Abstract
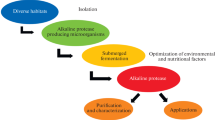
Alkaline proteases are well known to be significant industrial enzymes. This study focused on isolating the fungus producing proteases from, a typical Ethiopian food, Injera. Further, the process optimization for protease production using response surface methodology (RSM) and the characterization of the acquired protease were investigated. The 18S rDNA gene sequence homology of the fungus isolate revealed that it was Aspergillus sp. Further, it was deposited in NCBI GenBank with accession number MK4262821. Using the isolate, owing to maximize the protease production, the independent process parameters, temperature, pH, and sucrose concentration were optimized using RSM followed by a genetic algorithm (GA). Based on the statistical approach by RSM-GA optimization, maximum enzyme activity (166.4221 U/ml) was found at 30.5 °C, pH 8.24, and 0.316% sucrose concentration. Also, the crude cocktail of enzymes acquired from optimal condition was partially purified using ammonium which showed that the increased activity by 1.96-fold. Considerably, the partially purified enzyme exhibited stable performance during the temperature range 30–60 °C, pH range 7–10, and NaCl concentration of 0.5–2 mM. Also, the antioxidant activity, degree hydrolysis for the protein, Michaelis–Menten (M–M) kinetic parameters, and activation energy were determined for the obtained enzyme cocktail. It showed that the M–M kinetic parameters, Km (5.54 mg/ml), and Vmax (24.44 mg/ml min) values were observed. Using Arrhenius law, the value of activation energy for the enzyme cocktail was determined as 32.42 kJ/mol.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Nowadays, the applications of proteases in production industries have become markedly increasing. Proteases (EC: 3.4.21–24 and 99; peptidyl peptide hydrolases) are enzymes that catalyze the cleavage of the peptide bond in proteins (CO–NH), liberating small chains of amino acids called peptides [1]. Proteases are commonly obtained from diversified sources such as plants, animals, and microorganisms. Microbial proteases are among the most important hydrolytic enzymes that have been studied extensively due to their fast growth, economical advantage, and the possibility of genetic manipulation [2]. Alkaline protease accounts for at least 25% of the total worldwide sales of enzymes, of which two-thirds of proteases produced commercially are of microbial origin [3]. They have a long history of applications in different industries, such as waste management, silver recovery, detergent, dairy, baking, textile, leather, and pharmaceuticals [4]. Fungi proteolytic enzyme is isolated from several natural and artificial environmental sources, and they are so far the most important group of enzymes produced commercially because they can produce a high amount of protease that have various applications and also the downstream processing is easy [2, 5]. As protease is gaining importance for its technical and economic advantages in various industries, finding and developing an optimized process for the production of protease enzyme will add significant potential advantage in the bioprocessing industries. Thus, this research validates the need to explore native fungal isolate screened from a decaying Injera, Ethiopian traditional food, as a source for isolation of fungal proteases enzyme. The influence of sucrose concentration, temperature, and pH on the production of protease was determined via submerged fermentation and the variables have been optimized through response surface methodology (RSM). Response surface methodology (RSM) is a proven and effective tool for sequential analysis to study process modeling and optimization. It gives sufficient information to test the interaction effects among the process variables with a reasonable number of experiments [6,7,8]. Additionally, it uses mathematical and analytical means to identify interrelationships between numerical input variables to obtain their optimum values [8]. Proteolytic activity was determined against temperature, pH, and NaCl concentration, since numerous biotechnological applications of proteases necessitate salt stability thermal resistance and stability at alkaline pH.
Materials and Methods
Materials and Chemicals
Unless specified otherwise, all fine chemicals and media components were obtained from HiMedia Laboratories Private Limited (Addis Ababa, Ethiopia) and Merck (Addis Ababa, Ethiopia), and were of analytical reagent grade. Sucrose was sourced from Sigma Chemical (St. Louis, MO).
Isolation and Identification of Protease-Producing Fungus
With some modification to the method of Chauhan et al. [9], different fungi were isolated from rotting or decaying Injera. A fresh Injera was prepared and kept open at room temperature for 5 days to produce a moldy decaying Injera. Moldy samples were collected from the surface of the decaying Injera and the dilution plate method was used to grow fungi on potato dextrose media. Serially diluted samples were plated on potato dextrose agar media and the petri dishes were kept at 30 °C in an incubator for 5 days until mycelial growth was observed. The grown fungi were microscopically identified using the lactophenol cotton blue staining method. Identification of fungal species was carried out using available reports and reference microscopic slides [9]. Further, the isolated fungi were screened for their proteolytic activities. Protease-producing fungi were identified by the presence of a clear zone in the skimmed milk agar around the colonies [10]. Skimmed milk agar (SMA) media containing casein was also used for screening purposes. The fungi showing the presence of a clear zone around the colony were considered as a positive for further characterization of protease enzyme source. The fungal colony from the largest zone of clearance was selected for further studies.
Molecular Identification of Fungi
A pure fungus isolate that had a high ability to hydrolyze casein in the skimmed milk agar (SMA) plate was identified via 18S rRNA gene sequencing [11]. For this purpose, mycelia were collected and centrifuged at 4000 rpm for 15 min at 4 °C. Further, the DNA was extracted using the protocol given by the manufacturer of the GeneJET Genomic DNA Purification Kit. The purified DNA was subjected to PCR amplification using a pair of 5′-TCCGTA GGTGAACCTGCGG-3′ and 5′-TCCTCCGCTTATTGATATGC-3′ forward and reverse primers for 18S rRNA gene amplification and sequencing [12]. The sequence data were investigated in the GenBank database using the BLAST program available in the National Center for Biotechnology Information. The obtained unknown sequence was compared to all of the sequences available in the database to assess DNA similarities. Then, the nucleotide sequence of the isolated fungus was submitted to the GenBank and obtained accession number, MK4262821.
Inoculum Preparation and Determination of Central Level
With some modification to the existing method given by Chimbekujwo et al. and Anandan et al. [13, 14], the inoculum was prepared by scattering the spores from a 7-day-old cold fungal slant culture in 0.1% Tween 80 solution with a sterile inoculation loop in aseptic condition. The suspension was adjusted with sterile distilled water until the optical density at 530 nm was 1.5–1.6 and it is labeled as the seed culture. Before proceeding with Central Composite Design (CCD), one-variable-at-a-time (OVAT) experiments were carried out to know the central level of the three parameters (sucrose concentration, temperature, and pH), which were used to study the interaction effect. All OVAT experiments were conducted in triplicates and their mean values were assessed. Based on this, the independent sequence of protease production was carried out by adjusting to the initial pH 7 and varying sucrose concentration while keeping fixed level of temperature. Similarly, OVAT experiments were applied to determine the central level of other parameters. The parameters were set in the range of 0.1 to 0.6% (w/v) for sucrose concentration, 5–10 for initial pH value, and 25–50 °C for temperature. Then, protease production was done by adding 2 ml of the seed culture in 200 ml Erlenmeyer flask containing 100 ml of sterilized medium containing (0.5 g casein, 0.05 g yeast extract, 1 g K2HPO4, 0.2 g MgSO4.7H2O, 0.25 NaCl, 0.05 g peptone, and 100 ml distilled water) [15]. Finally, after mixing, it was gently shaken at 30 °C and 200 rpm in a shaking incubator for 3 days [16]. The culture medium was centrifuged at 5000 rpm for 20 min at 4 °C to remove the fungal mycelia and medium debris present. The supernatant was collected as a crude enzyme solution and was used to estimate the protease activity.
Proteases Enzyme Assay
Protease activity was measured using casein digestion method of Keay and Bernard S. Wildi [17]. Adopting this method, 1 ml of culture supernatant, which was pre-incubated at 30 °C for 2 min, further, was mixed with 1 ml of 1% (w/v) casein in 0.1 M phosphate buffer (pH 7). The reaction was incubated for 10 min at 30 °C and it was terminated by the addition of 2 ml of 0.4 M trichloroacetic (TCA) acid. Then the solution was incubated for 10 min at room temperature followed by centrifugation at 5000 rpm at 4 °C for 10 min to remove any insoluble from the samples. The filtration of 1 mL of the samples was mixed with 5.0 ml of 0.4 M Na2CO3 and for best results; 1 mL of Folin’s reagent (1 ml of Folin reagent mixed with 2 ml of distilled water) was added immediately afterward. Sodium carbonate was added to regulate any pH drop created by the addition of the Folin’s reagent. The tubes were allowed to stand for 20 min at 30 °C and then the absorbance was measured at 660 nm in UV–Vis Spectrophotometer. The same procedure was used to prepare the blank; however, 1 ml of 1% (w/v) casein was added only after the reaction was stopped. One unit of protease activity was defined as the amount of enzyme that released 1 μg of amino acid equivalent to tyrosine per min under the assay conditions.
Protease Production Based on Central Composite Design Experiments
To investigate the interaction effect of temperature, pH, and sucrose concentration on the production of the protease enzyme, the experiment design was decided using the central composite design (CCD) method. Temperature, pH, and sucrose concentrations were studied at five levels with six replicates at a midpoint with a total of twenty experimental runs. A second-order polynomial equation was applied for the analysis of protease enzyme activity and the data were fitted in the equation by multiple regression procedure. The model equation (Eq. 1) was used to generate the counterplot:
where Y (protease activity U/ml) is the predicted response and A, B, and C are the temperature, pH, and sucrose concentrations, respectively. β0 is an intercept; β1, β2, and β3 are linear coefficients; β11, β22, and β33 are squared coefficients; and β12, β13, and β23 are interaction coefficients [18]. Table 1 shows the levels of the three independent variables studied. The levels of the variables were set based on the information obtained from the earlier OVAT characterization study.
The Statistical Analysis
ANOVA (analysis of variance) was used to estimate the statistical parameters. The optimum values of the selected variables were obtained by solving the regression equation and by analyzing the response surface contour plots. The plotting of the response surface was performed using Design-Expert 7.0 software.
Validation of the Model
The validity of the chosen quadratic model, predicted by the point prediction feature of Design-Expert 7 software, was confirmed experimentally [6]. The experiments were run in triplicate.
Partial Purification of the Crude Enzyme
Protein precipitation was done by salting out using ammonium sulfate fractionation with uniform and gentle stirring. The crude enzyme was saturated up to 40–80% ammonium sulfate, which was added slowly under continuous stirring at 4 °C. The precipitated protein was separated by centrifugation at 5000 rpm and 4 °C for 15 min. The resultant pellets were dissolved in 0.5 M potassium phosphate buffer, which was then dialyzed for 24 h at 4 °C in a refrigerator to remove excess salt. This was followed by centrifugation of the resulting solution at 5000 rpm and 4 °C and the supernatant was tested for protease activity [19].
Characterization of Partially Purified Protease
By assaying the enzyme at different pH values, the activity and stability of protease were examined. The buffers used were phosphate (pH 5.0–8.0), glycine–NaOH (pH 8.5–10.5), and NaHCO3–NaOH (pH 9.6–12); the concentration of glycine–NaOH was 0.1 M, which was within 0.5 differences using 1% casein as substrate. The optimal temperature of the enzyme activity and stability was investigated by following the protease assay method as described by Keay and Wildi [17], using 1% casein (w/v) as a substrate in 0.5 M phosphate at pH 8. The experiment was conducted by incubating the reaction mixture at temperatures ranging from 30 to 80 °C with 10 °C interval. The temperature stability of the enzyme was determined by measuring the residual activities by following the modified procedure of Pant et al. [20]. The enzyme was appropriately diluted with 0.5 M phosphate (pH 8) and then the aliquot was incubated at temperatures ranging from 45 to 65 °C for 1 h in the incubator. The ionic strength of the alkaline protease was determined by adding 0–3 m M (within 0.5 mM interval) of NaCl to the reaction mixture and then by carrying out a standard protease assay. As per the procedure described by Phrommao et al. [21], the stability of protease in the different concentrations of NaCl was examined by preparing 0–3 mM NaCl (within 0.5 mM intervals).
Determination of Antioxidant Activity and Degree Hydrolysis
The antioxidant activity of the observed enzymes was determined using 2,2-diphenyl-1-picrylhydrazyl (DPPH) with the radical scavenging method as described by Bougatef et al. [22]. A volume of 50 μL of the proteases was mixed with 50 μL of 99.5% ethanol and 30 μl of 0.02% DPPH in 99.5% ethanol. The mixture was kept at room temperature in the dark for 60 min. The reduction of the DPPH was measured at 540 nm using a microplate reader (Diatek Instruments, DR-200BModel). The DPPH radical scavenging activity was determined using Eq. 2:
The degree of hydrolysis of the proteases was calculated as described by Adler-Nissen [23]. Mixtures of 0.5 mL of diluted proteolytic enzymes and 0.250 mL of 0.01% (w/v) 2,4,6-trinitrobenzene sulfonic acid (TNBSA) in 0.1 M sodium bicarbonate pH 8.5 were incubated for 30 min at 37 °C. A volume of 200 μl of each mixture was taken and the absorbance measured at 415 nm using a microplate reader (Diatek Instruments, DR-200B Model). The degree of hydrolysis (DH) value can be calculated using Eq. 3:
where p is the number of hydrolyzed peptide bonds and ptot is the total number of peptide bonds per protein equivalent (the value of ptot for casein and gelatin are 8.2 and 11.1, respectively). The P-value can be calculated as the relationship given below (Eq. 4):
where [cysteine-NH2]—milliequivalents of cysteine amine groups per gram of protein (Eq. 5):
where X is the weight of protein sample (g), V is the volume reaction solution (L), and f is the dilution factor. For casein, α = 1.039 and β = 0.383. For gelatin, α = 0.796 and β = 0.457, can be considered [24].
Michaelis–Menten (M–M) Kinetics
Owing to its significant commercial applications, it is important to investigate the kinetic characterization of observed enzymes. Determination of kinetic parameters, Km, and Vmax provides a clear picture for designing enzymatic reactors [24, 25]. To investigate the kinetic parameters, a known concentration of enzyme (1 mL) was added with different concentrations of casein [0, 2, 4, 8, and 10 (mg/mL)] as substrate. The reaction mixture was incubated at predetermined conditions (pH 8 using 1% w/v in 50 mM Tris–HCl buffer and 45 °C). Protease activities were measured as described earlier. Km and Vmax values were calculated using Eq. 6:
where V is reaction rate and S is substrate concentration.
Based on the Lineweaver–Burk (L–B) plot, the graph of [1/V] vs [1/S] was being made to determine Km and Vmax [24, 25].
Determination of Activation Energy
The value of activation energy (Ea) for the proteases produced by Aspergillus sp. was determined by incubating enzymes with 1% casein at different temperatures ranging from 30 to 50 °C [25, 26]. Arrhenius Law (Eq. 7) was used to determine the value of Ea from the slope of the plot of [1/T] vs [ln (protease activity)], where Ea = − slope × R. R (Gas constant) = 8.314 (J/Kmol), and T refers absolute temperature (K) [25, 26]
Results
Screening Based on the Proteolytic Activity of Fungi
Screening of the four fungi isolates showed a positive result on SMA. The zone of clearance (mm) of each isolated fungus is given in Table 2. Strain coded S-1 was selected for further study to produce protease by submerged fermentation due to the maximum capacity of hydrolyzing the casein present in the SMA (Fig. S1).
Molecular Identification of the Fungus
The 18S rRNA sequencing data revealed that the strain was Aspergillus species. Its sequence was deposited in the GenBank database with accession number MK426282 and coded as SVP03.
Response Investigation and Interpretation Study
The combined effect of the parameters was studied at an incubation time of 3 days with 200 rpm shaking speed in the shaker incubator. The experiment was conducted using CCD, and the experimental and predicted results for enzyme activity are given in Table 3.
Statistical Analysis
Table 4 lists other statistics used to compare models. The statistical summary for each model is given in Table 4. A quadratic model in comparison to a cubic model is suggested because it has a higher value of adjusted R2 and predicted R2 and the cubic model is aliased. Table 5 shows the outcomes of the second-order response surface model fitting in the form of ANOVA. The lack-of-fit of the model is not significant with an F-value of 1.41 and Prob > F of 0.3582 indicates the experimental data were accurately fitted by the model. Moreover, the goodness-of-fit of the model was tested by analyzing the coefficients R2, Adj R2, and Pred R2.
In this case, R2 is 0.9888, which indicates a high correlation between the experimental and predicted values and demonstrates that only 1.12% of the total variation was not explained by the fitted model. The Pred R2 of 0.94 is in reasonable agreement with the Adj R2 of 0.97. The obtained ratio of 26.244 indicates an adequate signal. The coefficient of variation is 4.89%. Table 6 shows the P-value and F-value of the variables included in each model. Values of Prob. > F less than 0.05 indicates model terms are significant. The regression model prepared to predict the production of protease enzyme (enzyme activity) by the isolate Aspergillus sp. is given in Eq. 8 in terms of coded values. The second-order polynomial model was used to express the enzyme activity (Y) as a function of independent variables.
Interaction Study
A three-dimensional (3D) response surface curve was plotted to understand the interaction effect of the three components required for maximizing the yield of protease enzyme production. The response surfaces obtained were convex, signifying that there were distinct optimum operating situations. Figure 1a shows the effect of temperature and pH on the production of protease. It was observed that the production of protease was increased on increasing both temperature and pH, and high activity was noted at the pick point. The peak point of the activity was declined on further increasing temperature and pH. However, as shown in Fig. 1a, the red color indicates the optimum range for both of them for protease production.
Figure 1b shows the effect of temperature and sucrose concentration on the mean pH level. As shown in the figure, the maximum enzyme activity was obtained at the midpoint of the design factors, whereas at the extreme point of interaction low level of enzyme activity was recorded. Figure 2c shows the response surface plot with the interaction effect of pH and sucrose concentration at fixing the temperature on its midpoint. For both factors, as we go from the lower level to the midpoint, an increase in the enzyme activity was observed, and maximum protease production was obtained at the mean of both sucrose concentration and pH. However, when the level values increase from the average value, the production of enzyme was declined. A genetic algorithm (GA) is used to solve both constrained and unconstrained optimization problems. It is developed based on the principles of natural genetics and natural selection. This algorithm follows Darwin’s theory of survival of the fittest and works by mimicking the conditions of the natural environment in which the process of evolution occurs. The best point in the population evolves toward the best solution. In this study, the RSM model was used as the fitness function for GA to optimize the maximum enzyme activity (U/ml). The parameters used in GA-based optimization are chromosome length = 30, population size = 30, crossover fraction = 0.8, mutation scale = 0.2, and the number of generations = 50. The GA toolbox of MATLAB 8.1 (The Mathworks, Inc., MA, USA) was used in the optimization studies. The fitness value vs generation and current best individual vs a number of variables obtained from the genetic algorithm are illustrated in Fig. S2. The best fitness and mean fitness values were found to be the same and it corresponds to the enzyme activity of 166.971%. The optimal parameter values were found to be sucrose concentration 0.316% w/v, initial pH of 8.24, and temperature 30.5 °C.
Validation of the Model
After optimization, triplicate experiments were performed using the predicted optimized process conditions. At these conditions, the mean percentage of enzyme activity 166.184 U/ml was obtained.
Partial Purification of the Crude Enzyme
The protease from Aspergillus sp. SPV03 was precipitated using ammonium sulfate and the active pellet was dialyzed. The purified protease showed a 1.96-fold increase in enzyme activity from the crude enzyme.
Characterization of the Partially Purified Enzyme
Effect of pH on the Activity and Stability
Aspergillus sp. SPV03 protease was active in a broad pH range of 5.5–12. It showed more than 87% of its maximum activity at a pH range of 6.0–11.5, with optimum activity at pH8, as indicated by the peak shown in Fig. 2a. The activity of protease from Aspergillus sp. SPV03 was found to increase gradually with the increase in the pH range from 5.0 to 8.0 and a slight drop-in activity beyond pH 8.0. The strain showed its maximum protease activity 94.26%, 98.62%, 97.93%, 95.63%, and 92.74% at pH values 7, 7.5, 8.5, 9, and 9.5, respectively. The protease activity sharply fell at pH above 11. Minimum relative activity was noted at pH 5 (20.056%) and pH 12.0 (40.47%). The stability profile of the protease enzyme shows an increment from pH 5.0 to 8.0 and a stability zone from the pH value of 7.0 to 9.5 shows more than 91% residual activity was retained after 1 h of incubation. Beyond pH 8.0, it starts to decrease and reaches 92.68% at pH 8.5. In general, the protease enzyme was stable between pH 7.5 and 9.0 (0.5 intervals) and reached 94.65%, 99.25%, 92.63%, and 90.60% of its residual activity. Its minimum residual activity was attained at a pH value of 5.0, that is, 11.24% of its original activity. As shown in Fig. 2b, alkaline protease enzyme in assaying buffers shows a stability profile between pH values of 7.0 and 10.0.
Effect of Temperature on Enzyme Activity and Stability
The protease enzyme was found to be very active within a wide temperature range between 30 and 60 °C with maximum relative activity at 50 °C, as shown as the peak shown in Fig. 3a. Beyond 60 °C, the relative activity declined sharply with a minimum relative activity of 31.47% at 80 °C.
Thermal stability of alkaline protease was tested at pH 8.0, as shown in Fig. 3b. At 50 °C, it shows high thermal stability with more than 90% of residual activity within 1 h of incubation. The protease showed thermal stability within an incubation time range of 10–40 min greater than 80% of its residual activity for assaying at 30–60 °C. A temperature beyond 60 °C has low thermal stability.
Effect of NaCl Concentrations on Enzyme Activity and Stability
Maximum relativity activity of proteases was found at 1 mM of NaCl, as shown in Fig. 4a. From 0.5 to 2 mM NaCl, the relative activity was more than 85%. At high concentration (> 2 mM) of NaCl, its activity sharply declined and the least activity was observed at 3 mM (8.01%) NaCl. Maximum stability after 1 h of incubation was obtained at 1 mM NaCl. More than 86% stability was achieved in the range of 0.5–2 mM NaCl concentration as shown in Fig. 4b. An optimal protease activity founded at 10% (w/v) NaCl.
Antioxidant Activity and Degree Hydrolysis
To explore the potential of the partially purified proteases obtained from Aspergillus sp. for protein hydrolysis, casein and gelatin were used. As given in Fig. 5a, the results showed the degree of hydrolysis (DH) of casein was significantly higher than gelatin. For the DPPH assay, the gelatin and casein hydrolyzates prepared with the proteases obtained from Aspergillus sp. showed the greatest radical scavenging as 66% in the casein hydrolyzates (Fig. 5b).
Kinetic Parameters and Activation Energy
An L-B plot was used to calculate the kinetic constants (Fig. 5c). From the observations, it was found that the Km and Vmax values of acquired enzymes were 5.54 mg/mL and 24.44 mg/mL min, respectively. The Arrhenius plot (Fig. 5d) within the temperature range from 30 to 50 °C exhibits a linear relationship. From the Arrhenius plot, the activation energy was calculated to be 32.42 kJ/mol that necessary to conduct the hydrolysis of casein.
Discussion
The current study demonstrated the isolation of fungus that produces protease enzyme from decayed injera and it also demonstrates the enhancement of protease production via using RSM. Stability of protease at alkaline pH, higher temperature, and with different concentrations of salt are preferred at different industry sectors, such as detergent formation. This study revealed the utilization of decayed injera for isolation of protease-producing fungi and the characterization of the enzyme.
The 18S rRNA sequencing data discovered that the fungal strain that had maximum inhibition zone in the SMA was the Aspergillus species. The most successful method to identify the exact species of microbes is to use PCR amplification of target sequences within the rRNA gene complex [27, 28].
The regression model obtained after the ANOVA indicated that the value of the quadratic model is highly significant (P-value < 0.0001). A large R2 value shows that the regression has accounted for a large proportion of the total variability in the observed value of Y that favors the regression equation model [7]. Adeq precision measures the signal-to-noise ratio, and a ratio greater than 4 is desirable [6]. This model can be used to navigate the design space. PRESS (predicted sum of squares) measures how well a particular model fits each point in the design and a higher value demonstrates the fitness of the model. Low values of coefficient of variation (CV% 4.89) show reliability and good precision of the experiments because it is the error expressed as a percentage of the mean. Values of Prob. > F less than 0.05 indicates model terms are significant. The P-values greater than 0.10 indicate the model terms are not significant [6].
Different studies conducted on the effect of carbon source on protease production reported that sucrose gives a high yield of protease production [29, 30]. Temperature influences both the growth rate and the amount of protease production because a microorganism’s metabolism and the fermentation process is affected by the environmental temperature [31, 32]. Most fungi protease are temperature sensitive; therefore, they have reduced protease production activities at extreme temperature [3]. A higher temperature is found to have some opposing effects on the metabolic activities of microorganisms and inhibits the growth of fungus [33]. Other studies on protease also confirm our study result, which reported a maximum protease production at 30 °C [34,35,36]. Different studies [37, 38], reported similar results as obtained in the present study on optimal pH for the production. In a little deviation, D. G. Franco et al. [39] reported that the peak pH for protease production was 6.5. Thus, the experimental results showed that protease production is affected by the production temperature and depends on the strain. The activities of protease-producing fungi and other microbes depend on the pH of the culture because it strongly affects many enzymatic processes and the transport of various components across cell membranes [40]. The validation analysis results corroborated the predicted values and hence reflected the applicability of RSM-GA to optimize the process of enzyme production, showing the effectiveness of the model.
Most of the industrial alkaline process works at alkaline pH (8 to 10). Thus, Aspergillus sp. SPV03 could be used for these industrial applications. In this regard, proteases with similar biochemical characteristics have been described for A. flavipes NRRL 295 [41]. Furthermore, similar studies were also obtained for protease activity from Aspergillus fumigatus TKU003, Aspergillus ustus, Aspergillus flavus AP2, and Aspergillus nidulans HA-10 [38].
The enzyme was found to be thermally unstable above the incubation time of 40 min at 60 °C. This phenomenon could be due to the denaturation of the enzyme at high temperatures for a long exposure. The temperature and stability profile of the purified protease confirmed that the enzyme was thermostable. Consequently, this enzyme could find prospective application in commercial, such as in food processing agent as most food industries, such as brewing and baking, that use proteases need their processes to be carried out at temperatures nearly 50 and 60 °C [15]. Prior studied data for the characterization of proteases confirmed 40 °C as optimal temperature for protease activity from Aspergillus Parastics [42] and Conidiobolus Coronatus [43].
Numerous metal ions have been described to have an impact on protease activity. The study done by Racheal et al. [15] stated that the enhancement of the activity of protease using NaCl salt. Only minimal enzymatic activity was manifest in the time off of salt [44].
A similar performance of DH has been observed for casein hydrolysis using an immobilized A. oryzae protease [45]. DPPH radical is a stable free radical. In ethanol, it shows maximal absorbance at 517 nm. It has been widely used to test the ability of natural compounds to act as free radical scavengers. This can be considered for assessing antioxidant potentials. The hydrolysis might be releasing the antioxidant peptides increasing the antioxidant activity of the enzymes [41].
The kinetic constants, Km and Vmax, designate the affinity of an enzyme with substrate. The Km value helps get the relation between the rates of product formation from the availability of substrates relative to substrate concentration [41]. The Km of an enzyme is an inverse measure of its affinity to the substrate. The Vmax value is a characteristic feature of an enzyme at a defined concentration of substrate. A previously studied data by Murthy and Naidu reported Km values of 3 mg/ml for a protease form A. oryzae strains [46]. Additionally, Janser et al. stated a wide range of Km (0.44–1.92 mg/mL) when Aspergillus niger was grown on different substrates [47]. Arrhenius plot suggested that this cocktail of proteolytic enzymes have single conformations up to the transition temperatures. Ortiz et al. [41] reported activation energy of the 31.66 kJ/mol for azo-casein hydrolysis using proteases from A. giganteus NRRL 10 within a temperature range of 30–46 °C. This value was similar to the protease obtained from Aspergillus sp. that is extracted from the rotten Injera.
Conclusion
In this study, the proteases-producing fungi, which can be derived from decaying Injera was isolated and identified as Aspergillus sp. Using the isolate, the protease production was optimized for temperature, pH, and sucrose concentrations by RSM and GA using CCD design. The optimum condition for the production of protease enzyme was found to be a temperature of 30.5 °C, pH 8.24, and 0.316% (w/v) sucrose concentration with an enzyme activity of 166.4221 U/ml. Maximum enzyme stability was obtained at pH 8.0, and the alkaline protease was found to be very active at temperatures between 30 and 60 °C, with maximum stability at 50 °C. It was stable at 0.5–2 mM NaCl concentration. Further, the antioxidant activity and degree hydrolysis for the protein hydrolyzates were calculated. From the M–M kinetics, the parameters, Km and Vmax values, were observed to be 5.54 mg/ml and 24.44 mg/ml min, respectively. Using Arrhenius law, the value of activation energy for the acquired enzyme cocktail was determined as 32.42 kJ/mol. It is well proven that the cocktail of the protease enzyme has high potential value in bio-based industries. However, it is required to investigate indifferent aspects, such as process optimization in small-scale bioreactor and use of other cheap alternative energy sources.
Data Availability
The DNA sequence can be found at repository of GenBank, the link given below. The experimental data observed have been presented in terms of graphical illustrations. MK426282.
References
Brix K, Stöcker W (2013) Proteases: structure and function. Springer, Vienna
Tremacoldi CR, Carmona EC (2005) Production of extracellular alkaline proteases by Aspergillus clavatus. World J Microbiol Biotechnol 21:169–172. https://doi.org/10.1007/s11274-004-2724-0
Rao MB, Tanksale AM, Ghatge MS, Deshpande VV (1998) Molecular and biotechnological aspects of microbial proteases. Microbiol Mol Biol Rev 62:597–635
Nadeem F, Mehmood T, Naveed M, Shamas S, Saman T, Anwar Z (2020) Protease production from Cheotomium globusum through central composite design using agricultural wastes and its immobilization for industrial exploitation. Waste Biomass Valoriz 11:6529–6539. https://doi.org/10.1007/s12649-019-00890-9
De Souza PM, de Assis M et al (2015) A biotechnology perspective of fungal proteases. Braz J Microbiol 46:337–346. https://doi.org/10.1590/S1517-838246220140359
Kidus Tekleab W, Beyan SM, Balakrishnan S, Admassu H (2020) Chicken feathers based keratin extraction process data analysis using response surface-box-Behnken design method and characterization of keratin product. Curr Appl Sci Technol 20:163–177. https://doi.org/10.14456/cast.2020.6
Venkatesa PS, Girma G, Gizachew AK, Surafel BM, Ramesh G (2019) Biosolubilization of Cr (VI) from tannery sludge: process modeling, optimization, rate kinetics and thermodynamics aspects. Int J Recent Technol Eng 8:4808–4816. https://doi.org/10.35940/ijrte.d7771.118419
Puri S, Beg QK, Gupta R (2002) Optimization of alkaline protease production from Bacillus sp. by response surface methodology. Curr Microbiol 44:286–290. https://doi.org/10.1007/s00284-001-0006-8
Chauhan NM, Gutama AD, Aysa A (2019) Endophytic fungal diversity isolated from different agro-ecosystem of Enset (Ensete ventericosum) in Gedeo zone, SNNPRS, Ethiopia. BMC Microbiol 19:1–10. https://doi.org/10.1186/s12866-019-1547-y
Omrane Benmrad M, Mechri S, Zaraî Jaouadi N et al (2019) Purification and biochemical characterization of a novel thermostable protease from the oyster mushroom Pleurotus sajor-caju strain CTM10057 with industrial interest. BMC Biotechnol 19:1–18. https://doi.org/10.1186/s12896-019-0536-4
Hinrikson HP, Hurst SF, Aguirre LDE, Morrison CJ (2005) Molecular methods for the identification of Aspergillus species. Med Mycol Suppl 43:129–137. https://doi.org/10.1080/13693780500064722
Raja HA, Miller AN, Pearce CJ, Oberlies NH (2017) Fungal identification using molecular tools: a primer for the natural products research community. J Nat Prod 80:756–770. https://doi.org/10.1021/acs.jnatprod.6b01085
Chimbekujwo KI, Ja’afaru MI, Adeyemo OM (2020) Purification, characterization and optimization conditions of protease produced by Aspergillus brasiliensis strain BCW2. Sci Afr 8:e00398. https://doi.org/10.1016/j.sciaf.2020.e00398
Anandan D, Marmer WN, Dudley RL (2007) Isolation, characterization and optimization of culture parameters for production of an alkaline protease isolated from Aspergillus tamarii. J Ind Microbiol Biotechnol 34:339–347. https://doi.org/10.1007/s10295-006-0201-5
Racheal OO, Ahmed ATF, Ndigwe EV, Morakinyo SD (2015) Extraction, purification and characterization of protease from Aspergillus Niger isolated from Yam Peels. Int J Nutr Food Sci 4:125. https://doi.org/10.11648/j.ijnfs.20150402.11
dos Santos Aguilar JG, Sato HH (2017) Microbial proteases: production and application in obtaining protein hydrolysates. Food Res Int 103:253–262. https://doi.org/10.1016/j.foodres.2017.10.044
Keay L, Wildi BS (1970) Proteases of the genus Bacillus. I. Neutral proteases. Biotechnol Bioeng 12:179–212. https://doi.org/10.1002/bit.260120205
Hussain F, Kamal S et al (2017) Alkaline protease production using response surface methodology, characterization and industrial exploitation of alkaline protease of Bacillus subtilis sp. Catal Lett 147:1204–1213. https://doi.org/10.1007/s10562-017-2017-5
Shankar S, Rao M, Laxman RS (2011) Purification and characterization of an alkaline protease by a new strain of Beauveria sp. Process Biochem 46:579–585. https://doi.org/10.1016/j.procbio.2010.10.013
Pant G, Prakash A et al (2015) Production, optimization and partial purification of protease from Bacillus subtilis. J Taibah Univ Sci 9:50–55. https://doi.org/10.1016/j.jtusci.2014.04.010
Phrommao E, Yongsawatdigul J, Rodtong S et al (2011) A novel subtilase with NaCl-activated and oxidant-stable activity from Virgibacillus sp. SK37. BMC Biotechnol 11:65. https://doi.org/10.1186/1472-6750-11-65
Bougatef A, Hajji M, Balti R, Lassoued I, Triki-ellouz Y, Nasri M (2009) Antioxidant and free radical-scavenging activities of smooth hound (Mustelus mustelus) muscle protein hydrolysates obtained by gastrointestinal proteases. Food Chem 114:1198–1205. https://doi.org/10.1016/j.foodchem.2008.10.075
Adler-nissen J (1979) Determination of the degree of hydrolysis of food protein hydrolysate by trinitrobenzensulfonic acid. J Agric Food Chem 27:1256–1262. https://doi.org/10.1021/jf60226a042
Nielsen PM, Petersen D, Dambmann C (2001) Improved method for determining food protein degree of hydrolysis. J Food Sci 66:642–646. https://doi.org/10.1111/j.1365-2621.2001.tb04614.x
Shankar J, Lall BM, Jadhav SK, Tiwari KL (2017) Parameter’s optimization and kinetics study of α-amylase enzyme of Bacillus sp. MB6 isolated from vegetable waste. Process Biochem 52:123–129. https://doi.org/10.1016/j.procbio.2016.10.005
Melikoglu M, Lin CSK, Webb C (2013) Kinetic studies on the multi-enzyme solution produced via solid state fermentation of waste bread by aspergillus awamori. Biochem Eng J 80:76–82. https://doi.org/10.1016/j.bej.2013.09.016
Souza PM, Aliakbarian B, Filho E et al (2015) Kinetic and thermodynamic studies of a novel acid protease from Aspergillusfoetidus. Int J Biol Macromol 81:17–21. https://doi.org/10.1016/j.ijbiomac.2015.07.043
Bhargavi SD, Praveen VK, Anil Kumar M, Savitha J (2018) Comparative study on whole genome sequences of Aspergillus terreus (soil fungus) and Diaporthe ampelina (Endophytic Fungus) with reference to lovastatin production. Curr Microbiol 75:84–91. https://doi.org/10.1007/s00284-017-1353-4
Sinha P, Singh RK, Srivastva R, Sharma R, Tiwari SP (2013) Characterization and optimization of alkaline protease enzyme produced by soil borne bacteria. Trends Life Sci (Solapur, India) 2:38–46
Wang HT, Hsu JT (2005) Optimal protease production condition for Prevotella ruminicola 23 and characterization of its extracellular crude protease. Anaerobe 11:155–162. https://doi.org/10.1016/j.anaerobe.2004.10.007
Pekkarinen AI, Jones BL, Niku-Paavola M-L (2002) Purification and properties of an alkaline proteinase of Fusarium culmorum. Eur J Biochem 269:798–807. https://doi.org/10.1046/j.0014-2956.2001.02697.x
Bhunia B, Basak B, Mandal T, Bhattacharya P, Dey A (2013) Effect of pH and temperature on stability and kinetics of novel extracellular serine alkaline protease (70kDa). Int J Biol Macromol 54:1–8. https://doi.org/10.1016/j.ijbiomac.2012.11.024
Pietikäinen J, Pettersson M, Bååth E (2005) Comparison of temperature effects on soil respiration and bacterial and fungal growth rates. FEMS Microbiol Ecol 52:49–58. https://doi.org/10.1016/j.femsec.2004.10.002
Gupta A, Khare SK (2007) Enhanced production and characterization of a solvent stable protease from solvent tolerant Pseudomonas aeruginosa PseA. Enzyme Microb Technol 42:11–16. https://doi.org/10.1016/j.enzmictec.2007.07.019
Agrawal D, Patidar P, Banerjee T, Patil S (2005) Alkaline protease production by a soil isolate of Beauveria felina under SSF condition: parameter optimization and application to soy protein hydrolysis. Process Biochem 40:1131–1136. https://doi.org/10.1016/j.procbio.2004.03.006
Elibol M, Moreira AR (2005) Optimizing some factors affecting alkaline protease production by a marine bacterium Teredinobacter turnirae under solid substrate fermentation. Process Biochem 40:1951–1956. https://doi.org/10.1016/j.procbio.2004.07.010
Sandhya C, Sumantha A, Szakacs G, Pandey A (2005) Comparative evaluation of neutral protease production by Aspergillus oryzae in submerged and solid-state fermentation. Process Biochem 40:2689–2694. https://doi.org/10.1016/j.procbio.2004.12.001
Sharma KM, Kumar R, Panwar S, Kumar A (2017) Microbial alkaline proteases: optimization of production parameters and their properties. J Genet Eng Biotechnol 15:115–126. https://doi.org/10.1016/j.jgeb.2017.02.001
Franco DG, Spalanzani R, Lima E et al (2017) Biochemical properties of a serine protease from Aspergillus flavus and application in dehairing. Biocatal Biotransform 35:249–259. https://doi.org/10.1080/10242422.2017.1322584
Moon S, Parulekar SJ (1990) A parametric study of protease production in batch and fed-batch cultures of Bacillus firmus. Biotechnol Bioeng 37:467–483. https://doi.org/10.1002/bit.2603370509
Ortiz GE, Noseda DG, Ponce Mora MC, Recupero MN, Blasco M, Albertó E (2016) A comparative study of new Aspergillus strains for proteolytic enzymes production by solid state fermentation. Enzyme Res 2016:1–11. https://doi.org/10.1155/2016/3016149
Tunga R, Shrivastava B, Banerjee R (2003) Purification and characterization of a protease from solid state cultures of Aspergillus parasiticus. Process Biochem 38:1553–1558. https://doi.org/10.1016/S0032-9592(03)00048-7
Bhosale SH, Rao MB, Deshpande VV, Srinivasan MC (1995) Thermostability of high-activity alkaline protease from Conidioboluscoronatus (NCL 86.8.20). Enzyme Microb Technol 17:136–139. https://doi.org/10.1016/0141-0229(94)00045-S
Patel R, Dodia M, Singh SP (2005) Extracellular alkaline protease from a newly isolated haloalkaliphilic Bacillus sp.: production and optimization. Process Biochem 40:3569–3575. https://doi.org/10.1016/j.procbio.2005.03.049
Geat S, Baib H, Yuan H, Zhang L (1996) Continuous production of high degree casein hydrolysates immobilized proteases in column reactor by immobilized protease in column reactor. J Biotechnol 50:61–170. https://doi.org/10.1016/0168-1656(96)01561-1
Murthy PS, Naidu MM (2010) Protease production by Aspergillusoryzae in solid-state fermentation utilizing coffee by-products. World Appl Sci J 8:199–205
Janser R, Ohara A, Nishide T et al (2015) A new approach for proteases production by Aspergillus niger based on the kinetic and thermodynamic parameters of the enzymes obtained. Biocatal Agric Biotechnol 7:199–207. https://doi.org/10.1016/j.bcab.2014.12.001
Acknowledgements
The authors thank Addis Ababa Science & Technology University, Addis Ababa, Ethiopia for laboratory support.
Author information
Authors and Affiliations
Contributions
SMB isolated the microbes and carried out the experiments. SVP was involved in experimental plan and drafting. TKM contributed to data kinetics and final draft formatting. MTG characterized the enzymes.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest to disclose.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Mustefa Beyan, S., Venkatesa Prabhu, S., Mumecha, T.K. et al. Production of Alkaline Proteases using Aspergillus sp. Isolated from Injera: RSM-GA Based Process Optimization and Enzyme Kinetics Aspect. Curr Microbiol 78, 1823–1834 (2021). https://doi.org/10.1007/s00284-021-02446-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-021-02446-4