of two protein spots, which are homologous to cell division protein FtsY (46 and 47) in banana EC indicated their prominent role in splitting the cellular components and uniformly dividing across the newly formed cells. During SE in citrus, cell division proteins were highly induced which are known to be linked with cell division and elongation (Pan et al. 2009).
Endogenous hormone secretion proteins
In our present study, auxin and cytokinin biosynthesis proteins were identified to be uniquely expressed in EC. Indole-3-pyruvate monooxygenase YUCCA3 (YUC3, spot 20), an auxin biosynthesis enzyme, was highly accumulated in EC which leads to increase the level of free 3-IAA (Zhao et al. 2001). This finding is supported by Liu et al. (2013) who evidenced that biosynthesis of auxin through YUCs is involved in embryogenesis and induction of somatic embryos. Adenylate isopentenyltransferase (AIPT, spot 16) is a key enzyme involved in the de novo biosynthesis of cytokinins which was abundantly expressed in EC. The adenine types of cytokinins are the most abundant ones found in plants. In Arabidopsis, cytokinin biosynthesis mediated by AIPT is responsible for maintenance of apical dominance in shoot meristems (Yeh et al. 2011). Our results suggested that endogenous hormones like auxin (YUC 3) and cytokinin (AIPT) are highly induced during embryonic induction by acting along with exogenous hormones like 2,4-d, 3-IAA and NAA, during the initial process of SE.
Conclusion
This is the very first initiative that reveals the molecular mechanisms at protein level that occurs in banana during SE, the process of transition from vegetative state to embryogenic state. High-resolution 2DE along with mass spectrometry directs the identification of many differentially expressed proteins which are either directly/indirectly involved in the process of SE. The present study showed that various biochemical and molecular process are affected during early embryogenesis. The extensively varied protein profile of IMFB compared to the pattern of its resultant callus samples revealed that gene reprogramming occurred during the process of cell dedifferentiation. In total, 65 protein spots were differentially expressed, of which 29 candidate proteins were uniquely in EC and 13 were highly abundant in EC than NEC and IMFB. The differentially expressed protein accumulation was high (65%) in EC. This indicated that the induction of EC is associated with the better capability of controlling oxidative stress by regulating ROS scavenging system through cationic peroxidase. This further protects the structure of proteins along with HSPs, thereby triggering signal transduction. High accumulation of indole-3-pyruvate monooxygenase and adenylate isopentenyltransferase, the precursors of auxin and cytokinin growth regulators in EC revealed their important role in early embryogenic induction in banana. The overexpression of ACP, CCOMT and PEI in NEC might be one of the few possible reasons for compact and waxy nature which inhibits the embryogenesis potential. Further functional analysis of the differentially expressed unknown proteins identified in this study will give a clear representation for a better understanding of the developmental plasticity of SE in banana.
Author contribution statement
The authors have made the following declarations regarding their contributions: MK and SU conceived the design of the experiments. MK, KPS and SU sampled plant materials. MK, SB, MM analyzed the proteomics data. SU, SB, MSS and MMV monitored the research work. MK, SU, SB, MMV contributed in writing the manuscript. All authors read and approved the final manuscript. This is a part of MK’s PhD work.
Abbreviations
- ACN:
-
Acetonitrile
- CHAPS:
-
3-[(3-Cholamidopropyl) dimethylammonio] propanesulfonic acid
- DAPI:
-
4′,6-Diamidino-2-phenylindole
- DTT:
-
Dithiotreitol
- EC:
-
Embryogenic calli
- 3-IAA :
-
Indole-3-acetic acid
- IAA:
-
Iodoacetamide
- IMFB:
-
Immature male flower bud
- MALDI-TOF:
-
Matrix-assisted laser desorption ionization time-of-flight
- MS:
-
Mass spectrum
- NAA:
-
Naphthaleneacetic acid
- NEC:
-
Non-embryogenic calli
- PEM:
-
Pro-embryogenic mass
- ROS:
-
Reactive oxygen species
- SE:
-
Somatic embryogenesis
- 2DE :
-
Two-dimensional gel electrophoresis
References
Akbar A (2011) Somatic embryogenesis through cell suspension culture in Indian commercial cultivars of Banana (Musa spp.) Thesis PhD, Bharathidasan University, Tiruchirappalli
Anil VS, Rao KS (2000) Calcium mediated signaling during sandalwood somatic embryogenesis. Role for exogenous calcium as second messenger. Plant Physiol 123:1301–1311
Backhausen JE, Vetter S, Baalmann E, Kitzmann C, Scheibe R (1998) NAD-dependent malate dehydrogenase and glyceraldehydes 3-phosphate dehydrogenase isoenzymes play an important role in dark metabolism of various plastid types. Planta 205:359–366
Baldwin TC, Domingo C, Schindler T, Seetharaman G, Stacey N, Roberts K (2001) DcAGP1, a secreted arabinogalactan protein, is related to a family of basic proline-rich proteins. Plant Mol Biol 45:421–435
Barberini S, Savona M, Raffi D, Leonardi M, Pistelli L, Stochmal A, Vainstein A, Pistelli L, Ruffoni B (2013) Molecular cloning of SoHPPR encoding a hydroxyphenylpyruvate reductase and its expression in cell suspension cultures of Salvia officinalis. Plant Cell Tissue Organ Culture. doi:10.1007/s11240-013-0300-8
Blum H, Beier H, Gross HJ (1987) Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis 8:93–99
Boutilier K, Offringa R, Sharma VK, Kieft H, Ouellet T, Zhang LM, Hattori J, Liu CM, van Lammeren AAM, Miki BLA et al (2002) Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell 14:1737–1749
Brukhin V, Gheyselinck J, Gagliardini V, Genschik P, Grossniklaus U (2005) The RPN1 subunit of the 26S proteasome in Arabidopsis is essential for embryogenesis. Plant Cell 17:2723–2737
Canovas FM, Gaudot ED, Recorbet G, Jorrin J, Mock HP, Rossignol M (2004) Plant proteome analysis. Proteomics 4:285–298
Carpentier SC, Witters E, Laukens K, Deckers P, Swennen R, Panis B (2005) Preparation of protein extracts from recalcitrant plant tissues: an evaluation of different methods for two dimensional gel electrophoresis analysis. Proteomics 5:2497–2507
Carpentier SC, Witters E, Laukens K, Van Onckelen H, Swennen R, Panis B (2007) Banana (Musa spp.) as a model to study the meristem proteome: acclimation to osmotic stress. Proteomics 7:92–105
Chen L, Zhong HY, Kuang JF, Li JG, Lu WJ, Chen JY (2011) Validation of reference genes for RT-qPCR studies of gene expression in banana fruit under different experimental conditions. Planta 234:377–390
Cordewener J, Booij H, Zandt DH, Engelen VF, Kammen A, Vries DS (1991) Tunicamycininhibited carrot somatic embryogenesis can be restored by secreted cationic peroxidase isoenzymes. Planta 184:478–486
Correia S, Vinhas R, Manadas B, Lourenco AS, Verissimo P, Canhoto JM (2012) Comparative proteomic analysis of auxin-induced embryogenic and nonembryogenic tissues of the solanaceous tree Cyphomandra betacea (Tamarillo). J Proteome Res 11:1666–1675
Dudits D, Bogre L, Gyorgyey J (1991) Molecular and cellular approaches to the analysis of plant embryo development from somatic cells in vitro. J Cell Sci 99:475–484
Feher A, Pasternak TP, Dudits D (2003) Transition of somatic plant cells to an embryogenic state. Plant Cell Tissue Organ Culture 74:201–228
Ganapathi TR, Suprasanna P, Bapat VA, Kulkarni VM, Rao PS (1999) Somatic embryogenesis and plant regeneration from male flower buds in banana. Curr Sci 76:1228–1230
Garavaglia BS, Thomas L, Zimaro T, Gottig N, Daurelio LD, Ndimba B, Orellano EG, Ottado J, Gehring C (2010) A plant natriuretic peptide-like molecule of the pathogen Xanthomonas axonopodis pv. Citri causes rapid changes in the proteome of its citrus host. Plant Biol 10(51):1–10
Heck GR, Perry SE, Nichols KW, Fernandez DE (1995) AGL15, a MADS domain protein expressed in developing embryos. Plant Cell Online 7:1271
Hoenemann C, Richardt S, Kruger K, Zimmer AD, Hohe A, Rensing SA (2010) Large impact of the apoplast on somatic embryogenesis in Cyclamen persicum offers possibilities for improved developmental control invitro. Plant Biol 10(77):1–17
Hu X, Liu L, Xiao B, Li D, Xing X, Kong X, Li D (2010) Enhanced tolerance to low temperature in tobacco by over-expression of a new maize protein phosphatase 2C, ZmPP2C2. Plant Physiol 167:1307–1315
Hurkman WJ, Tanaka CK (1986) Solubilization of plant membrane proteins for analysis by two-dimensional gel electrophoresis. Plant Physiol 81:802–806
Hussein S, Ibrahim R, Kiong ALP (2006) Somatic embryogenesis: an alternate method for in vitro micropropagation. Iran J Biotechnol 4:156–161
Imin N, Jong FD, Mathesius U, Noorden GV, Saeed NA, Wang XD, Rose RJ, Rolfe BG (2004) Proteome reference maps of Medicago truncatula embryogenic cell cultures generated from single protoplasts. Protemics 4:1883–1896
Ito D (2012) Functional analysis of Arabidopsis Nudix hydrolases having CoA and guanosine-3,5,-tetraphosphate (ppGpp) specific pyrophosphohydrolase activities. Thesis PhD, Kinki University
Ji A, Geng X, Zhang Y, Yang H, Wu G (2011) Advances in somatic embryogenesis research of horticultural plants. Am J Plant Sci 2:727–732
Kintzios S, Nikolaou A, Skoula M (1999) Somatic embryogenesis and invitro rosmarinic acid accumulation in Salvia officinalis and S. fruticosa leaf callus cultures. Plant Cell Rep 18:462–466
Kuroda H, Takahashi N, Shimada H, Seki M, Shinozaki K, Matsui M (2002) Classification and expression analysis of Arabidopsis F-box-containing protein genes. Plant Cell Physiol 43:1073–1085
Laemmli UK (1970) Cleavage of structural protein during the assembly of the head of bacteriophage T4. Nature 227:680–685
Lippert D, Zhuang J, Ralp S, Ellis DE, Gilbert M, Olafson R, Ritland K, Ellis B, Douglas CJ, Bohlmann J (2005) Proteome analysis of early somatic embryogenesis in Picea glauca. Proteomics 5:461–473
Liu H, Xie WF, Zhang L, Valpuesta V, Ye ZW, Gao QH, Duan K (2013) Auxin biosynthesis by the YUCCA6 flavin monooxygenase gene in woodland strawberry. J Integr Plant Biol 56:350–363
Lotan T, Ohto M, Yee KM, West MAL, Lo R, Kwong RW, Yamagishi K, Fischer RL, Goldberg RB, Harada JJ (1998) Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93:1195–1205
Ma L, Xie L, Lin G, Jiang S, Chen H, Li H, Taka T, Samaj J, Xu C (2012) Histological changes and differences in activities of some antioxidant enzymes and hydrogen peroxide content during somatic embryogenesis of Musa AAA cv. Yueyoukang 1. Sci Hortic 144(2012):87–92
Marsoni M, Bracale M, Espen L, Prinsi B, Negri AS, Vannini C (2008) Proteomic analysis of somatic embryogenesis in Vitis vinifera. Plant Cell Rep 27:347–356
Mittler R (2002) Oxidative stress, antioxidants and stress tolerance. Trends Plant Science 7:405–410
Neuhoff V, Arnold N, Taube D, Ehrhardt W (1988) Improved staining of proteins in polyacrylamide gels including isoelectric focusing gels with clear background at nanogram sensitivity using Coomassie Brilliant Blue G-250 and R-250. Electrophoresis 9:255–262
Nievesi N, Nieto MS, Blanco MA, Sanchez M, Gonzalez A, Gonzalea JL, Castillo R (2003) Biochemical characterization of embryogenic and non embryogenic calluses of sugarcane. In Vitro Cell Dev Biol 39:343–345
Nolan KE, Irwanto RR, Rose RJ (2003) Auxin up-regulates MtSERK1 expression in both Medicago truncatula root-forming and embryogenic cultures. Plant Physiol 133:218–230
Overvoorde PJ, Grimes HD (1994) The role of calcium and calmodulin in carrot somatic embryogenesis. Plant Cell Physiol 35:135
Owiti J, Grossmann J, Gehrig P, Dessimoz C, Laloi C, Hansen MB, Gruissem W, Vanderschuren H (2011) iTRAQ-based analysis of changes in the cassava root proteome reveals pathways associated with post-harvest physiological deterioration. Plant J 67:145–156
Palmieri L, Picault N, Arrigoni R, Besin E, Palmieri F, Hodges M (2008) Molecular identification of three Arabidopsis thaliana mitochondrial dicarboxylate carrier isoforms: organ distribution, bacterial expression, reconstitution into liposomes and functional characterization. J Biochem 410:621–629
Pan Z, Guan R, Zhu S, Deng X (2009) Proteomic analysis of somatic embryogenesis in valencia sweet orange (Citrus sinensis Osbeck). Plant Cell Rep 28:281–289
Pan X, Yanga X, Linb G, Zoua R, Chena H, Samaj J, Xua C (2011) Ultrastructural changes and the distribution of arabinogalactan proteins during somatic embryogenesis of banana (Musa spp. AAA cv. ‘Yueyoukang 1′). Physiol Plant 142:372–389
Papaevgeniou N, Chondrogianni N (2014) The ubiquitin proteasome system in Caenorhabditis elegans and its regulation. Redox Biol 2:333–347
Patena LF, Refuerzo LRC, Barba RC (2002) Somatic embryogenesis and plantlet regeneration in mango (Mangifera indica L.). In vitro cell. Dev Biol 38:173–177
Ravi I, Uma S (2011) Phenotyping banana and plantains for adaptation to drought, in drought phenotyping in crops: from theory to practice. In: CGIAR Generation Challenge Programme, c/o CIMMYT, Mexico, pp 417–430
Ribeiro LO, Paiva LV, Padua MS, Santos BR, Alves E, Stein VC (2012) Morphological and ultrastructural analysis of various types of Banana callus, cv. Prata ana. Acta Sci 34:423–429
Rode C, Lindhorst K, Braun HP, Winkelmann T (2011) From callus to embryo: a proteomic view on the development and maturation of somatic embryos in Cyclamen persicum. Planta. doi:10.1007/s00425-011-1554-1
Rodrigues EP, Torres AR, Batista JSS, Huergo L, Hungria M (2012) A simple, economical and reproducible protein extraction protocol for proteomics studies of soybean roots. Genet Mol Biol 35:348–352
Sales EK, Butardo NG (2014) Molecular analysis of somaclonal variation in tissue culture derived bananas using MSAP and SSR markers. Int J Biol Biomol Agricult Food Biotechnol Eng 8(6):615–622
Scheible WR, Pauly M (2004) Glycosyltransferases and cell wall biosynthesis: novel players and insights. Curr Opin Plant Biol 7:285–295
Schoofs H (1997) The origin of embryogenic cells in Musa. Thesis PhD, KU Leuven, Leuven
Sharifi G, Ebrahimzadeh H, Ghareyazie B, Gharechahi J, Vatankhah E (2012) Identification of differentially accumulated proteins associated with embryogenic and non-embryogenic calli in saffron (Crocus sativus L.). Proteome Sci 10(3):1–15
Sharma SK, Millam S, Hedley PE, McNicol J, Bryan GJ (2008) Molecular regulation of somatic embryogenesis in potato: an auxin led perspective. Plant Mol Biol 68:185–201
Sidha M, Suprasanna P, Bapat VA, Kulkarni UG, Shinde BN (2006) Developing somatic embryogenic culture system and plant regeneration in banana. BARC Newslett 285:153–161
Singh A, Pandey GK (2012) Protein phosphatases: a genomic outlook to understand their function in plants. J Plant Biochem Biotechnol. doi:10.1007/s13562-012-0150-1
Singh HP, Uma S, Selvarajan R, Karihaloo JL (2011) Micropropagation for production of quality banana planting material in Asia–Pacific. Asia–Pacific Consortium on Agricultural Biotechnology (APCoAB), New Delhi
Smertenko A, Bozhkov PV (2014) Somatic embryogenesis: life and death processes during apical and basal patterning. J Exp Bot 65:1343–1360
Strosse H, Domergue R, Panis B, Escalant JV, Cote F (2003) Banana and plantain embryogenic cell suspension. In: INIBAP technical guidelines 8, Montpellier
Sun L, Wu Y, Zou H, Su S, Li S, Shan X, Xi J, Yuan Y (2013) Comparative proteomic analysis of the H99 inbred maize (Zea mays L.) line in embryogenic and non-embryogenic callus during somatic embryogenesis. Plant Cell Tissue Organ Culture 113:103–119
Suprasanna P, Desai NS, Nishanth G, Ghosh SB, Laxmi N, Bapat VA (2004) Differential gene expression in embryogenic, non-embryogenic and desiccation induced cultures of sugarcane. Sugar Technol 6:305–309
Tan EC, Karsani SA, Foo GT, Wong SM, Rahman NA, Khalid N, Othman S, Yusof R (2012) Proteomic analysis of cell suspension cultures of Boesenbergia rotunda induced by phenylalanine: identification of proteins involved in flavonoid and phenylpropanoid biosynthesis pathways. Plant Cell Tissue Organ Culture. doi:10.1007/s11240-012-0188-8
Tang H, Ren Z, Krczal G (2000) Somatic embryogenesis and organogenesis from immature embryo cotyledons of three sour cherry cultivars (Prunus cerasus L.). Sci Hortic 83:109–126
Torres LF, Diniz LEC, Livramento KGD, Freire LL, Paiva LV (2015) Gene expression and morphological characterization of cell suspensions of Coffea Arabica L. cv. Catigua MG2 in different cultivation stages. Acta Physiol Plant 37:175
Tsai HL, Lue WL, Lu KJ, Hsieh MH, Wang SM, Chen J (2009) Starch synthesis in Arabidopsis is achieved by spatial cotranscription of core starch metabolism genes. Plant Physiol 151:1582–1595
Uhrig RG, Labandera AM, Moorhead GB (2013) Arabidopsis PPP family of serine/threonine protein phosphatases: many targets but few engines. Trends Plant Sci 18:505–510
Uma S, Lakshmi S, Saraswathi MS, Akbar A, Mustaffa MM (2012) Plant regeneration through somatic embryogenesis from immature and mature zygotic embryos of Musa accuminata ssp. Burmannica. In vitro Cell Dev Biol Plant 48:539–545
Verdeil JL, Hocher V, Huet C, Grosdemange F, Escoute J, Ferriere N, Nicole M (2001) Ultrastructural changes in coconut calli associated with the acquisition of embryogenic competence. Annu Bot 88:9–18
Vierstra RD (1996) Proteolysis in plants: mechanisms and functions. Plant Mol Biol 32:275–302
Vinodhana NK, Ganesan NM (2013) Studies on embryogenic competence and regeneration potential with relation to anthocyanin biosynthesis in cotton (G. hirsutum). Plant Gene Trait 4:53–59
Wang X, Niu Q, Teng C, Li C, Mu J, Chua NH, Zuo J (2009) Overexpression of PGA37/MYB118 and MYB115 promotes vegetative-to-embryonic transition in Arabidopsis. Cell Res 19:224–235
Wang X, Shia L, Linb G, Pana X, Chena H, Wua X, Taka T, Samaj J, Xua C (2013) A systematic comparison of embryogenic and non-embryogenic cells of banana (Musa spp. AAA): ultrastructural, biochemical and cell wall component analyses. Sci Hortic 159:178–185
Wisniewska A, Grabowska A, Bogiel A, Tagashira N, Zuzga S, Woycicki R, Przybecki Z, Malepszy S, Filipecki M (2012) Identification of genes up-regulated during somatic embryogenesis of cucumber. Plant Physiol Biochem 50:54–64
Wu MC, Tseng KC, Huang TH, Chang HM (2002) Pectinesterase inhibitor in rubbery Banana (Musa sapientum L.). J Food Sci 67:1337–1340
Xu C, Zhao L, Pan X, Samaj J (2011) Developmental localization and methylesterification of pectin epitopes during somatic embryogenesis of banana (Musa spp. AAA). PLoS One 6(8):e22992. doi:10.1371/journal.pone.0022992
Yang X, Zhang X (2010) Regulation of somatic embryogenesis in higher plants. Crit Rev Plant Sci 29:36–57
Yang X, Zhang X, Yuan D, Jin F, Zhang Y, Xu J (2012) Transcript profiling reveals complex auxin signalling pathway and transcription regulation involved in dedifferentiation and redifferentiation during somatic embryogenesis in cotton. Plant Biol 12(110):1–19
Yeh CS, Hsieh LS, Yang CC, Lee PD (2011) Molecular characterization of isopentenyltransferase (BoAIPT1) from Bambusa oldhamii expressed in Escherichia coli. Bot Stud 52:249–256
Yuffa AM, Garcia EG, Nieto MS (1993) Comparative study of protein electrophoretic patterns during embryogenesis in Coffea arabica cv. Catimor. Plant Cell Rep 13:197–202
Zaffagnini M, Fermani S, Costa A, Lemaire SD, Trost P (2013) Plant cytoplasmic GAPDH: redox post-translational modification and moonlighting properties. Front Plant Sci 4:1–18
Zavattieri MA (2010) Induction of somatic embryogenesis as an example of stress-related plant reactions. Electron J Biotechnol 13:1–14
Zhang J, Ma H, Chen S, Ji M, Perl A, Kovacs L, Chen S (2009) Stress response proteins’ differential expression in embryogenic and non-embryogenic callus of Vitis vinifera L. cv. Cabernet Sauvignon—a proteomic approach. Plant Sci 177:103–113
Zhao Y, Christensen SK, Fankhauser C, Cashman JR, Cohen JD, Weigel D, Chory J (2001) A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291:306–309
Zuo J, Niu QW, Frugis G, Chua NH (2002) The WUSCHEL gene promotes vegetative to embryonic transition in Arabidopsis. Plant J 30:349–359
Acknowledgements
Authors acknowledge the Director, NRCB for her support to conduct this research program at National Research Centre for Banana, Trichy. We are thankful to Dr. A. Akbar, Mr. K. Arun, Mr. G. Kannan, Mrs. K. Udhayanjali, Mrs. S. Lakshmi, Mr. A. Chandrasekar, Mr. A. S. Saravanakumar, Mrs. G. Valarmathi, Mr. K. Raja and Mr. Sriram Vishwanathan of NRCB for their support and critical comments while carrying out the work, data analysis and preparation of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflict of interest.
Additional information
Communicated by R. Wells.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kumaravel, M., Uma, S., Backiyarani, S. et al. Differential proteome analysis during early somatic embryogenesis in Musa spp. AAA cv. Grand Naine. Plant Cell Rep 36, 163–178 (2017). https://doi.org/10.1007/s00299-016-2067-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-016-2067-y