Abstract
To investigate the rice root proteome, we applied the PEG fractionation technique combined with two-dimensional gel electrophoresis which rendered more well-separated protein spots. Out of the 295 chosen proteins, 93 were identified by MALDI-TOF mass spectrometry. The proteins were classified as relating to metabolism (38.7%), reactive oxygen species (ROS)-related proteins (22.5%), protein processing/degradation (8.6%), stress/defense (7.5%), energy (6.5%) and signal transduction (5.4%). The high percentage of ROS-related proteins found in rice root brings us to assess the roles of ROS on rice root growth. Treatment with ROS quenching chemicals such as reduced glutathione (GSH), diphenyleneiodonium (DPI) and ascorbate inhibited root growth dose-dependently. Forty-nine proteins identified were either up- or down-regulated by GSH treatment, of which 14 were ROS-related proteins, such noticeably modulated ones as glutathione-S-transferase (GST), superoxide dismutases (SOD) and l-ascorbate peroxidases. The protein levels of four GSTs (NS4, 8, 56 and 57), three APXs (NS46, 49 and 50) and MnSOD (NS45) were strongly reduced by GSH treatment but slightly reduced by ascorbate and DPI. Ascorbate and DPI strongly inhibited expression levels of a catalase A (NP23) and an APX (NS65) but did not affect APXs (NS46, 49 and 50) protein levels. Northern analysis demonstrated that changes in transcript levels of five genes––GST (NS4), GST (NS43), Mn-SOD (NS45), APX (NS50) and APX (NS46/49) in response to ROS quenching chemicals were coherent with patterns shown in two-dimensional electrophoresis analyses. Taken together, we suggest that these proteins may take part in an important role in maintaining cellular redox homeostasis during rice root growth.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The rice root system plays important roles in nutrient uptake (Teo et al. 1995), lodging tolerance (Terashima et al. 1994), drought tolerance (Azhiri-Sigari et al. 2000), and yield of growing plants (Morita et al. 1988). Therefore it is of interest to determine how roots detect levels of water and nutrients in soil and adapt to their architecture. Many studies have used genetic approaches to identify cellular functions in roots (Lopez-Bucio et al. 2003). A few studies have been undertaken using proteomic approaches to understand biological function of rice root so far (Tanaka et al. 2004, 2005).
Understanding the biological functions of genes requires more than simply identifying them genetically and biochemically. Since the rice genome sequence became publicly available (International rice genome sequencing project 2005), proteomics has emerged as a tool to study molecular or cellular functions of rice proteins. Such studies have been applied for understanding protein expression, posttranslational modification, and protein–protein interactions in roots of rice (Komatsu and Konishi 2005; Tanaka et al. 2004; Koller et al. 2002), maize (Hochholdinger et al. 2005) and cassava (Manihot esculenta Crantz) (Sheffield et al. 2006). Proteins identified in these studies primarily functioned in metabolism or defense/stress mechanisms. Interestingly, a previous report categorized many reactive oxygen species (ROS)-related proteins as belonging to the defense/stress group. These proteins play an important role not only in influencing defense/stress but also in controlling cellular redox states (Halliwell 2006).
In recent years, ROS have emerged as important regulators of leaf and root development. It was initially thought that ROS, such as hydroxyl radicals (OH•), superoxide anion (O2 −) and hydrogen peroxide (H2O2), were toxic by-products of respiration and photosynthesis. However, other evidence suggests that ROS play significant roles in intracellular signaling in radish seed germination (Schroeder and Mori 2004), interaction with biotic and abiotic environments in various plants (Blokhina et al. 2003), and auxin signal transport and gravitropism in maize (Joo et al. 2001; Kwak et al. 2006). There is also evidence that ROS are required during leaf growth. H2O2 functions in the lignifying xylem of the first internode of Zinnia elegans to stiffen cell walls as growth ceases and cells differentiate (Ros-Barcelo et al. 2002). ROS in leaves and roots of maize are necessary for cellular extension and expansion (Rodriguez et al. 2002; Liszkay et al. 2004). The Arabidopsis root hair defective mutant rhd2 encoding NADPH oxidase shows a 20% decrease in both ROS levels and root growth compared to wild-type plants (Foreman et al. 2003; Renew et al. 2005). Both genetic and biochemical studies indicate that ROS function in plant growth and development. However, little is known about either modulation of ROS-related proteins associated with root growth or the effect of ROS quenching chemicals on root growth.
In this study, we used two-dimensional electrophoresis (2-DE) based proteome analyses to obtain an understanding of relationship between ROS and expression levels of ROS-related proteins during early stage of rice root growth. We analyzed effects of ROS quenching chemicals such as reduced glutathione (GSH; γ-glutamylcysteinyl glycine), ascorbate and diphenyleneiodonium (DPI) on root growth. ROS quenching chemicals not only differentially modulated ROS-related proteins but also root growth. These data represent the first proteomic approach showing that ROS and their related proteins may implicate important functions in root growth.
Materials and methods
Plant growth conditions and measurements
Mature rice seeds (Oryza sativa L. cv. Jinheung) were obtained from the National Yeongnam Agricultural Experimentation Station. Dehulled seeds were sterilized in 70% ethanol for 10 min and then in 3% sodium hypochlorite for 20 min. Sterilized seeds were grown on MS Phytagel® medium at 28°C. Root growth was measured from 7-day-old roots treated with GSH (at 250, 500 μM or 1 mM), DPI (at 500 nM, 1 or 2 μM) or ascorbate (at 1, 2.5 or 5 mM).
Detection of H2O2 release
The ROS released from rice roots were observed on agar containing 25 mM 2′, 7′,-dichlorofluorescin (DCFH), a fluorescent ROS indicator (Schopfer et al. 2001). Agar was prepared by adding an appropriate volume of 25 mM DCFH in ethanol to a 1% (w/v) agar solution in 20 mM phosphate buffer, pH 6.0, to obtain a 10 μM DCFH mixture. Roots were incubated in 1 mM GSH or H2O for 2 h and then embedded in DCFH-containing agar supplemented with the same medium. Epifluorescence was observed under a fluorescence microscope equipped with a UV light.
Protein extraction and 2-DE analysis
Rice root proteins were extracted from 7-day-old roots grown in Phytagel® containing MS medium treated with H2O, 1 mM GSH, 2 μM DPI, or 2.5 mM ascorbate. Proteins were extracted using Mg/NP-40 buffer containing 0.5 M Tris–HCl (pH 8.3), 2% v/v NP-40, 20 mM MgCl2, 1 mM phenyl methyl sulfonyl fluroride (PMSF) and fractionated with PEG 4000, following the method described by Kim et al. (2001, 2003a). The isoelectric focusing (IEF) gel mixture consisted of a 4.5% w/v acrylamide, 9.5 M urea, 2% v/v NP-40, and 2.5% v/v pharmalytes (pH 3–10:pH 5–6:pH 5–8 at a ratio of 1:2.5:3.5 [neutral condition]; pH 3–10:pH 7–9:pH 5–8 at a ratio of 1:3.5:2.5 [basic condition] [Amersham Pharmacia Biotech]). Each sample of total extract (150 μg) was mixed with IEF sample buffer and loaded onto an 18-cm IEF tube gel (Kim et al. 2001). The second dimension was carried out on SDS-PAGE as described by Leamml (1970) using 12% polyacrylamide gels. 2-DE gels were silver-stained by the method of Blum et al. (1987). Image acquisition was achieved using a transmissive scanner (PowerLook III, UMAX). Pixel depth was 16 bit, resolution was 300 dpi; brightness and contrast were set to default. Gel images were exported as TIFF files from the scanner, and gel spots were automatically detected using ImageMaster 2D Platinum software (Amersham Biosciences). The intensity of each spot was then normalized as an average of the intensity of spots on the gel. Statistic analyses were created between each control groups and corresponding treated groups. In the statistic sets, the Student’s t test and significance level of 95% were chosen.
MALDI-TOF mass spectrometry
Gel spots digested with trypsin were analyzed using a Voyager-DE STR (matrix-assisted laser desorption ionization-time-of-flight) MALDI-TOF mass spectrometer (PerSeptive Biosystems). Digestion mixtures were remelted using a solution of 93% water, 5% acetonitrile and 2% trifluoroacetic acid (TFA). The samples were sonicated for 5 min and centrifuged for 2 min. The matrix solution [dissolved α-cyano-4-hydroxycinnamic acid (Sigma) in acetone (40 mg/ml) and nitrocellulose in acetone (20 mg/ml)], the nitrocellulose solution and isopropanol were mixed 100:50:50 (Kim et al. 2004). Two microliters of the sample was added to 2 μl of the peptide sample solution, and 1 μl of this was spotted immediately onto a MALDI plate and left for 5 min. The MALDI plate was then washed with 0.1% v/v TFA. Parent ion masses were measured in the reflectron/delayed extraction mode with an accelerating voltage of 20 kV, a grid voltage of 76.000%, a guide wire voltage of 0.010%, and a delay time of 150 ns. Des-Arg1-bradykinin (m/z 904.4681) and angiotensin 1 (m/z 1296.6853) were used as a two-point internal standard for calibration. Peptides were selected in the mass range of 500–3,000 Da. For data processing, the PerSeptive-Grams software package was used. Database searches were performed using Protein Prospector (http://prospector.ucsf.edu).
cDNA cloning and Northern blot analyses
GST (NS4 and NS43), SOD (NS45) and APX (NS50 and NS46/49) sequences identified from public databases were used to design primers used to amplify cDNAs from a rice cDNA library. The primer pairs were as follows: GST (NS4), forward primer (5′-atctcaagaacaagagcgag-3′) and reverse primer (5′-catgacttcaacagcttgtc-3′); GST (NS43), forward primer (5′-atgtaccaacaaagtgcagg-3′) and reverse primer (5′-gattgatagagcgtcaggtc-3′); Mn-SOD (NS45), forward primer (5′-atggcgctccgcacgctg-3′) and reverse primer (5′-agcagtcgcattttcgatcacctc-3′); APX(NS50), forward primer (5′-atgggcagcaagtcgtaccc-3′) and reverse primer (5′-ttcctcagcaaatcccagttc-3′); APX (NS46/49), forward primer (5′-atggctaagaactaccccgtc-3′) and reverse primer (5′-agcatcagcgaaccccagttc-3′). PCR products were cloned into the Gateway system™ (Invitrogen) and sequenced. Total RNA extracted from 3- and 6-day-old roots grown in Phytagel® containing MS medium supplemented with 1 mM GSH, 2.5 mM ascorbate, and 2 μM DPI. Each RNA sample (20 μg) was blotted onto nylon membranes, hybridized with 32P-labeled 5 cDNA probes, and washed at high stringency. Equal loading of total RNA was verified by staining rRNA with ethidium bromide. Blots were exposed to X-ray film at −70°C for 1 day.
Results and discussion
Identification of rice root proteins by prefractionation followed by 2-DE
For comprehensive analysis of rice root proteome, we used the PEG fractionation (supernatant/pellet) technique combined with neutral (pI, 4∼7) and basic (pI, 6∼9) IEF gels to obtain more discrete spots from rice roots (Kim et al. 2001, 2003a, 2004). Gels were stained with silver to provide high sensitivity. Using ImageMaster 2D Platinum analysis software, more than 1,600 protein spots from four different sets of gels were reproducibly detected through three replicates. As shown in Fig. 1, gel spots were well resolved, and little streaking was observed from four different sets of gels for one sample (neutral supernatant, NS; neutral pellet, NP; basic supernatant, BS; basic pellet, BP). This separation method allows us to display many proteins synchronously.
Two-DE analysis of PEG-fractionated proteins from rice root. Protein samples (150 μg) from the 15% (w/v) PEG supernatant (a, c) and pellet (b, c) fractions were applied to neutral (pI 4–7) (a, b) and basic (pI 6–9) range IEF gels (c, d), followed by 2-DE and silver staining. Identified were 93 proteins indicated by numbered arrows. Molecular masses in kDa are indicated at left and pI values at the top
Protein spots from the gels of each root sample were subjected to the in-gel digestion and analyzed by MALDI-TOF mass spectrometry. Database searches were performed using Protein Prospector and blast homology searching. Ninety-three proteins among 295 spots chosen from the four sets were identified; 42 proteins in the NS, 16 proteins in the NP, 15 proteins in the BS and 20 proteins in the BP fractions (Fig. 1; Table 1). Identified proteins were classified into nine groups according to the functional categories established by Bevan et al. (1998). The functional groups were represented by metabolism (38.7%) followed by ROS-related proteins (22.5%), protein processing/degradation (8.6%), stress/defense (7.5%), energy (6.5%), signal transduction (5.4%), chaperone/heat shock (3.2%), development (2.2%) and unknown function (5.4%) (Fig. 2).
The largest group identified, composed of 36 proteins, was associated with metabolism (Table 1). Among 36 proteins, malate dehydrogenase (MDH; NS30, NS34, NS38 and BP54), glyceraldehyde 3-phosphate dehydrogenase (GAPDH; BS34, BS37, BS43, BP26, BP35 and BP41) and enolase (NS15 and NS17) were identified as major protein spots. These proteins are important in several metabolic pathways in plants. MDH is involved in catalyzing the reversible reduction of oxaloacetate to malate in TCA cycle (Miller et al. 1998). GAPDH catalyzed the regeneration of NAD+, which is required for continued glycolysis and ATP production (Yang et al. 1993). Enolase (2-phospho-d-glycerate hydratase) is an integral enzyme in glycolysis. It catalyzes the interconversion of 2-phosphoglycerate to phosphoenolpyruvate. Recently, these proteins were also detected by proteomic analyses in the primary root of maize (Hochholdinger et al. 2004, 2005) and cassava (Manihot esculenta Crantz) (Sheffield et al. 2006). These results indicate that the identified proteins involved in metabolism are likely critical for generating energy during root growth.
The ROS related-proteins comprised the second most abundant group (22.5%). The 21 ROS-related proteins that we identified included glutathione S-transferases (GST; NS4, NS8, NS43, NS44, NS56, NS57 and NP78), ascorbate peroxidases (APX; NS46, NS49, NS50, NS65 and NP85), catalase isozymes (CAT; NP23, BS13, BP4 and BP5), superoxide dismutases (SOD; NS45, NS68 and NS71), GSH-dependent dehydroascorbate reductase 1 (DHAR1; NS42) and glutathione reductase (GR; BS2) (Table 1; Fig. 1). Komatsu’s group previously reported that the most abundant proteins in rice roots functioned in defense (26%) and metabolism (21%), where their classification included ROS-related proteins in defense (Tanaka et al. 2005). In that study, 14 of 73 proteins (19%) as second-most abundant group were ROS-related ones, consistent with the proportion observed in our study. In contrast to rice, ROS-related proteins constituted 12.5% in cassava (Sheffield et al. 2006) and 6.2% maize root (Hochholdinger et al. 2005), respectively. Differences in these proportions between rice and maize or cassava roots could be related to different growth conditions such as water-logged or aerobic upland conditions, respectively.
Third largest protein functional group consisted of eight proteins involved in protein processing and degradation. 26S protease regulatory proteins (NP9, NP10 and BP30) and 20S proteasome subunit proteins (NP74, NP88, NP81, BP18 and BP20) were collectively found in the PEG pellet fraction (Table 1; Fig. 1b, d). The 26S proteasome, multi-catalytic complex proteins comprising 20S core and 19S regulatory particles, plays an important role in protein degradation and processing during growth and development. Recently, the HALTED ROOT (HLR) gene, which encodes a subunit of the 26S proteasome, was identified as essential to maintain cellular organization and normal shoot and root apical meristem activities required to initiate new growth in young seedlings at the tips of roots and shoots in Arabidopsis (Ueda et al. 2004). These data suggest that proteasome-dependent proteolysis may be involved in root growth.
Among 93 identified protein spots, 19 different proteins spots were found in multiple spots, most of which consisted of two to four isoforms and the identical protein name but do not possess the same amino acid sequence. It is likely that such different isoforms proteins come from the same gene family and are most likely derived from gene duplication (Ostergaard et al. 2002). All the proteins found in multiple spots showed different characteristic property to each other either in their pI values or molecular mass or both. The discrepancies in pI and molecular mass might be due to posttranslational modification, protein processing, or degradation. Protein disulfide isomerase (PDI) with three isoforms (NS1, NS2 and NS3) was a typical example of posttranslational modification showing higher molecular mass (∼57 kDa) than theoretical molecular mass (33.4 kDa) by ubiquitination (Kim et al. 2003a, b) (Fig. 1; Table 1).
Effect of GSH on ROS quenching and root growth
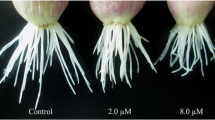
The ROS-related proteins in rice roots have been identified more than other plants such as maize and cassava (Hochholdinger et al. 2005; Sheffield et al. 2006). We used an ROS quenching chemical, GSH, to see if they involve in root growth and development. It is known that GSH can effectively scavenge hydrogen peroxide (H2O2) via APX by generating ascorbate through dehydroascorbate reductase conjugated with glutathione reductase (May et al. 1998). We treated roots with GSH (250, 500 μM or 1 mM) for 3 days and then measured root length. Root length significantly decreased by approximately 50% in the presence of 1 mM GSH compared to untreated controls (Fig. 3a, b) and H2O2 levels detected by fluorescence using 2′, 7′,-dichlorofluorescin (DCFH) were also decreased in rice seedling roots grown on an agar medium (Schopfer et al. 2001) (Fig. 3c). These data indicate that GSH-mediated decreases in ROS may inhibit root growth. DPI (2 μM) and ascorbate (2.5 mM) treatment also inhibited root growth approximately 50% compared to untreated control (data not shown). DPI is a compound which binds in the reaction center of flavoproteins such as NADPH oxidases inhibiting their activity (O’Donnell et al. 1993). The growth of wild-type root hairs is inhibited by DPI treatment in Arabidopsis (Foreman et al. 2003). Since ascorbate, known as an antioxidant, removes ROS, it follows that low ROS availability in roots may affect root growth. In addition, GSH is a physiological regulator of many thiol-disulphide exchange reactions (Fang et al. 2002). Ascorbate can regulate cell division by influencing progression from G1 to S phase of the cell cycle and cell expansion (Smirnoff 1996, 2000). DPI can deplete intracellular GSH and therefore disrupt cellular glutathione homeostasis (Pullar and Hampton 2002). These results suggested that root growth might be inhibited by ROS quenching chemicals which inhibited ROS generation in root.
Effect of GSH on root growth and H2O2 production on the root surface. a Rice roots were treated with the GSH (250, 500 μM or 1 mM) for 3 days and their growth compared with untreated plants. Values are the means ± SE of five independent experiments. b Shown are roots following treatment with 1 mM GSH. c DCFH fluorescence in roots treated with 1 mM GSH for 2 h in aerated media and then embedded in the same medium containing 1% (w/v) agar and 10 μM DCFH. After 30 min, fluorescent images were taken under UV light
Differential proteomic analysis of rice roots proteins after GSH, ascorbate and DPI treatment
To determine whether ROS-related proteins are indeed differentially expressed in response to different ROS quenching chemicals, we compared protein profiles displayed on 2-DE gels in the presence of GSH, ascorbate and DPI (Fig. 4). Using ImageMaster 2D Platinum program, we detected changes of 49 protein levels among 93 identified ones after GSH treatment (Table 1). Among them, 14 ROS-related proteins which modulated by ROS quenching chemicals were divided into two groups (type I and type II) according to their modulation. These modulated proteins were calculated using probability of the differences being statistically significant based on the Student’s t test. Most of the significant difference in modulated proteins gave P values of <0.05. Expression levels of the type I group—GST (NS4, NS8, NS56 and NS57), APX (NS46, NS49 and NS50), Mn-SOD (NS45), DHAR (NS42) and CAT-A (NP23)—were significantly reduced in GSH-treated samples compared with controls but were only slightly altered by ascorbate or DPI treatment except for GST (NS4), DHAR (NS42) and CAT-A (NP23) (Fig. 5). In the type II group, GST (NS43), Cu–Zn SOD (NS71), APX (NS65) and Cu–Zn SOD (NS68) were either unchanged or up-regulated by GSH treatment, but APX (NS65) was decreased by DPI or ascorbate treatment (Fig. 5). However, SOD (NS68) expression was increased up to 1.5-fold by GSH treatment (Table 1).
Modulation of rice root proteins by ROS quenching chemicals analyzed with 2-DE. Close-up views of ROS-related proteins showing differences in expression of proteins mediated by GSH (1 mM), ascorbate (2.5 mM) or DPI (2 μM) treatment. Protein samples (150 μg) from the PEG supernatant and pellet fractions were separated on a 2-DE neutral gel (pI 4–7) and silver stained. The number of each protein spot corresponds to its listing in Table 1
Quantification of expression levels of 14 ROS-related proteins. The intensities of 14 modulated protein spots from samples after treatment with ROS quenching chemicals were recorded as digitalized images using ImageMaster 2D Platinum and compared to controls. The mean relative expression level of three replicate samples is shown in the histograms based on relative protein intensities compared with background levels. Error bars indicate the standard deviation. Significant difference was P < 0.05 by Student’s t test
To confirm whether the protein expression profile analyzed by 2-DE reflects transcript levels, we performed Northern analyses using gene-specific probes from cDNAs encoding five proteins [GST (NS4), GST (NS43), Mn-SOD (NS45), APX (NS50) and APX (NS46/49)] following treatment with ROS quenching chemicals. In 6-day-old root seedlings, transcripts of GST (NS4), Mn-SOD (NS45), APX (NS50) and APX (NS46/49), but not GST (NS43) were reduced to a greater extent by GSH than by ascorbate treatment (Fig. 6). By contrast, transcript levels of GST (NS4), Mn-SOD, APX (NS50) and APX (NS46/49) decreased in 3-day-old roots following DPI treatment but recovered in 6-day-old roots (Fig. 6). These results confirm that these genes were indeed differently regulated by ROS quenching chemicals, consistent with those in 2-DE.
Northern analysis of transcripts encoding GST (NS4 and NS43), APX (NS50 and NS46/49) and SOD (NS45) proteins. Rice roots were treated with ROS quenching chemicals (GSH, Ascorbate or DPI) during seeding growth. Root samples were harvested at 3 or 6 days, and total RNA was extracted. Each RNA sample (20 μg) was blotted onto nylon membranes, hybridized with 32P-labeled probes, and washed at high stringency. Equal loading was verified by staining rRNA with ethidium bromide. Blots were exposed to X-ray film at −70°C for 1 day
Some functions of GSTs are to detoxify cytotoxic substrates and protect cells against oxidative damage (Marrs 1996). Interestingly, expression levels of several GST isoforms differentially responded to ROS quenching chemicals. GST (NS43) was little affected by GSH, ascorbate and DPI treatment compared to GSTs; NS4, NS8, NS56 and NS57 (Figs. 4, 5). Plant GSTs are abundant proteins encoded by a highly divergent, ancient gene family and divided into four sequence-related classes, namely, the phi (F), zeta (Z), tau (U) and theta (T) class (Edwards et al. 2000; Gong et al. 2005). From an NCBI database search, we found that the NS43 spot belonged to the F-class and the other four were of the U-class. Pau’s group reported that expression of F class GST in mustard was little affected by 5 mM GSH treatment (Gong et al. 2005). In Arabidopsis and maize, F-class GSTs with similar 3D structures have been analyzed by X-ray crystallography (Reinemer et al. 1996; Neuefeind et al. 1997). F-class GSTs have a conserved GSH-binding site (G-site) located in the N-terminus, which is specific for GSH and facilitate formation of the catalytically active thiolate anion of GSH. It is speculated that rice GSTs in accordance with a different structural group may be differentially expressed following GSH treatment. However, further study is needed to clarify that the effect of GST on root growth may be associated with the endogenous pool of GSH.
Four GST isoforms (NS4, NS8, NS56 and NS57), which were derived from the same cDNA sequence, were highly decreased by GSH than ascorbate and DPI treatment compared to that of control (Figs. 4, 5). All the protein spots were found in different pI values, thus forming a train of spots horizontally (Fig. 4). Recently, shifts in GST pIs between acidic and basic forms observed in proteomic analysis of Arabidopsis have been shown to be due to oxidation of methionine residues in response to bacterial inoculation; these changes have been identified by peptide MS fingerprints and MS/MS sequence analyses (Jones et al. 2004). The difference in expression level and shifts in pI observed in our gels among four GSTs (NS4, NS8, NS56 and NS57) following treatment with ROS scavengers may be due to oxidation of methionine residues in GST similar to the situation in Arabidopsis. However, their biological functions remain unknown.
We also identified three SODs (NS45, NS68 and NS71) whose expression differed following GSH treatment (Fig. 4, 5). NS45, NS68 and NS71 spots were identified as mitochondrial Mn SOD, chloroplastic Cu–Zn SOD and cytoplasmic Cu–Zn SOD using NCBI domain search, respectively. NS68 was increased but NS71 was little changed by GSH compared to NS45 (Fig. 4, 5). Ascorbate and DPI had little influence on protein expression levels of three SODs compared to GSH treatment. Our observations that GSH induced the level of Cu–Zn SOD (NS68) are in agreement with the finding for Nicotiana Cu–Zn SOD which was found to be up-regulated by GSH treatment (Herouatr et al. 1993). However, expression of Mn SOD (NS45) found to be down-regulated by GSH treatment in contrast to that of Cu–Zn SOD (NS68). In both Pinus sylvestris L. and human, Cu–Zn SOD and Mn SOD were down-regulated by thiols (Suzuki et al. 1993; Wingsle and Karpinski 1996). These results suggest that different expression patterns may result from different aspects of plant physiology affected by ROS quenching chemicals during root growth (Fang et al. 2002; Smirnoff 2000; Pullar and Hampton 2002).
The present data suggest that ROS-scavenging activities of these three ROS quenching chemicals differentially regulate protein expression levels and patterns because GSH, ascorbate and DPI have other cellular functions in addition to ROS scavenging. In summary, proteomic analysis of rice root not only showed functional protein profiles in root growth, but also modulation of ROS-related proteins, 14 out of 49 proteins (28.6%) under GSH treatment. This implicated that ROS-related proteins may have an important role in regulating ROS levels or homeostasis, which in turn may affect rice root growth.
Conclusions
To understand effect of ROS during rice root growth, we have carried out for the first time to identify ROS responsive proteins in rice root growth by proteomic analyses using PEG prefractionation combined with two IEF systems. Four sets of 2-DE gels of each sample replicated, enhanced reliability of the result obtained. Through MALDI-TOF analyses, we found that metabolism and ROS-related proteins were assigned as predominant group in the rice root proteome. To identify the role of ROS during rice root growth, we treated various concentrations of GSH, ascorbate and DPI concentrations. Rice root treated with GSH (1 mM), ascorbate (2.5 mM) and DPI (2 uM) inhibited root growth and ROS production which was identified by DCFH fluorescence. Fourteen proteins out of 49, modulated by GSH treatment, were ROS-related proteins. GSH predominantly reduced expression levels of ROS-related proteins. However, an isoform of GST (NS43) and APX (NS65) were little affected. The differential modulations of ROS-related proteins among different isoforms by different ROS quenching chemicals suggest that ROS-related proteins may play either regulation of ROS levels or unknown specific functions depending on each reactive oxygen to maintain cellular redox homeostasis within cells during root growth.
References
Azhiri-Sigari T, Yamauchi A, Kamoshita A, Wade LJ (2000) Genotypic variation in response of rainfed lowland rice to drought and rewatering. Plant Prod Sci 3:180–188
Bevan M, Bancroft I, Bent E, Love K, Goodman H, Dean C, Bergkamp R, Dirkse W, van Staveren M, Stiekema W, Drost L, Ridley P, Hudson SA, Patel K, Murphy G, Piffanelli P, Wedler H, Wedler E, Wambutt R, Weitzenegger T, Pohl TM, Terryn N, Gielen J, Villarroel R, De Clerck R, Van Montagu M, Lecharny A, Auborg S, Gy I, Kreis M, Lao N, Kavanagh T, Hempel S, Kotter P, Entian KD, Rieger M, Schaeffer M, Funk B, Mueller-Auer S, Silvey M, James R, Montfort A, Pons A, Puigdomenech P, Douka A, Voukelatou E, Milioni D, Hatzopoulos P, Piravandi E, Obermaier B, Hilbert H, Dusterhoft A, Moores T, Jones JDG, Eneva T, Palme K, Benes V, Rechman S, Ansorge W, Cooke R, Berger C, Delseny M, Voet M, Volckaert G, Mewes HW, Klosterman S, Schueller C, Chalwatzis N (1998) Analysis of 1.9 Mb of contiguous sequence from chromosome 4 of Arabidopsis thaliana. Nature 391:485–488
Blokhina O, Virolainen E, Fagerstedt KV (2003) Antioxidants, oxidative damage and oxygen deprivation stress: a review. Ann Bot (Lond) 91:179–194
Blum H, Beier H, Gross HJ (1987) Improved silver staining of plant proteins, RNA, and DNA in polyacrylamide gels. Electrophoresis 8:93–99
Edwards R, Dixon DP, Walbot V (2000) Plant glutathione S-transferase: enzymes with multiple functions in sickness and in health. Trends Plant Sci 5:193–198
Fang YZ, Yang S, Wu G (2002) Free radical, antioxidants, and nutrition. Nutrition 18:872–879
Foreman J, Demidchik V, Bothwell JHF, Mylona P, Miedema H, Torres MA, Linstead P, Costa S, Brownlee C, Jones JDG, Davies JM, Dolan L (2003) Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 422:442–446
Gong H, Jiao Y, Hu WW, Pua EC (2005) Expression of glutathione-S-transferase and its role in plant growth and development in vivo and shoot morphogenesis in vitro. Plant Mol Biol 57:53–66
Halliwell B (2006) Reactive species and antioxidants. Redox biology is a fundamental theme of aerobic life. Plant Physiol 141:312–322
Herouatr D, Montagu MV, Inze D (1993) Redox-activated expression of the cytosolic copper/zinc superoxide dismutase gene in Nicotiana. Proc Natl Acad Sci 90:3108–3112
Hochholdinger F, Guo L, Schnable PS (2004) Lateral roots affect the proteome of the primary root of maize (Zea mays L.). Plant Mol Biol 2004:397–412
Hochholdinger F, Woll K, Guo L, Schnable PS (2005) The accumulation of abundant soluble proteins changes early in the development of the primary roots of maize (Zea mays L.). Proteomics 5:4885–4893
International rice genome sequencing project (2005) The map-based sequence of the rice genome. Nature 436:793–800
Jones AME, Thomas V, Truman B, Lilley K, Mansfield J, Grant M (2004) Specific changes in the Arabidopsis proteome in response to bacterial challenge: differentiating basal and R-gene mediated resistance. Phytochemistry 65:1805–1816
Joo JH, Bae YS, Lee JS (2001) Role of auxin-induced reactive oxygen species in root gravitropism. Plant Physiol 126:1055–1060
Kim ST, Cho KS, Jang YS, Kang KY (2001) Two-dimensional electrophoretic analysis of rice proteins by polyethylene glycol fractionation for protein arrays. Electrophoresis 22:2103–2109
Kim ST, Cho KS, Yu S, Kim SG, Hong JC, Han CD, Bae DW, Nam MH, Kang KY (2003a) Proteomic analysis of differentially expressed proteins induced by rice blast fungus and elicitor in suspension-cultured rice cells. Proteomics 3:2368–2378
Kim ST, Kim HS, Kim HJ, Kim SG, Kang SY, Lim DB, Kang KY (2003b) Prefractionation of protein samples for proteome analysis by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Mol Cells 16:316–322
Kim ST, Kim SG, Hwang DH, Kang SY, Kim HJ, Lee BH, Lee JJ, Kang KY (2004) Proteomic analysis of pathogen-responsive proteins from rice leaves induced by rice blast fungus, Magnaporthe grisea. Proteomics 4:938–949
Koller A, Washburn MP, Lange BM, Andon NL, Deciu C, Haynes PA, Hays L, Schieltz D, Ulaszek R, Wei J, Wolters D, Yates III JR (2002) Proteomic survey of metabolic pathways in rice. Proc Natl Acad Sci 99:11969–11974
Komatsu S, Konishi H (2005) Proteome analysis of rice root proteins regulated by gibberellin. Genomics Proteomics Bioinformatics 3:132–142
Kwak JM, Nguyen V, Schroeder JI (2006) The role of reactive oxygen species in hormonal responses. Plant Physiol 141:323–329
Leamml UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophgae. Nature 227:680–685
Liszkay A, van der Zalm E, Schopfer P (2004) Production of reactive oxygen inermediates (O2 •−, H2O2, and •OH) by maize roots and their role in wall loosening and elongation growth. Plant Physiol 136:3144–3123
Lopez-Bucio J, Cruz-Ramirez A, Herrera-Estrella L (2003) The role of nutrient availability in regulating root architecture. Curr Opin Plant Biol 6:280–287
Marrs KA (1996) The function and regulation of glutathione S-transferase in plants. Annu Rev Plant Physiol Plant Mol Biol 47:127–158
May MJ, Vernoux T, Leaver C, Montagu MV, Inze D (1998) Glutathione homeostasis in plants: implications for environmental sensing and plant development. J Exp Bot 49:649–667
Miller SS, Driscoll BT, Gregerson RG, Gantt JS, Vance CP (1998) Alfalfa malate dehydrogenase (MDH): molecular cloning and characterization of five different forms reveals a unique nodule-enhanced MDH. Plant J 15:173–184
Morita S, Suga T, Yamazaki K (1988) The relationship between root length density and yield in rice plants. Jpn J Crop Sci 57:438–443
Neuefeind T, Huber R, Dasenbrock H, Prade L, Bieseler B (1997) Crystal structrure of herbicide-detoxifying maize glutathione S-transferase-I in complex with lactoylglutathione: evidence for an induced-fit mechanism. J Mol Biol 274:446–453
O’Donnell BV, Tew DG, Jones OT, England PJ (1993) Studies on the inhibitory mechanism of iodonium compounds with special reference to neutrophil NADPH oxidase. Biochem J 290:41–49
Ostergaard O, Melchior S, Roepstorff P, Svensson B (2002) Initial proteome analysis of mature barley seeds and malt. Proteomics 2:733–739
Pullar JM, Hampton MB (2002) Diphenyleneiodonium triggers the efflux of glutathione from cultured cells. J Biol Chem 277:19402–19407
Reinemer P, Prade L, Hof P, Neuefeind T, Huber R, Zettl R, Palme K, Schell J, Koelln I, Bartunik HD, Bieseler B (1996) Three-dimensional structure of glutathione S-transferase from Arabidopsis thaliana at 2.2 Å resolution: structural characterization of herbicide-conjugating plant glutathione S-transferases and a novel active site architecture. J Mol Biol 255:289–309
Renew S, Heyno E, Schopfer P, Liszkay A (2005) Sensitive detection and localization of hydroxyl radical production in cucumber roots and Arabidopsis seedlings by spin trapping electron paramagnetic resonance spectroscopy. Plant J 44:342–347
Rodriguez AA, Grunberg KA, Taleisnik EL (2002) Reactive oxygen species in the elongation zone of maize leaves are necessary for leaf extension. Plant Physiol 129:1627–1632
Ros-Barcelo A, Pomar F, Lopez-Serrano M, Martinez P, Pedreno MA (2002) Developmental regulation of the H2O2-producing system and of a basic peroxidase isoenzyme in the Zinnia elegans lifnifying xylem. Plant Physiol Biochem 40:325–332
Schopfer P, Plachy C, Frahry G (2001) Release of reactive oxygen intermediates (superoxide radicals, hydrogen peroxide, and hydroxyl radicals) and peroxidase in germinating radish seeds controlled by light, gibberellin, and abscisic acid. Plant Physiol 125:1591–1602
Schroeder JI, Mori IC (2004) Reactive oxygen species activation of plant Ca2+ channels. A signaling mechanism in polar growth, hormone transduction, stress signaling, and hypothetically mechanotransduction. Plant Physiol 135:702–708
Sheffield J, Taylor N, Fauquet C, Chen S (2006) The cassava (Manihot esculenta Crantz) root proteome: protein identification and differential expression. Proteomics 6:1588–1598
Smirnoff N (1996) The function and metabolism of ascorbic acid in plants. Ann Bot 78:661–669
Smirnoff N (2000) Ascorbate acid: metabolism and functions of a multi-facetted molecule. Curr Opin Plant Biol 3:229–235
Suzuki H, Matsumori A, Matoba Y, Kyu BS, Tanaka A, Funita J, Sasayama S (1993) Enhanced expression of superoxide dismutase messenger RNA in viral myocarditis: SH-dependant reduction of its expression and myocardial injury. J Clin Invest 6:2727–2733
Tanaka N, Konishi H, Khan MMK, Komatsu S (2004) Proteome analysis of rice tissues by two-dimensional electrophoresis: an approach to the investigation of gibberellin regulated proteins. Mol Genet Genomics 270:485–496
Tanaka N, Mitsui S, Nobori H, Yanagi K, Komatsu S (2005) Expression and function of proteins during development of the basal region in rice seedling. Mol Cell Proteomics 4:796–808
Teo YH, Beyrouty CA, Norman RJ, Gbur EE (1995) Nutrient uptake relationship to root characteristics of rice. Plant Soil 171:297–302
Terashima K, OgataT, Akita S (1994) Eco-physiological characteristics related with lodging tolerance of rice in direct sowing cultivation. Jpn J Crop Sci 63:34–41
Ueda M, Matsui K, Ishiguro S, Sano R, Wada T, Paponov I, Palme K, Okada K (2004) The HALTED ROOT gene encoding the 26S proteasome subunit RPT2a is essential for the maintenance of Arabidopsis meristems. Development 131:2101–2111
Yang Y, Kwon HB, Peng HP, Shih MC (1993) Stress response and metabolic regulation of glyceraldehyde-3-phosphate dehydrogenase gene in Arabidopsis. Plant Physiol 101:209–216
Wingsle G, Karpinski G (1996) Differential redox regulation by glutathione of glutathione reductase and CuZn-superoxide dismutase gene expression in Pinus sylvestris L. needles. Planta 198:151–157
Acknowledgments
This work was supported by Grant No. CG1122 from the Crop Functional Genomic Center; by a grant from KOSEF/MOST to the Environmental Biotechnology National Core Research Center (to S.G. Kim and S.T. Kim); and by scholarships from the Brain Korea 21 Program, Ministry of Education and Human Resources Development, Korea (to S.G. Kim, Y. Wang).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by W.T. Kim.
Rights and permissions
About this article
Cite this article
Kim, S.G., Kim, S.T., Kang, S.Y. et al. Proteomic analysis of reactive oxygen species (ROS)-related proteins in rice roots. Plant Cell Rep 27, 363–375 (2008). https://doi.org/10.1007/s00299-007-0441-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-007-0441-5