Abstract
Aspergillus tamarii expresses an extracellular alkaline protease that we show to be effective in removing hair from cattle hide. Large quantities of the enzyme will be required for the optimization of the enzymatic dehairing process so the growth conditions for maximum protease expression by A. tamarii were optimized for both solid-state culture on wheat bran and for broth culture. Optimal protease expression occurred, for both cultural media, at initial pH 9; the culture was incubated at 30 °C for 96 h using a 5% inoculum. The crude enzyme was isolated, purified and characterized using MALDI TOF TOF. The alkaline protease was homologous to the alkaline protease expressed by Aspergillus viridinutans.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The use of pure proteolytic enzymes for the removal of hair from animal hides dates back to the early 1900s [24] and has been an ongoing area of research at the Eastern Regional Research Center since the 1950s [6]. The major roadblocks to the commercialization of an enzymatic dehairing process have been the cost of the proteolytic enzymes and the inability of the enzymes to remove ‘fine’ hairs from the animal hide [10]. The former could be resolved if sufficient quantities of inexpensive enzymes were available to allow researchers to study the enzymatic dehairing in detail. Until recently, most proteases used in enzymatic dehairing studies were bacterial in origin. It would be advantageous to use a fungal protease as fungal expression systems are capable of producing larger quantities of enzymes than bacterial expression systems. Filamentous fungi, such as Aspergillus, have been the organism of choice for large scale production of bulk industrial enzymes, as the fungi can be grown on relatively inexpensive (agricultural waste) media and the fungi can secrete bulk quantities of enzymes [3]. A proteolytic enzyme that had been isolated from Aspergillus tamarii was used to dehair goat skins [7]. The A. tamarii had been isolated from a tannery waste field and grown on wheat bran (solid-state fermentation). The authors of the paper did not discuss the optimization of the growth conditions of the A. tamarii. This particular isolate of A. tamarii was not available to us so a strain of A. tamarii (NRRL 20818) was obtained from the National Center for Agricultural Utilization Research and was screened for proteolytic activity. The challenges to develop an efficient enzymatic dehairing system for cattlehides—initially the ease, low cost, and high yield of enzyme production—and a history of success with this enzyme for dehairing small skins led us to conduct this research. The results from this study will subsequently be applied to determine the effectiveness of the enzyme on hair removal and the concomitant maintenance of the integrity of the grain layer of the cattlehide.
We performed a set of initial experiments with the A. tamarii strain to determine if it, indeed, produces a protease. A recent article reported that the addition of either glucose, wheat bran or soybean meal increased the enzyme production of A. tamarii grown in broth culture [4]. We now have optimized the growth conditions, for both solid state and broth fermentation, for maximum enzyme production.
Materials and methods
Materials
Wheat bran was obtained from a local feed store in 50-lb bags, divided into 2-lb bags and stored at 4 °C before use. Czapek Agar was obtained from Oxoid (Basingstoke, England), peptone and yeast extract were obtained from Difco (Detroit, MI, USA), and non-fat milk powder was obtained from a local grocery store. All other chemicals were obtained from Sigma (St. Louis, MO, USA) and were used without further purification. All solutions were prepared in sterile distilled H2O.
Microorganism
Aspergillus tamarii NRRL 20818 was plated on Czapek Dox, grown at 30 °C, and then stored at 4 °C. Czapek Dox agar slants, incubated for 7 days, were used for the preparation of the inoculum.
Preparation of inoculum
Ten milliliters of distilled water containing 0.1% Tween-80 was transferred to a sporulated (7 day old) Czapek Dox agar slant culture of A. tamarii. The spores were dislodged using an inoculation needle, under aseptic conditions, and the suspension to be used as the inoculum was adjusted with sterile distilled water until the optical density at 530 nm was 1.5–1.6. The resultant inoculum (5 mL) was added to the solid substrate (100 g wheat bran) or liquid medium (100 mL broth culture medium).
Optimization of solid-state fermentation (SSF) and broth fermentation growth conditions
The various process parameters influencing enzyme production were optimized individually and independently of the others and the optimized conditions were used in all subsequent experiments in sequential order. For the optimization, either the same solid-state fermentation growth conditions or the same broth culture growth conditions were used. For solid-state conditions, wheat bran was inoculated with 5% A. tamarii inoculum and the substrate incubated for 96 h at 30 °C. For broth conditions, the basal broth medium was composed of 0.5% glucose, 0.5% peptone, 0.25% skimmed milk, 0.25% yeast extract, 0.1% Na2HPO4 and 1% NaNO3 (all w/v) in distilled water, pH 9.0. The basal medium was inoculated with a 5% A. tamarii inoculum and the inoculated broth was incubated on a rotary shaker at 180 rpm for 7 days at 30 °C.
For enzyme production from solid-state fermentation, first the initial moisture content was optimized by adjusting the initial moisture content (30, 40, 50, 65, 75 and 100%) of the wheat bran with distilled water. Then the pH of the media (both wheat bran and broth) was adjusted to 5, 6, 7, 8, 9,10, 11 or 12 using either sterilized 5 N HCl or 10% w/v Na2CO3. The optimization of the incubation times was established from run times of 24, 48, 72, 96, 120, 144, 168, and 192 h. The incubation temperature was varied (20, 25, 27, 30, 35, 40 or 50 °C) to study the temperature effect on enzyme production. Not only does the initial inoculum level play a role in how rapidly the culture reaches steady state, but it may also play a role in enzyme production, so various inoculum levels (1, 2, 5, 10, 15, 20 or 25%) were used. Lastly the amount of substrate for the solid-state fermentation could also play a role, so the amount of wheat bran placed in a 500-mL Erlenmeyer flask was varied (10, 20, 30, 45, 50, 75 or 100 g) to determine if there was an effect on enzyme production.
For the optimal production of protease the A. tamarii may require additional sources of carbon and nitrogen in its growth medium. Therefore the growth was supplemented (1% w/w) with the carbon sources starch, sucrose, lactose maltose, glucose or citrate and nitrogen sources peptone, skimmed milk, yeast extract, ammonium nitrate, ammonium sulfate or sodium nitrate. The incubations were done at 30 °C with all other conditions at the optimal levels determined previously. As glucose, peptone and ammonium nitrate showed a positive influence on enzyme production by A. tamarii, the levels of enrichment by these three supplements were optimized. Additionally phosphorus is required by A. tamarii; this was optimized by the addition of KH2PO4 (0.25, 0.5, 1.0, 2.0, 3.0, 4.0, or 5.0% w/v) to the substrate.
Isolation of alkaline protease
The extracellular enzyme secreted by A. tamarii, grown on wheat bran, was isolated by extraction of the bran (50 g) with Tris–HCl buffer (0.02 M, pH 8.5, 2 × 50 mL). The extracts were filtered through several layers of cheese cloth, combined and centrifuged (10,000 × g, 15 min, 4 °C). The supernatant was carefully decanted off the solid pellet and was used for the enzyme assay. The enzyme secreted by A. tamarii was isolated by filtration of the broth through several layers of cheese cloth; the filtrates were combined and centrifuged (10,000 × g, 15 min, 4 °C). The supernatant was decanted from the solid pellet and was used for the enzyme assay.
The crude solid enzyme used for the enzymatic dehairing experiments was obtained by precipitation from the supernatant by the slow addition of cold (0 °C) acetone, with stirring, until the solution was 0.8:1 acetone: water (v/v). After storage overnight at 4 °C the mixture was centrifuged (10,000 × g, 20 min, 4 °C) and the supernatant was decanted off of the solid pellet. The solid pellet was dissolved in a minimal amount of 20 mM Tris–HCl buffer (pH 8.5, 2 mM CaCl2). Dialysis of the resulting solution (Spectra/POR 6, MWCO 1,000) was against the same buffer. Lyophilization of the dialyzed solution yielded the solid crude enzyme, which was stored at −20 °C.
Enzyme assays
The proteolytic activity of the enzyme was determined by the modified method of Anson [1], using 1% casein as the substrate. One unit of protease activity is the amount of enzyme required to release 1 μg of tyrosine under standard assay conditions. The units of specific activity are activity/mg of protein. Peptidase activity was determined by measuring the amount of p-nitroanilide released from the substrate, benzoyl-d,l-arginine-p-nitroanilide (BAPNA), as described by Strongin et al. [28]. Determination of esterolytic activity used benzoyl-l-arginine ethyl ester (BAEE) as the substrate using the method described in [1]. The activity was determined by monitoring the increase in optical density at 254 nm.
Purification of crude alkaline protease
Ammonium sulfate was added (80% saturation) to the crude enzyme obtained after centrifugation of the extracts of the A. tamarii culture extract. The resulting solution was cooled at 4 °C overnight. The resulting precipitate was collected by centrifugation (rcf (xg), 20 min). The solid material was collected and dissolved in a minimal amount of 20 mM Tris/HCl buffer, (pH 8.0 containing 2 mM CaCl2). The resulting solution was dialyzed (Spectra/POR 6 MWCO = 3.5 kDa) against the same buffer (with repeated changes) used to dissolve the sample for 24 h and then the solution was lyophilized. Further purification of the enzyme was achieved using ion-exchange chromatography. The lyophilized enzyme was loaded on an Express-Ion exchanger D free base column (2.5 × 22 cm) that had been equilibrated with 20 mM Tris/HCl, pH 8.0, containing 2 mM CaCl2. The enzyme was eluted with a linear gradient of 0–0.5 M NaCl in the same buffer used to equilibrate the column. Fractions of 5 mL each were collected at a flow rate of 30 mL/h. Fractions exhibiting protease activity were pooled, dialyzed against 20 mM Tris–HCl, pH 8.0 and then lyophilized. The enzyme was also purified by the use of size-exclusion chromatography. The enzyme was further purified on a Sephadex G-100 column (2 × 65 cm) equilibrated with 2 mM Tris/HCl, pH 8.0, and eluted with the same buffer containing 0.5 M NaCl. Fractions of 3 mL each were collected at a flow rate of 15 mL/h. The active fractions were pooled, dialyzed against the Tris/HCl buffer and lyophilized. This purified enzyme was then used in further analysis.
Characterization of alkaline protease
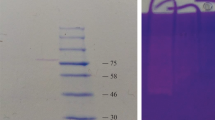
The molecular weight of the alkaline protease was determined by three different methods. The molecular mass was determined by gel filtration through a column of Sephadex G-100 that had been calibrated previously with standard marker proteins BSA (67 kDa), ovalbumin (45 kDa), chymotrypsin (25 kDa) and ribonuclease A (17.7 kDa); Blue dextran was used to determine the void volume. Additionally, the molecular weight was determined using SDS/PAGE and SDS gel electrophoreses.
Gelatin zymography was performed in polyacrylamide slab gels containing SDS and gelatin (0.1%) as a co-polymerized substrate, as described by Heussen and Dowdle [11]. After electrophoresis, the gels were washed in Triton X-100 (2.5% w/v) for 1 h at 4 °C to remove the SDS, incubated in 0.1 M Tris/HCL buffer, pH 8.0, for 4 h at 45 °C, and then stained with Coomassie Brilliant Blue. The activity band was observed as a clear colorless area depleted of gelatin in the gel against the blue background.
The optimum pH for protease activity was determined by incubating the reaction mixture at pH values of 6.0–13.0 at 30 °C. The pH stability was determined by measuring the residual activity of the enzyme after 30 min pre-incubation of the enzyme in buffers ranging from pH 6.0 to 13.0 at 30 °C.
The optimum temperature for enzyme activity was determined by conducting the assay at temperatures ranging from 20 to 80 °C (in 20 mM borate buffer, pH 8.0). The thermostability of the enzyme was measured after preincubation of the enzyme.
The effects of metal ions, enzyme activators and enzyme inhibitors were also studied. After incubation for 20 min at 30 °C with the various metal ions, inhibitors and activators, the residual activity was determined using standard assay conditions. Likewise the substrate specificity of the A. tamarii protease was determined against various synthetic and native substrates.
Mass spectrometry and protein identification
Matrix-Assisted Laser Desorption/Ionization mass spectrometry with automated tandem time of flight fragmentation of selected ions (MALDI-TOF/TOF) was run on the trypsin-digested alkaline protease. Spectra were acquired using a 4700 Proteomics Analyzer mass spectrometer (Applied Biosystems, Framingham, MA, USA) in the positive reflection mode with a 200 Hz Nd-YAG 355 nm laser. Reported spectra were the average of 1,000 acquired spectra in the MS mode or 2,500 in the MS/MS mode. Collision-induced dissociation (CID) with air as the collision gas at approximately 1 × 10−6 Torr, and a 1 keV acceleration voltage were used for obtaining the MS/MS spectra of selected peptides. Conversion of time of flight to mass (Da) for the mono-isotopic ions, [M + H]+, was based on calibration of the instrument with a peptide standard calibration kit (Applied Biosystems) that contained the following peptides: des-Arg1-bradykinin (m/z 904.4681), angiotensin I (m/z 1,296.6853), Glu1-fibrinopeptide B (m/z 1,570.6774), ACTH (clip 1–17) (m/z 2,903.0867), ACTH (clip 18–39) (m/z 2.465.1989), and ACTH (clip 7–38) (m/z 3,657.9294). The MS/MS time of flight calibration was obtained from the CID-produced fragments of Glu1-fibrinopeptide B. Peptides resulting from the trypsin digestion were extracted using a C18 ZipTip, washed with water containing 0.1% trifluoroacetic acid (TFA), re-extracted with acetonitrile–water (50:50)–0.1% TFA, and mixed with a recrystallized α-cyano-4-hydroxycinnamic acid matrix solution (5 mg/mL, acetonitrile–water (50:50)–0.1% TFA) to a final concentration between 100 fmol to 1 pmol/μL. Approximately 0.6–0.7 μL of the peptide-matrix solution was spotted in the MALD/I target plate. Peptide mass fingerprints and MS/MS of selected peptides were combined and queried against a primary sequence database using a Mascot search engine-associated GPS Explorer program (Applied Biosystems). Reported protein(s) from database searches from putative peptide sequences are within a ≥95% confidence interval.
Results and discussion
Optimization of growth parameters
Effect of initial moisture content (solid-state fermentation)
The initial moisture content of the wheat bran played a role in A. tamarii protease production. The optimal enzyme production occurs at a moisture content of 65% (1.43 × 103 U/g) with suppression of enzyme production at lower moisture concentrations (Fig. 1). A previously published paper reported that, for filamentous fungi such as A. tamarii, the optimal moisture content of the substrate varied between 50 and 75% [18]. For low initial moisture content, the substrate would dry out and inhibit the growth of the fungus. If the moisture content is too high (>65%), the enzyme production drops and because water fills the inter-particle space, the amount of oxygen available for fungal growth is reduced [5].
Effect of initial pH on enzyme production
The pH of the culture medium affects many enzymatic processes, enzyme production, and cell transport across membranes such as the expression of an extracellular protease [17]. The maximum A. tamarii protease production occurred at pH 9.0 for A. tamarii grown both on wheat bran (1.46 × 104 U/g) and in broth culture (7.4 × 103 U/10 mL). The initial pH appears to have a broad range (7–10) for nearly optimal protease production for broth culture while for solid-state culture the optimal enzyme production occurred over a much narrower range (Fig. 2).
Effect of incubation time
The incubation period for maximum A. tamarii protease production was 96 h for both solid state and broth culture (Fig. 3). The decrease in the protease production after 96 h was due to the denaturation/decomposition of the protease.
Effect of incubation temperature
The optimal incubation temperature for the A. tamarii protease production was 30 °C for both solid-state and broth culture. The solid-state fermentation is more sensitive to temperature variation than the broth culture (Fig. 4). Not only does the temperature regulate the synthesis of the enzyme, but possibly the secretion of the enzyme by changing the properties of the cell wall [25].
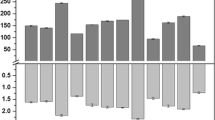
Plot of enzymatic activity versus inoculum size. The % inoculum is v/v of the original isolated spores suspended in base and diluted to the % shown. The inoculum was prepared by adding 10 mL of distilled water containing 0.1% Tween-80 to a sporulated (7-day old) Czapek Dox agar slant culture of A. tamarii. The spores were dislodged using an inoculation needle, under aseptic conditions, and the suspension was used as the inoculum. Dilutions of the suspension were prepared by adding the appropriate amount of sterile distilled H2O. The units for the y axis are U/g for the enzyme expressed by A. tamarii grown on the wheat bran (filled diamond) and U/10 mL for the enzyme expressed by A. tamarii grown in broth (filled square)
Effect of the inoculum size
The level of inoculation is one of the key factors for microbial growth and activity for solid-state fermentation. Large inoculum levels proved to be inhibitory in nature [27]. A. tamarii protease production appears to be more sensitive when the culture is grown on wheat bran than when grown in broth (Fig. 4). The optimal protease production occurred with a 5% inoculum.
The effect of supplemental carbon and nitrogen sources
The supplemental carbon sources had only a minimal effect on protease production by A. tamarii when grown on wheat bran; the addition of the glucose resulted in a 10% increase in enzyme production. The addition of maltose and sucrose increased the enzyme production by roughly 4%. Likewise, for broth culture of A. tamarii, the addition of sucrose resulted in a 7% increase in protease production; the addition of maltose and sucrose resulted in a 4% increase in enzyme production by A. tamarii (Fig. 5).
The addition of nitrogen sources does not affect A. tamarii protease production. The one exception is peptone, which increased protease production 12% when A. tamarii was grown on wheat bran and 6.7% when A. tamarii was grown in broth culture. Peptone can be a source of both carbon as well as nitrogen and is used as a nitrogen source for those organism that cannot assimilate inorganic nitrogen [9]. The addition of yeast extract to the A. tamarii culture grown on wheat bran increased the protease production by 7.5% (Fig. 6).
We studied, in greater detail, the effects of selected supplements on the A. tamarii protease production. We chose glucose, peptone, NH4NO3, and phosphorus (KH2PO4) for A. tamarii grown on wheat bran and for A. tamarii grown in broth culture we also enriched the media with skim milk. The optimal enrichment of NH4NO3 and phosphorus was 0.5% for wheat bran culture and of glucose and peptone was 1% (Fig. 7). Higher concentrations of these supplemental materials appear to inhibit protease production. The addition of simple sugars to cultures of A. tamarii, grown on sugar cane bagasse or corn cob, caused severe catabolic repression of xylanase. A. tamarii grown on wheat bran was resistant to catabolic repression, though the xylanase production was less than the amount produced by the control [27]. Similar results where observed for A. tamarii grown in broth culture. Maximum protease production occurred when the broth was enriched with glucose, peptone or skimmed milk enrichment at the 1% level (Fig. 8). Likewise maximum protease production occurred when the broth culture was supplemented with 0.5% NH4NO3. The addition of phosphate appears to increase the protease production as the maximum protease production occurred at 0.25% enrichment, but we did not run a control for phosphorus enrichment experiments (Fig. 9). Inhibition of protease production occurred at higher phosphate enrichment levels. Inhibition of phytase production in A. ficuum grown on canola meal occurred at higher phosphate enrichment levels [8]. Likewise phosphate concentrations greater than 0.004% inhibited phytase production by A. ficuum grown in broth culture [26].
Characterization of the protease
The purification of the A. tamarii protease by ammonium sulfate fractionation, ion exchange chromatography followed by size exclusion chromatography resulted in a 21-fold purification from the culture broth (Table 1) with an 8% yield.
Both the PAGE and SDS/PAGE gels contained only a single band at 45 KDa, from which we conclude that the purified protein was homogeneous. The molecular weight was in good agreement with the molecular weight obtained by gel filtration chromatography (∼47 KDa). The gelatin zymography gel contained only one zone of proteolytic activity, which confirmed the homogeneity observed in both the PAGE and SDS/PAGE gels.
We found the protease to be active over a broad pH range, with maximum activity at pH 8.5. This is in good agreement with other alkaline proteases, isolated from various sources, which have their maximum proteolytic activity in the range of pH 8–9 [2, 14, 19]. The A. tamarii protease was found to be relatively stable over a pH range of 7.5–11.0.
The optimum temperature for A. tamarii protease activity is around 45 °C. This is slightly higher than the optimum temperature for TKU003 protease (37–42 °C), a serine protease isolated from A. fumigants [13], and lower than the optimum temperature (60 °C) for a metalloprotease also isolated from A. fumigatus [15].
The inhibition/enhancement of proteolytic activity was determined for a number of metal ions (Table 2) and protease inhibitors (Table 3). Both Ca2+ and Mg2+ enhanced the proteolytic activity of the enzyme while Cu2+ and Hg2+ inhibited the proteolytic activity. Fe2+, Zn2+ and Mn2+ suppressed the proteolytic activity of the enzyme. PMSF suppressed the protease’s activity, suggesting that the A. tamarii protease is a serine protease [29, 30]. Inhibitors of thiol protease (iodoacetic acid and p-chloromercuribenzoate) and metalloprotease (o-phenanthroline and EDTA) had no effect on enzyme activity.
We next determined the specificity of the protein against a number of substrates (Table 4). The protease showed greatest activity against casein and the lowest against hemoglobin. The alkaline serine proteases isolated from B. stearothermophilus and Bacillus sp KSM-K16 [12, 20] exhibited identical substrate specificity. The A tamarii protease was also incapable of hydrolyzing the fibrous proteins collagen, elastin and keratin. The protease showed low peptidase activity on BAPNA and could not hydrolyze the ester substrate BAEE.
The MALDI/TOF TOF data confirmed that the enzyme was an alkaline protease. The protease is homologous with the alkaline protease fragment (MW 35,154 Da) isolated from Aspergillus fumigatus [16, 22, 23]. A unique peak (mw = 1,208.6316) was found that corresponds to the sequence GAPWGLGSISHK (Ion Score 100), which corresponds to the 128–139 segment of the alkaline serine protease from A. fumigatus. This was not unexpected as there is a high degree of homology among the alkaline serine proteases isolated from fungal sources [21].
Conclusions
We have identified the extracellular protease expressed by A. tamarii as an alkaline serine protease that shows strong homology to other fungal alkaline serine proteases. The growth conditions were optimized and most of the conditions where the same for both broth culture and solid-state culture (wheat bran): pH 9, incubation temperature 30 °C, incubation time 96 h, and inoculum 5%. The moisture content of the wheat bran should be 65%. Supplementation of either media with sucrose and peptone (1%) increases the A. tamarii’s protease production. We will scale up the solid-state fermentation on wheat bran (currently we are using approximately 1 kg wheat bran). The amounts of protease produced in broth culture are significantly smaller than produced from the solid-state fermentation, but the amounts are greater than those obtained from a bacterial system. Application of the enzyme as a cattlehide dehairing agent will be reported in a separate publication.
References
Anson ML (1938) The estimation of pepsin, papain and cathepsin with hemoglobin. J Gen Physiol 22:79–89
Aoyama M, Yasuda M, Nakachi K, Kobamoto N, Oku H, Kato F (2000) Soybean-milk-coagulating activity of Bacillus pumilus derives from a serine proteinase. Appl Microbiol Biotechnol 53:390–395. doi:10.1007/s002530051631
Bergquist PL, Te’o VS Jr, Gibbs MD, Cziferszky ACE, DeFaria FP, Azevedo MO, Nevalainen KMH (2002) Production of recombinant bleaching enzymes from thermophilic microorganisms in fungal hosts. Appl Biochem Biotechnol 98–100:165–176
Boer CG, Peralta RM (2000) Production of extracellular protease by Aspergillus tamarii. J Basic Microbiol 40:75–81
Bogar B, Szakacs G, Pandey A, Sabu A, Linden JC (2003) Production of phytase by Mucor racemosus in solid-state fermentation. Biotechnol Prog 19:312–319. doi:10.1021/bp020126v
Cordon TC, Jones HW, Clarke ID, Naghski J (1958) Microbial and other enzymes as depilatory agents. Appl Microbiol 6:293–297
Dayanandan A, Kanagaraj J, Sounderraj L, Govindaraju R, Suseela Rajkumar G (2003) Application of an alkaline protease in leather processing: an ecofriendly approach. J Clean Prod 11:533–536. doi:10.1016/S0959-6526(02)00056-2
Ebune A, Al-Asheh S, Duvnjak Z (1995) Effects of phosphate surfactants and glucose on phytase production and hydrolysis of phytic acid in canola meal by Aspergillus ficuum during solid-state fermentation. Bioresour Technol 54:241–247. doi:10.1016/0960-8524(95)00133-6
Gerhardt P, Murray RGE, Wood WA, Kreig NR (eds) (1994) Methods for general and molecular bacteriology. American Society for Microbiology, Washington, DC, p 157
Gehring AG, DiMaio GL, Marmer WN, Mazenko C (2002) Unhairing with proteolytic enzymes derived from Streptomyces griseus. J Am Leather Chem Assoc 97:406–411
Heussen C, Dowdle EB (1980) Electrophoretic analysis of plasminogen activity in polyacrylamide gels containing sodium dodecyl sulfate and copolymerized substrates. Anal Biochem 102:196–202
Kobayashi T, Hakamada Y, Adachi S, Hitomi J, Yoshimatsu T, Koike K, Kawai S, Ito S (1995) Purification and properties of an alkaline protease from alkalophilic Bacillus sp. KSM-K16. Appl Microbiol Biotechnol 43:473–481
Larcher G, Bouchara JP, Annaix V, Symoens F, Chabasse F, Tronchin G (1992) Purification and characterization of a fibrinogenolytic serine proteinase from Aspergillus fumigatus culture filtrate. FEBS Lett 308:65–69
Manachini PL, Fortina MG, Parini C (1988) Thermostable alkaline protease produced by Bacillus thermoruber—a new species of Bacillus. Appl Microbiol Biotechnol 28:409–413. doi:10.1007/BF00268205
Markaryan A, Morozova I, Yu H, Kolattukudy PE (1994) Purificationand characterization of an elastinolytic metalloprotease from Aspergillus fumigatus and immunoelectron microscopic evidence of secretion of this enzyme by the fungus invading the murine lung. Infect Immun 62:2149–2157
Monod M, Togni G, Rahalison L, Frenk E (1990) Isolation and characterisation of an extracellular alkaline protease of Aspergillus fumigatus. J Med Microbiol 35:23–28
Moon PJ, Parulekar SJ, Tsai S-P (1998) Competitive anion transport in desalting of mixtures of organic acids by batch electrodialysis. J Memb Sci 141:75–89. doi:10.1016/S0376-7388(97)00292-5
Pandey A, Soccol CR, Rodriguez-León JA, Nigam P (2001) Solid-state fermentation in biotechnology, 1st edn. Asiatech Publishers Inc., New Delhi
Peek K, Daniel RM, Monk C, Parker L, Coolbear T (1992) Purification and characterization of a thermostable proteinase isolated from Thermos strain RT41A. Eur J Biochem 207:1035–1044. doi:10.1111/j.1432-1033.1992.tb17140.x
Rahman RNZA, Razak CN, Ampon K, Basri M, Yunus WMZW, Salleh AB (1994) Purification and characterization of a heat-stable alkaline serine protease from Bacillus stearothermophilus F1. Appl Microbiol Biotechnol 40:822–827
Rao MB, Tanksale AM, Ghate MS, Deshpande VV (1998) Molecular and biotechnological aspects of microbial proteases. Microbiol Mol Biol Rev 62:597–635
Reichard U, Buttner S, Eiffert H, Staib F, Rüchel R (1990) Purification and characterisation of an extracellular serine proteinase from Aspergillus fumigatus and its detection in tissue. J Med Microbiol 33:243–251
Reichard U, Monod M, Rüchel R (1995) Molecular cloning and sequencing of the gene encoding an extracellular aspartic proteinase from Aspergillus fumigatus. FEMS Microbiol Lett 130:69–74
Röhm O (1913) A new unhairing process. Collegium 374–377
Satyanarayana T (1994) Production of bacterial extra-cellular enzymes by solid-state fermentation, 1st edn. Wiley Eastern Ltd, New Delhi
Shieh TR, Ware JH (1968) Survey of microorganisms for the production of extracellular phytase. Appl Microbiol 16:1348–1351
de Souza DF, de Souza CGM, Peralta RM (2001) Effect of easily metabolizable sugars in the production of xylanase by Aspergillus tamarii in solid-state fermentation. Proc Biochem 36:835–838
Strongin AYA, Izotova LS, Abramov ZT, Gorodetsky DI, Ermakova LM, Baratova LA, Belyanova LP, Stepanov VM (1978) Intracellular serine protease of Bacillus subtilis: sequence homology with extracellular subtilisins. J Bacteriol 133:1401–1411
Tsuchida O, Yamaga Y, Ishizuka T, Arai T, Yamada Y, Takeuchi M, Ichishima E (1986) An alkaline proteinase of an alkalophilic Bacillus sp. Curr Microbiol 14:7–12
Yamagata Y, Ichishima E (1989) A new alkaline proteinase with pI 2.8 from an alkalophilic Bacillus sp. Curr Microbiol 19:259–264
Acknowledgments
The authors acknowledge the valuable assistance of Laurie Fortis and Alberto Nuñez in obtaining and analyzing the MALDI/TOF TOF data.
Author information
Authors and Affiliations
Corresponding author
Additional information
Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
Rights and permissions
About this article
Cite this article
Anandan, D., Marmer, W.N. & Dudley, R.L. Isolation, characterization and optimization of culture parameters for production of an alkaline protease isolated from Aspergillus tamarii . J Ind Microbiol Biotechnol 34, 339–347 (2007). https://doi.org/10.1007/s10295-006-0201-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-006-0201-5