Abstract
Two dimensional gel electrophoresis combined with matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) was employed to study the somatic embryogenesis (SE) in Valencia sweet orange (Citrus sinensis Osbeck). Twenty-four differentially expressed proteins were identified at five time points of citrus SE (0, 1, 2, 3, 4 weeks after embryo initiation) covering globular, heart/torpedo and cotyledon-shaped embryo stages. The general expression patterns for these proteins were consistent with those appeared at 4 weeks of citrus SE. The most striking feature of our study was that five proteins were predicted to be involved in glutathione (GSH) metabolism and anti-oxidative stress, and they exhibited different expression patterns during SE. Based on that oxidative stress has been validated to enhance SE, the preferential representation for anti-oxidative proteins suggests that they could have a developmental role in citrus SE. Some proteins involved in cell division, photosynthesis and detoxification were also identified, and their possible roles in citrus SE were discussed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Somatic embryogenesis (SE), based on plant totipotency, is the development switch of somatic cell to embryo (Zimmerman 1993). SE can serve as a model system to study the molecular, cytological, physiological and developmental events of plant embryogenesis with accessible experimental manipulation (Zimmerman 1993). Furthermore, for many woody plant species, embryogenic cultures are common materials for genetic transformation, and SE is a primary pathway of indirect plantlet regeneration for these biotechnologies. However, embryogenic capacity is limited to certain genotypes and often lost during long-term in vitro conservation, which hinders relevant applications. Understanding the mechanism of SE process will be helpful for successful utilization.
It is believed that analyses of gene expression during SE can provide information for better understanding of this process (Zimmerman 1993; Chugh and Khurana 2002; Fehér et al. 2003; Stasolla et al. 2004). Increasingly studies are focused on characterizing genes specifically expressed during this process (Chugh and Khurana 2002; Fehér et al. 2003). Numerous genes involved in or associated with SE such as SERK (Schmidt et al. 1997), LEAFY COTYLEDON (Meinke 1992), DcAGP1 (Baldwin et al. 2001), BABY BOOM (Boutilier et al. 2002) and WUSCHEL (Zuo et al. 2002) were identified. Moreover, transcriptomic studies were applied to investigate global gene expression in SE (Thibaud-Nissen et al. 2003; Stasolla et al. 2004). Recently, proteomics were employed to study SE in several plant species including Medicago truncatula (Imin et al. 2004, 2005), Picea glauca (Lippert et al. 2005), Cyclamen persicum (Winkelmann et al. 2006) and Vitis vinifera (Milena et al. 2008). Since proteins directly influence cellular biochemistry and provide a more accurate analysis of cellular changes during growth and development (Chen and Harmon 2006), identification of proteins associated with somatic embryo development may provide mechanistic insight onto SE.
Citrus is one of the most important fruits in the world. However, citrus cultivar improvement through conventional genetic breeding is hindered by several factors inherent to the citrus species such as a long juvenile period, polyembryony and male/female sterility. SE provided an avenue for citrus genetic improvement due to its vital role in biotechnological tools such as tissue culture, protoplast fusion and genetic transformation which may, to some degree, overcome these obstacles associated with sexual reproduction. Research on citrus SE over the past decades mainly focused on physiological description and improvements in culturing technologies (Francesco et al. 1998; Stefania et al. 2002; Mukaddes and Kemal 2006). Very little is known about the molecular process involved in citrus SE.
This study aims to identify proteins differentially expressed during citrus SE using a proteomic approach. A number of proteins have been shown to be preferentially expressed during certain stages of citrus SE, validating this method to probe the molecular process of citrus embryo development and providing a basis for future genetic/biochemical studies on the roles of some of the candidate genes/proteins in citrus SE.
Materials and methods
Plant materials, culture treatment and sample collection
Embryogenic callus of Valencia sweet orange (C. sinensis) was maintained on MT basal medium (Murashige and Tucker 1969), and subcultured at one-month intervals. For somatic embryo initiation, embryogenic callus was cultured on glycerol-added medium where sucrose was replaced by 2% (w/v) glycerol (Liu and Deng 2002). Samples were collected at 1, 2, 3, and 4 weeks after somatic embryo initiation. The cultures were sequentially subcultured on glycerol-containing medium, and somatic embryos at globular, heart/torpedo and cotyledon-shaped stages were selected under microscope when they were visible.
Protein extraction
Protein was extracted according to a modified method described previously (Dai et al. 2007). Sample (1 g) was ground to powder with liquid nitrogen. The powder was suspended in 10 ml extraction buffer containing 50 mM Tris-HCl pH 7.5, 20 mM KCl, 1% CHAPS, 2% (v/v) beta-mercaptoethanol, 100 μl protease inhibitor cocktail (BS384, BBI), then incubated for 20 min on ice. Supernatant was collected after centrifugation at 10,000×g for 15 min at 4°C, followed by adding three volumes of 12.5% trichloroacetic acid to precipitate proteins on ice for 2 h. Proteins were pelleted by centrifugation at 10,000×g for 15 min at 4°C, and then resuspended in cold acetone containing 0.07% β-mercaptoethanol. The mixture was kept at −20°C for 1 h before centrifugation at 10,000×g for 15 min at 4°C. Protein pellet was washed twice with cold acetone, lyophilized by vacuum, and stored at −80°C until use. Before 2-DE, protein pellets were dissolved in a lysis buffer (8 M urea, 2 M thiourea, 4% CHAPS, 13 mM DTT, 2% pharmalyte 3–10) and used immediately after debris was removed by centrifugation as above. The protein concentration was determined with the Bio-Rad protein assay kit based on the Bradford method using BSA as standard. Three independent protein extractions were performed.
2-DE
Samples were applied to 17 cm liner IPG strips (Bio-Rad, pH 4–7). After 14-h rehydration, isoelectric focusing was performed on PROTEAN IEF Cell (Bio-Rad) as following: 30 min at 250 V, 1 h at 1,000 V, 5 h to increase the voltage from 1,000 to 8,000 V, 7.5 h at 8,000 V. Gels were then subjected to 2 × 20 min equilibration, using buffer I and buffer II, which basically contains 6 M Urea, 50 mM Tris-HCl (pH 8.8), 30% glycerol, 2% (w/v) SDS, with addition of 2% (w/v) DTT in Buffer I and 2.5% (w/v) iodoacetamide in Buffer II, respectively. SDS-PAGE was performed with 12% acrylamide gels in the PROTEAN MINI 2 (Bio-Rad) for 5 hrs at 180 V. pI and molecular mass of the proteins were determined, respectively, by the liner pH arrangement of IPG strips and SDS-PAGE marker.
Staining and image analysis
Proteins were visualized with Coomassie Brilliant Blue (CBB) G250 according to Giovanni et al. (2004). Briefly, gels were fixed in a solution of 40% ethanol and 10% acetic acid for 30 min, washed with MilliQ water for 4 × 15 min, stained overnight in a CBB G250 solution (0.12% G250, 10% phosphate acid, 10% (NH4)2SO4, 20% methanol), and then destained with MilliQ water until the background was clean. Gels were imaged by UVP (Bio-Rad) under “Trans White” mode. Protein spots were detected by PDQuest software (Bio-Rad) and normalized to total spot quantity in valid spots under PPM (×1,000,000) scaling step. Spot quantity is the total intensity of a defined spot in a gel image. The intensity is the sum of intensities of the image pixels inside boundary. It is calculated during spot detection and Gaussian fitting, which is calculated by PDQuest software. After manual processing, the well-separated spots in all triplicate gels were submitted to ANOVA analysis (P < 0.05).
Tryptic digestion and protein identification
Protein spots were excised from gels, destained for 20 min in 30 mM potassium ferricyanide/100 mM sodium thiosulfate (1:1 v/v), and washed in Milli-Q water until the gels were destained. The spots were kept in 0.2 M NH4HCO3 for 20 min and then lyophilizated. Each spot was digested overnight in 2 μl 12.5 ng/μl trypsin in 0.1 M NH4HCO3. The peptides were extracted with 50% ACN, 0.1% TFA for three times. All mass spectra were acquired on an AutoFlex MALDI-TOF/TOF mass spectrometer with LIFT technology (Bruker Daltonics, Bremen, Germany). Tryptic digests were prepared on AnchorChip sample plate (Bruker Daltonics, Bremen, Germany) according to the manufacturer’s instructions. Both MS and MS/MS data were acquired with a N2 laser at 25-Hz sampling rate. PMF data and MS-MS data were combined using Flex Analysis and the data set was submitted to MASCOT program for protein identification. EST_viridiplantae viridiplantae_20080108 (86,321,490 sequences; 15,273,276,944 residues) and National Center for Biotechnology non-redundant (NCBInr 20080221) protein database (6,122,577 sequences; 2,096,230,148 residues) were searched against. The search was performed taking green plants as taxonomy, which contained 74,605,494 sequences (EST) and 473,596 sequences (NCBInr). The other parameters for searching were the enzyme of trypsin; one missed cleavage; fixed modifications of carbamidomethyl (C); variable modifications of oxidation (Met). Peptide tolerance of 100 ppm; fragment mass tolerance of ±0.5 Da; peptide charge of 1+ were selected. Only significant hits, as defined by the MASCOT probability analysis (P < 0.05), were accepted.
Results
Identification of differentially expressed proteins during citrus SE
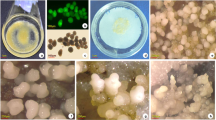
Somatic embryos were induced from embryogenic callus (Fig. 1a) cultured on MT medium containing glycerol. Massive globular embryos were produced after 3 weeks (Fig. 1b); globular, heart-shaped, torpedo and cotyledon embryos were emerged as a mixture in 4 weeks (Fig. 1c). Total proteins were extracted from embryogenic cultures at 0, 1, 2, 3 and 4 weeks and analyzed by 2-DE (Fig. 2a–e). The average number of spots varied with developmental stages, ranging from 746 (0 week) to 825 (3 weeks) spots. To construct a master gel, we chose one image of 0-week stage gel as a reference and then added the spots detected on the reference gels of the other stages. Finally, 928 spots were pooled to create this synthetic master gel (Fig. 2f). Relative quantity data of each spot in all triplicate gels was exported to excel and was submitted to a one-way ANOVA analysis. A total of 62 protein spots were found to be significantly changed (P < 0.05). All these spots were excised from CBB G250 stained gels, and were submitted to MALDI-TOF or MALDI-TOF-TOF after in-gel digestion. Because only limited citrus-specific protein sequences are available in the databases, we searched against EST databases combined with the non-redundant protein database at the NCBI. In total, 24 proteins were successfully identified (Table 1). These protein spots were marked in the master gel (Fig. 2f). Out of these 24 proteins, 11 were up-regulated, 9 down-regulated and 4 transiently up/down-regulated during the SE. Among these proteins, spot 7 and 20 were identified to be two forms of a same ferritin-3. These two proteins may undergo posttranslational modification because of their different pI and similar molecular mass. Three proteins (spot 16, 26 and 29) were identified as members of glutathione S-transferase families. As for spot 15 and 28, each was identified as a mixture of two different proteins (Table 1). In summary, 24 proteins were identified from the 22 protein spots.
Embryogenic callus of Valencia sweet orange at different time points (upper) and somatic embryos at different stages (lower). The panels represent a 0 WAI, b 3 WAI and c 4 WAI of embryo; d globular embryo, e heart-shaped embryo, f torpedo-shaped embryo, g cotyledon-shaped embryo. The WAI represents weeks after initiation. Bar = 1 mm
The development of somatic embryos from Valencia sweet orange as analyzed by 2-DE. The images represent the expressed protein profiles obtained from samples collected at a 0 WAI, b 1 WAI, c 2 WAI, d 3 WAI and e 4 WAI. The WAI represents weeks after initiation. The master gel (f) was constructed by adding the spots detected on the reference gels of other stages to one image of 0 WAI stage gel
Based on predicted biochemical functions, most of the 24 proteins are likely to be involved in oxidative stress responses, cell division, photosynthesis and detoxification. Specifically, five proteins may be associated with oxidative stress responses: one phospholipid hydroperoxide glutathione peroxidase (PHGPx, spot 18), one cysteine synthase (spot 23) and three isoforms of glutathione S-transferase GST, spot 16, 29, 26; spot 26 being a putative dehydroascorbate reductase also belonging to the GST family in plant (Dixon et al. 2002). Interestingly, as shown in Table 1, the expression patterns during SE varied for these anti-oxidative proteins. The abundance of the PHGPx (spot 18), the cysteine synthase (spot 23) and one of the GSTs (spot 16) gradually decreased, but that of the dehydroascorbate reductase (spot 26) increased to a higher level in the last two weeks of SE induction. For the GST identified in spot 29, it showed a transient down-regulation in the first 3 weeks but restored to the original level in the 4th week. Three proteins, i.e. two forms of tubulin (spot 15) and a putative DNA-damage-repair/toleration protein DRT102 (spot 17), are predicted to be involved in cell division and they exhibited gradual up-regulation during citrus SE. In addition, two forms of ferritin-3 (spot 7 and 20) (known to be involved in photosynthesis) were shown to be up-regulated in the late phase of citrus SE. A spermidine synthase (spot 11) showed transiently up-regulation in 2 weeks after SE induction, and a putative rhodanase (spot 9) was strongly up-regulated in the 4th week of SE.
Protein expression in globular, heart/torpedo and cotyledon-shaped embryo stages
Because the induction of SE from individual cell is not a synchronized process, a mixture of heterogeneously developed somatic embryos was often found in the starting citrus callus material four weeks after induction of SE. To further gain, the accurate protein expression associated with specific SE stages, we hand-picked out the embryos in globular, heart, torpedo and cotyledon-shaped stages under a microscope (Fig. 1d–g). Due to the difficulty in distinguishing heart-shaped and torpedo-shaped embryos, embryos at these two stages were pooled together, and designated as heart/torpedo-shaped embryos. Based on the position of spots in the 2-DE gel, expression levels of the above identified proteins extracted from the embryos of these three stages were compared with those extracted from the embryogenic callus (0 week). Due to the use of a different electrophoresis system, four protein spots (spot 2, 9, 18 and 19) ran out of the gels because of their small molecular mass. Thus, 18 protein spots corresponding to 20 proteins were investigated. The results summarized in Fig. 3 showed that the general expression patterns of these proteins were similar with the data collected from the time-course study of citrus SE (Table 1). In detail, 7 spots (spot 5, 12, 13, 16, 22, 23 and 29) and 9 spots (spot 7, 15, 17, 20, 24, 25, 26, 27, 29) found to be down-regulated and up-regulated, respectively, during the time-course of citrus SE, were indeed down-regulated and up-regulated in globular, heart/torpedo-shaped and cotyledon-shaped embryos comparing with embryogenic callus. Spot 11 and 21 were sustained in these three stages, consisting with the result that they were transiently up-regulated in the first 3 weeks but restored to the original level in the 4th week. Interestingly, within these three stages, the 7 down-regulated spots were sustained; two of the 9 up-regulated spots (spot 15 and 25) were gradually up-regulated and three (spot 20, 24 and 29) were transiently up-regulated (Fig. 3).
Expression of identified proteins in embryogenic callus, globular embryo, heart/torpedo-shaped embryo and cotyledon-shaped embryo. An independent experimental system was carried out, and 18 proteins were investigated according to the position of spots existed in 2-DE gel. Spot number and protein name were presented in the abscissa. The quantity of protein expression was presented in ordinate. The quantity of protein expression is the total intensity of a defined protein spot which is calculated by PDQuest software (see “Materials and methods” for details). Columns and bars represent the mean values and standard errors (n = 3), respectively
Discussion
SE is a method of regeneration for citrus in nature from nucellar cells and laboratories from embryogenic callus. SE provides an important approach of regeneration for citrus in nature from nucellar cells and laboratories from embryogenic callus. The process obviously involves extensive reprogramming of gene/protein expression (Chugh and Khurana 2002). In the present study, we have identified 24 proteins whose expression was shown to be altered during SE of in vitro-cultured citrus callus. Some of these proteins may play important roles in initiation of the SE process or the changes of their expression may reflect direct consequences of the cellular reprogramming in the cultured tissue during SE.
Preferential representation for anti-oxidative proteins implies a role in SE
Though accumulating data indicated that oxidative stress caused by radical oxygen species (ROS) enhanced SE in many plant species (Luo et al. 2001; Pasternak et al. 2002; Caliskan et al. 2004; Ganesan and Jayabalan 2004), few investigated the dynamic expression of oxidative stress-related proteins during SE.
The most striking feature of our results was that five differentially expressed proteins (spot 16, 18, 23, 26 and 29) are predicted to be involved in oxidative stress and metabolism of glutathione (GSH) which acts as a donor of reducing equivalents for the scavenging of ROS (May et al. 1998). Three of these proteins (spot 16, 18 and 23) were down-regulated during citrus SE, which agrees well with the speculation that oxidative stress stimulates cell dedifferentiation and promotes somatic embryo formation (Fehér et al. 2003). However, one anti-oxidative protein (spot 26) showed up-regulation in late stages of SE, while the last (spot 29) showed only transient down-regulation in the second and third week after SE induction. These two proteins (spot 26 and 29) and the one from spot 16 are classified as different isoforms of the glutathione S-transferase (GST). Because GSTs are known to catalyze conjugation of a broad range of substrates to GSH, they may be involved in many different stress responses, including those arising from pathogen attack, oxidative stress, and heavy-metal toxicity (Kathleen 1996). Thus, it is possible that some of these five anti-oxidative proteins identified in our analysis may play roles in regulation of the redox homeostasis critical for maintaining cell differentiation status (Phyllis 2007), and the redox changes resulting from the down-regulation of these proteins may trigger SE, whereas other antioxidative proteins may serve a different function(s) such as protecting the cultures cells from toxicity caused by long time in vitro-culturing.
It is interesting to note that one of the down-regulated anti-oxidative proteins (spot 18), a phospholipid hydroperoxide glutathione peroxidase (PHGPx), has been shown to be involved in defense against various stresses such as salt treatment, mechanical stimulation and pathogen attack (Holland et al. 1993; Depege et al. 1998; Levine et al. 1994). In citrus, PHGPx was validated to be up-regulated in response to high concentration of salt. It was purified and the enzyme activity was determined (Holland et al. 1993; Beeor-Tzahar et al. 1995). Furthermore, PHGPx is essential for embryogenesis in mammalian and the targeted disruption of the PHGPx gene can cause early embryonic lethality in mouse (Hirotaka et al. 2003). Thus, we propose that PHGPx in citrus may regulate certain cellullar signaling pathways associated with embryogenesis via its ROS scavenging activity.
Cell division-associated proteins accumulated to higher levels during SE
Embryogenesis is accompanied with a morphogenesis process during which the embryo differentiates through several distinct stages (globular, heart-shaped, torpedo and cotyledon stages). The morphogenesis is based on co-ordinated cell elongation and division. Thus, it is not surprising that we identified three proteins associated with cell division during citrus SE, two of which are tubulins (spot 15) and the third a putative DNA-damage-repair/toleration protein DRT102 (spot 17). Tubulins are known to be associated with cell division and cell elongation and played an important role in separation of the organelles and daughter chromosomes (mitosis). For example, some tubulins correlating with cell-size increase were shown to be up-regulated during the late stages of carrot SE (Cyr et al. 1987). Likewise, the two tubulin proteins identified in this analysis also showed gradual increase in the treated tissues over the whole period of the SE. DRT102 was first primitively isolated in E. coli on the basis of the ability to partially complement the UV-sensitivity of an E. coli mutant, and was suggested to encode a UV-specific excision repair enzyme w42x (Edward et al. 1998). Besides the role concerned with repair of UV-induced lesions, DNA repair plays vital roles in DNA replication and cell division (Edward et al. 1998). Our observation that a putative citrus DRT102 (spot 17) showed a four-fold increase in the late phases of citrus SE in comparison with the original embryogenic callus (Fig. 3) supports the speculation that DRT102 may participate in a late stage of morphogenesis of citrus SE.
Two forms of Ferritin-3 accumulated in mature embryos
As shown in Table 1 and Fig. 3, two forms of Ferritin-3 (spot 7 and spot 20) had higher expression levels in mature embryos. Ferritins are ubiquitous iron storage proteins that exist in plants, animals and micro-organisms. Lobreaux and Briat (1996) found that ferritin accumulated in mature pea seeds but degraded upon germination. The iron derived from ferritin degradation was necessary for seedling growth (Ragland and Theil 1993). These results suggest that ferritins play an important role in early plant development which might help establish the photosynthesis system and lead to the autotrophical state. Increasing reports also suggest that ferritins play a role in responses to broad physiological or pathologic stresses such as those induced by abscisic acid, oxidative damage and fungal infection (Pauline and Paolo 1996; Mária et al. 1999; Jean-Michel et al. 2001). Hence, we provide two possible explanations for the accumulation of ferritins in mature citrus somatic embryos: (1) When somatic embryos are formed and heterotrophically grown on medium, resembling zygotic embryos (seeds), ferritins are more actively synthesized and stored in mature embryos, providing a rich source of iron required for photosynthesis of early green seedlings; (2) Ferritins may help the cultured tissues better cope with the stresses occurring in the late phases of SE.
Spermidine synthase and rhodanese
Our analysis also identified a spermidine synthase (spot 11) to be up-regulated during the citrus SE. Early studies showed that spermidine (a polyamine) positively affected the embryogenic capability in Pinus radiate (Rakesh et al. 1999), Panax ginseng (Kevers et al. 2000), Picea rubens Sarg (Minocha et al. 2004) and citrus sinensis (Wu et al. 2008). Liu et al. (2005) reported that during SE of Valencia, spermidine increased with the development of somatic embryo and peaked at the globular embryo stage, suggesting that it may promote globular embryo formation. Our finding that a transient increase of a spermidine synthase preceded the appearance of globular embryo stage is in full agreement with the above speculation.
Another protein, a putative rhodanese (spot 9), was accumulated at a higher level in late phase of citrus SE. A rhodanese is a sulphur transferase that can transform cyanide into a less toxic compound, thiocyanate (Saidu 2004). The cyanide, which kills the cell by inhibiting cytochrome oxidase of the mitochondrial electron transport chain, is produced by hydrolysis of cyanogenic compounds and is also released as a co-product of ethylene biosynthesis (Irena and Renata 2006). During SE, ethylene is often detected at high levels in cultures and has negative effects on both number and quality of embryo formation (Stasolla and Yeung 2003; Kumar et al. 1989). It was believed that embryos cultured in the presence of high ethylene levels could prevent post-embryonic growth. Based on this information, we speculated that, in the late phase of citrus SE, cyanide arising from ethylene biosynthesis might have accumulated, and affected the post-embryonic growth. As a cell detoxification mechanism, rhodanese may thus be up-regulated to lower the cyanide level. Consistent with this idea, cyanide has been shown to stimulate the synthesis of rhodanese in Lotus corniculatus (Smith and Urbanska 1986). A strong up-regulation of a rhodanese during citrus SE implies a potential role of rhodaneses in maintaining embryogenic competence during somatic embryo development and maturation.
Conclusion
In this study, 24 differentially expressed proteins were identified during the process of SE in Valencia sweet oranges. Several proteins involved in anti-oxidative stress response (GSTs and a PHGPx), cell division (tubulins), photosynthesis (ferritins), and cyanide detoxification (rhodanese) were likely to be associated with SE. These data will provide important information for understanding the molecular process of SE in citrus. This investigation will facilitate further studies in improving germplasm conservation, genetic transformation and vegetative propagation.
Abbreviations
- 2-DE:
-
Two-dimensional gel electrophoresis
- ACN:
-
Acetonitril
- CBB:
-
Coomassie brilliant blue
- CHAPS:
-
3-[(3-Cholamidopropyl)dimethylammonio]propanesulfonic acid
- DTT:
-
dl-Dithiothreitol
- EST:
-
Expressed sequence tag
- GSH:
-
Glutathione
- GST:
-
Glutathione-S-transferase
- MALDI-TOF:
-
Matrix-assisted laser desorption ionization time-of-flight
- MS:
-
Mass spectrum
- PEBP:
-
Phosphatidylethanolamine-binding family protein
- PHGPx:
-
Phospholipid hydroperoxide glutathione peroxidase
- ROS:
-
Reactive oxygen species
- SE:
-
Somatic embryogenesis
- TFA:
-
Trifluoroacetic acid
References
Baldwin TC, Domingo C, Schindler T, Seetharaman G, Stacey N, Roberts K (2001) DcAGP1, a secreted arabinogalactan protein, is related to a family of basic proline-rich proteins. Plant Mol Biol 45:421–435
Beeor-Tzahar T, Ben-Hayyim G, Holland D, Faltin Z, Eshdat Y (1995) A stress-associated citrus protein is a distinct plant phospholipids hydroperoxide glutathione peroxidase. FEBS Lett 366:151–155
Boutilier K, Offringa R, Sharma VK, Kieft H, Ouellet T, Zhang LM, Hattori J, Liu CM, van Lammeren AAM, Miki BLA et al (2002) Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell 14:1737–1749
Caliskan M, Turet M, Cuming AC (2004) Formation of wheat (Triticum aestivum L.) embryogenic callus involves peroxidegenerating germin-like oxalate oxidase. Planta 219:132–140
Chen SX, Harmon AC (2006) Advances in plant proteomics. Proteomics 6:5504–5516
Chugh A, Khurana P (2002) Gene expression during somatic embryogenesis-recent advances. Curr Sci 86:715–730
Cyr RJ, Bustos MM, Guiltinan MJ, Fosket DE (1987) Developmental modulation of tubulin protein and mRNA levels during somatic embryogenesis in cultured carrot cells. Planta 171:365–376
Dai SJ, Chen TT, Chong K, Xue YB, Liu SQ, Wang T (2007) Proteomic identification of differentially expressed proteins associated with pollen germination and tube growth reveals characteristics of germinated Oryza sativa pollen. Mol Cell Proteomics 6:207–230
Depege N, Drevet J, Boyer N (1998) Molecular cloning and characterization of tomato cDNAs encoding glutathione peroxidase-like proteins. Eur J Biochem 253:445–451
Dixon DP, Davis BG, Edwards R (2002) Functional divergence in the glutathione transferase superfamily in plants. J Biol Chem 277:30859–30869
Edward JV, Helen LM, Ramachandran K, Ishita C, Bernard AK (1998) DNA repair in higher plants. Mutat Res 400:187–200
Fehér A, Pasternak TP, Dudits D (2003) Transition of somatic plant cells to an embryogenic state. Plant Cell Tissue Organ Cult 74:201–228
Francesco C, Maria CT, Fabio DP, Francesco GC (1998) Somatic embryogenesis and plant regeneration from undeveloped ovules and stigma/style explants of sweet orange navel group [Citrus sinensis (L.) Osb]. Plant Cell Tissue Organ Cult 54:183–189
Ganesan M, Jayabalan N (2004) Evaluation of haemoglobin (erythrogen): for improved somatic embryogenesis and plant regeneration in cotton (Gossypium hirsutum L. cv. SVPR 2). Plant Cell Rep 23:181–187
Giovanni C, Maurizio B, Luca M, Laura S, Gian Marco G, Barbara C, Paola O, Luciano Z, Pier GR (2004) Blue silver: a very sensitive colloidal coomassie G-250 staining for proteome analysis. Electrophoresis 25(9):1327–1333
Hirotaka I, Fumi H, Taro S, Kanae S, Yumi M, Makoto S, Takeo K, Michiko H, Kazunori H, Yasuhito N (2003) Early embryonic lethality caused by targeted disruption of the mouse PHGPx gene. Biochem Biophys Res Commun 305:278–286
Holland D, Ben-Hayyim G, Faltin Z, Camoin L, Strosberg AD, Eshdat Y (1993) Molecular characterization of salt-stress-associated protein in citrus: protein and cDNA sequence homology to mammalian glutathione peroxidases. Plant Mol Biol 21:923–927
Imin N, De Jong F, Mathesius U, van Noorden G, Saeed NA, Wang X, Rose RJ, Rolfe BG (2004) Proteome reference maps of Medicago truncatula embryogenic cell cultures generated from single protoplasts. Proteomics 4:1883–1896
Imin N, Nizamidin M, Daniher D, Nolan KE, Rose RJ, Rolfe BG (2005) Proteomic analysis of somatic embryogenesis in Medicago truncatula. Explant cultures grown under 6-benzylaminopurine and 1-naphthaleneacetic acid treatments. Plant Physiol 137:1250–1260
Irena S, Renata B (2006) Cyanide action in plants—from toxic to regulatory. Acta Physiol Plant 28:483–497
Jean-Michel P, Jean-François B, Stephane L (2001) Structure and differential expression of the four members of the Arabidopsis thaliana ferritin gene family. Biochem J 359:575–582
Kathleen AM (1996) The functions and regulation of glutathione S-transferases in plants. Annu Rev Plant Physiol Mol Biol 47:127–158
Kevers C, Gal NL, Monteiro M, Dommes J, Gaspar T (2000) Somatic embryogenesis of Panax ginseng in liquid cultures: a role for polyamines and their metabolic pathways. Plant Growth Regul 31:209–214
Kumar PP, Joy RW IV, Thorpe TA (1989) Ethylene and carbon dioxide accumulation and growth of cell suspension cultures of Picea glauca (white spruce). J Plant Physiol 135:592–596
Levine A, Tenhaken R, Dixon R, Lamb C (1994) H2O2 from oxidative burst orchestrates the plant hypersensitive disease resistance response. Cell 79:583–593
Lippert D, Zhuang J, Ralph S, Ellis DE, Gilbert M, Olafson R, Ritland K, Ellis B, Douglas CJ, Bohlmann J (2005) Proteome analysis of early somatic embryogenesis in Picea glauca. Proteomics 5:461–473
Liu JH, Deng XX (2002) Regeneration and analysis of citrus interspecific mixoploid hybrid plants from asymmetric somatic hybridization. Euphytica 125:13–20
Liu HY, Xiao LT, Lu XD, Hu JJ, Wu S, He CZ, Deng XX (2005) Changes in polyamine levels in Citrus sinensis Osb. cv. Valencia callus during somatic embryogenesis. J Plant Physiol Mol Biol 31(3):275–280
Lobreaux S, Briat JF (1996) Ferritin accumulation and degradation in different organs of pea during development. Biochem J 274:601–606
Luo JP, Jiang ST, Pan LJ (2001) Enhanced somatic embryogenesis by salicylic acid of Astralagus adsurgens Pall.: relationship with H2O2 production and H2O2 metabolizing enzyme activities. Plant Sci 161:125–132
Mária D, Gábor VH, Sholpan D, Katalin T, László S, Imre V, Balázs B, Zoltán K, Dénes D (1999) Plants ectopically expressing the iron-binding protein, ferritin, are tolerant to oxidative damage and pathogens. Nat Biotechnol 17:192–196
May MJ, Vernoux T, Leaver C, Van Montagu M, Inze D (1998) Glutathione homeostasis in plants: implications for environmental sensing and plant development. J Exp Bot 49:649–667
Meinke DW (1992) A homoeotic mutant of Arabidopsis thaliana with leafy cotyledons. Science 258:1647–1650
Milena M, Marcella B, Luca E, Bhakti P, Alfredo SN, Candida V (2008) Proteomic analysis of somatic embryogenesis in Vitis vinifera. Plant Cell Rep 27:347–356
Minocha R, Minocha SC, Long S (2004) Polyamines and their biosynthetic enzymes during somatic embryo development in red spruce (Picea rubens Sarg.). In Vitro Cell Dev Plant 40:572–580
Mukaddes K, Kemal NK (2006) The effects of some carbohydrates on growth and somatic embryogenesis in citrus callus culture. Sci Hortic 109:29–34
Murashige T, Tucker DPH (1969) Growth factors requirement of citrus tissue cultures. In: Chapman HD (ed) Proceedings of the International Citrus Symposium, vol 3. Riverside, California, pp 1155–1161
Pasternak T, Prinsen E, Ayaydin F, Miskolczi P, Potters G, Asard H, van Onckelen H, Dudits D, Fehe′r A (2002) The role of auxin, pH and stress in the activation of embryogenic cell division in leaf protoplast-derived cells of alfalfa (Medicago sativa L.). Plant Physiol 129:1807–1819
Pauline MH, Paolo A (1996) The ferritins’ molecular properties, iron storage function and cellular regulation. Biochim Biophys Acta 1275:161–203
Phyllis AD (2007) Effects of oxidative stress on embryonic development. Birth Defects Res C Embryo Today 81:155–162
Ragland M, Theil EC (1993) Ferritin and iron concentration during soybean nodule development. Plant Mol Biol 21:555–560
Rakesh M, Dale RS, Cathie R, Kevin DS, Subhash CM (1999) Polyamine levels during the development of zygotic and somatic embryos of Pinus radiate. Physiol Plant 105:155–164
Saidu Y (2004) Physicochemical features of rhodanese: a review. Afr J Biotech 3:370–374
Schmidt EDL, Guzzo F, Toonen MAJ, Devries SC (1997) A leucine-rich repeat containing receptor-like kinase marks somatic plant cells competent to form embryos. Development 124:2049–2062
Smith J, Urbanska KM (1986) Rhodanese Activity in Lotus corniculatus sensu-lato. J Nat Histol 20:1467–1476
Stasolla C, Yeung EC (2003) Recent advances in conifer somatic embryogenesis: improving somatic embryo quality. Plant Cell Tissue Organ Cult 74:15–35
Stasolla C, Bozhkov PV, Chu TM, van Zyl L, Egertsdotter U, Suarez MF, Craig D, Wolfinger RD, von Arnold S, Sederoff RR (2004) Variation in transcript abundance during somatic embryogenesis in gymnosperms. Tree Physiol 24:1073–1085
Stefania F, Fabio DP, Francesco C, Maurizio S (2002) Effect of 2, 4-DD and 4-CPPU on somatic embryogenesis from stigma and style transverse thin cell layers of Citrus. Plant Cell Tissue Organ Cult 68:57–63
Thibaud-Nissen F, Shealy RT, Khanna A, Vodkin LO (2003) Clustering of Microarray data reveals transcript patterns associated with somatic embryogenesis in soybean. Plant Physiol 132:118–136
Winkelmann T, Heintz D, Dorsselaer AV, Serek M, Braun HP (2006) Proteomic analyses of somatic and zygotic embryos of Cyclamen persicum Mill. reveal new insights into seed and germination physiology. Planta 224:508–519
Wu XB, Wang J, Liu JH, Deng XX (2008) Involvement of polyamine biosynthesis in somatic embryogenesis of Valencia sweet orange (Citrus sinensis) induced by glycerol. J Plant Physiol. doi:10.1016/j.jplph.2008.02.005
Zimmerman JL (1993) Somatic embryogenesis: a model for early development in higher plant. Plant Cell 5:1411–1423
Zuo JR, Niu QW, Frugis G, Chua NH (2002) The WUSCHEL gene promotes vegetative-to-embryonic transition in Arabidopsis. Plant J 30:349–359
Acknowledgments
We thank Prof. S. Xiao and Dr. Q. Xu for their critical review of the manuscript. We also thank Shanghai Applied Protein Technology Co. Ltd for the technology support. The research was financially supported by the National Natural Science Foundation of China (No. 30570973, 30830078) and the Ministry of Education of China (IRT0548).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by P. Lakshmanan.
Rights and permissions
About this article
Cite this article
Pan, Z., Guan, R., Zhu, S. et al. Proteomic analysis of somatic embryogenesis in Valencia sweet orange (Citrus sinensis Osbeck). Plant Cell Rep 28, 281–289 (2009). https://doi.org/10.1007/s00299-008-0633-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-008-0633-7