Abstract
Strain YA was newly isolated from an enrichment culture of river sediment and was identified as Janibacter sp. It was able to utilize dibenzofuran as the sole source of carbon and energy. Strain YA degraded > 90% of 1-chloro-dibenzo-p-dioxin (1-CDD) and > 80% of 2-chloro-dibenzo-p-dioxin in 18 hours with each initial concentration at 40 mg/L. A novel metabolite, 2-chloro-2′,6-dihydroxydiphenylether, was observed in 1-CDD degradation. From the metabolites detected by gas chromatography–mass spectrometry, strain YA was supposed to have at least two types of oxidation pathways in 1-CDD degradation.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Polychlorinated dibenzo-p-dioxins (DDs) and dibenzofurans (PCDD/DFs) are highly toxic compounds and are persistent in the environment. Bacterial activities in soil play some part in diminishing their concentrations. It is necessary to elucidate the degradation activity and pathway to understand their natural biodegradation and further enhance the degradation. Recently, several bacteria capable of using DD and/or DF as the sole source of carbon and energy have been isolated, and they also degrade chlorine-substituted DD and/or DFs. Regarding Gram-negative bacteria, Sphingomonas wittichii strain RW1 has been characterized. It can grow both DD and DF as the sole source of carbon and energy [18]. Some sphingomonad [2, 4] and several Pseudomonas strains can metabolize DD and DF [6, 11, 14]. Regarding Gram-positive bacteria, Janibacter [19, 20], Rhodococcus [7] and some Terrabacter strains [9, 15] have been reported to degrade DD and DF. Two types of dioxygenations are involved in the initial step of the degradation of dioxins. One is lateral dioxygenation, in which one of the aromatic rings is attacked at the lateral 1,2 or 2,3 positions, resulting in the formation of cis-dihydrodiols [1]. The other type of dioxygenation is angular dioxygenation, which takes place at the angular positions 4 and 4a adjacent to the ether bridge [10]. S. wittichii strain RW1 degraded several mono-chlorinated and dichlorinated DFs and DDs, but it did not degrade more highly chlorinated congeners. Most mono-chlorinated and dichlorinated DFs and DDs were degraded to the corresponding mono-chlorinated and dichlorinated salicylates and catechols, respectively, together with salicylate and catechol [16].
In this article, we describe the isolation and characterization of a novel bacterium, Janibacter sp. strain YA, which utilizes DF as the sole source of carbon and energy. It can degrade mono-chlorinated dibenzo-p-dioxin (1-CDD and 2-CDD) by resting cells. This is the first report of Janibacter spp. to degrade chlorinated DDs. Transformation of 1-CDD proceeded by way of both the angular and lateral dioxygenation pathways. Moreover, a novel metabolite, 2-chloro-2′,6-dihydroxydiphenylether (2-Cl-2′,6-DHDE) was observed, and possible degradation pathways are discussed.
Materials and Methods
Isolation of DF-utilizing bacteria
Strain YA was isolated from sediment of the Ayase River in Japan by an enrichment culture method using DF as the sole source of carbon and energy. DF dissolved in hexane was added to test tubes, and the hexane was evaporated. Then the mineral salt medium (MM = 2.2 g Na2PO4, 0.8 g KH2PO4, 3.0 g NH4NO3, 0.2 g MgSO4·7H2O, 10 mg FeSO4·7H2O, and 10 mg CaCl2·2H2O/l; 20 mg yeast extract was added to supplement trace metals) was added to the tube with silicon sponge cap. The amount of DF in the tube was set at 0.1 % (w/v). For the plate culture, 12 g/L gellan gum was used for solidification of MM, and DF was supplied in vapor form by placing solid DF on the lid of inverted Petri dishes. The cultivation was done at 25°C.
Identification of the bacterium
The nucleotide sequence of 16S rDNA was identified by the polymerase chain reaction (PCR) direct-sequencing method. The sequencing primers were 27f, f2L, 926f, f3L, r1L, r2L, r3L, and 1492r, corresponding to the positions 8-27, 518-536, 907-926, 1094-1112, 536-518, 821-803, 1111-1093, and 1510-1492 of the Escherichia coli 16S rDNA sequence, respectively. The sequencing reaction was analyzed with an ABI310 automatic DNA sequencer (Applied Biosystems, Foster City, CA). The sequence was compared with the DDBJ/GenBank/EMBL database by BLAST search. Sequence data were aligned with the CLUSTALW program, the distance was calculated according to the Kimura two-parameter method, and the phylogenetic tree was constructed by the neighbor-joining method.
Transformation of mono-chlorinated DDs
Transformation of 1-CDD and 2-CDD by strain YA was demonstrated by resting cells reaction. Resting cells reaction was carried out as follows. Strain YA was cultivated in 1-L Erlenmeyer flasks containing 0.1% (w/v) DF as the substrate. Residual crystals of DF were separated from the culture by sterilized glass filter (GF/D, Whatman, Kent, UK). Cells were harvested by centrifugation and washed twice with 50 mM phosphate buffer (PB, pH 7.0). The pellet was resuspended to PB at an 660 nm optical density of approximately 4. After incubation of this cell suspension for 2 hours to consume residual DF, 0.2 mg 1-CDD or 2-CDD from a stock solution (prepared in 1 mg/mL dimethylsulfoxide) was added to 5 mL cell suspension in screw-capped tubes. The consumption of DF was confirmed by gas chromatography–mass spectrometry (GC-MS) analysis (Fig. 3A). The tubes were incubated at 25°C and shaking at 250 rpm for 2, 4, 6, 9, 12, and 18 hours. For the estimation of degradation ratio, samples were extracted three times with the same volume of acidified ethyl acetate and were derivatized with N-methyl-N-trimethylsilyltrifluoroacetamide (MSTFA) to form trimethylsilyl (TMS) derivatives. GC-MS (GCMS-QP5050, Shimadzu, Tokyo, Japan) with a capillary column (DB-5MS, 0.25-mm inner diameter, 30-m length; J&W Scientific, Folsom, CA) was used for analyses. The initial temperature, 80°C, was maintained for 3 minutes, then temperature was increased to 250°C at a rate of 30°C/min and held at 250°C for 10 minutes. Helium was used as the carrier gas. Degradation ratio was calculated from the comparison of residual with initial amounts. Experiments were performed in duplicate. Control experiments were carried out with PB containing only 1-CDD or 2-CDD and heat-inactivated cells. We could not find any depletion of 1-CDD or 2-CDD in these control experiments. Transformation of non-chlorinated DD was also tested in the same manner.
Chemicals
DF was purchased from Kanto Kagaku (Tokyo, Japan). 1-CDD and 2-CDD were purchased from AccuStandard (New Haven, CT). DD, DHDE, and 3-chlorocatechol (3-CC) were purchased from Tokyo Kasei Kogyo (Tokyo, Japan). The derivatization reagent MSTFA was purchased from GL Sciences, Inc. (Tokyo, Japan). All chemicals were of analytical grade or of the highest purity available.
Nucleotide sequence accession number
Sequence data described in this article appear in the DDBJ/GenBank/EMBL databases under accession number AB110422.
Results
Taxonomic analysis of strain YA
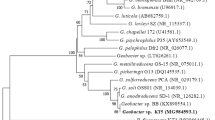
Strain YA is a Gram-positive, aerobic, and nonmotile coccus. Strain YA has 99.1% identity with J. brevis strain DSM 13953T from the result of 16S rDNA (1476 bp) sequence. J. brevis was reclassified as J. terrae [8]. A phylogenetic tree is shown in Fig. 1.
Transformation of mono-chlorinated DDs by resting cells
Because strain YA could not utilize chlorinated DDs as a sole source of carbon and energy, we tried to degrade DDs with resting cells reactions. Figure 2 shows 1-CDD and 2-CDD degradation by DF-grown resting cells of strain YA. The degradation percentage was calculated relative to control samples without strain YA. Approximately 80% of 40 mg/L 1-CDD was degraded in 9 hours, and > 90% was degraded after 18 hours of incubation. In contrast, 2-CDD was degraded > 80% after 18 hours of incubation, and the degradation was lower than that of 1-CDD.
Identified metabolites and proposed pathway of 1-CDD degradation
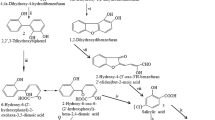
Total-ion chromatograms (TICs) of TMS-derivatized 1-CDD metabolites of strain YA after 0, 6, and 18 hours incubation are shown in Fig. 3. Four metabolites except 1-CDD were detected by GC-MS. None of the metabolites was found in the samples without strain YA. Metabolite A was identified as the TMS derivative of 3-CC with the same fragmentation pattern and retention time as those observed for the TMS derivative of authentic 3-CC. Metabolite B was strongly suggested as the TMS derivative of 2-Cl-2′,6-DHDE, with diagnostic peaks at m/z 310 (M+, 37Cl) (30%), m/z 308 (M+, 35Cl) (56%), m/z 295 (M+-CH3, 37Cl) (39%), m/z 293 (M+-CH3, 35Cl) (100%), m/z 275 (27%), m/z 265 (43%), m/z 258 (M+-CH3-Cl) (55%), m/z 243 (M+-CH3-Cl-CH3) (16%), and m/z 185 (M+-CH3-Cl-Si(CH3)3) (17%). This product had possible molecular ions whose mass unit corresponded to the calculated molecular weight of the TMS derivative of 2-Cl-2′,6-DHDE (inexplicably, one hydroxyl function was not derivatized). Furthermore, the isotope ratio of m/z 308 (M+, 35Cl) (56%) to m/z 310 (M+, 37Cl) (30%) showed that metabolite B contained one chlorine substituent. Metabolite C was identified as the TMS derivative of 2′-chloro-2,3,6′-trihydroxydiphenylether (2′-Cl-2,3,6′-THDE) or of 2′-Cl-2,5′,6′-THDE, with diagnostic peaks at m/z 470 (M+, 37Cl) (34%), m/z 468 (M+, 35Cl) (67%), m/z 367 (45%), m/z 365 (100%), m/z 330 (26%), and m/z 137 (15%). This fragment pattern showed strong fragment ions at M+-103, indicating that it corresponds to the TMS derivatives of THDE [3]. The mass unit of the possible molecular ion and isotope ratio of a chlorine substituent corresponded to mono-Cl-THDE. Furthermore, 5′-Cl-2,2′,3-THDE and 4′-Cl-2,2′,3-THDE from 2-CDD degradation had similar patterns of fragment ions [3]. Metabolite D was identified as the TMS derivative of di-hydroxylated 1-CDD. This product had the mass unit of the possible molecular ion and isotope ratio of a chlorine substituent corresponded to TMS derivative of di-hydroxylated 1-CDD.
(I) Total ion chromatograms of 1-CDD metabolites after 0, 6, and 18 hours of incubation. (II): Mass spectrum of the metabolites produced from 1-CDD by strain YA. Identified as (A) 3-CC (TMS derivative); (B) 2-Cl-2′,6-DHDE (TMS derivative); (C) 2′-Cl-2,3,6′-THDE or 2′-Cl-2,5′,6′-THDE (TMS derivative); (D) di-hydroxylated 1-CDD (TMS derivative). 1-CDD, 1-chloro-dibenzo-p-dioxin; 3-CC, 3-chlorocatechol; DHDE, dihydroxydiphenylether; THDE, trihydroxydiphenylether; TMS, trimethylsilyl.
Formation of 2,2′-DHDE from DD
Although the formation of 2-Cl-2′,6-DHDE from 1-CDD was strongly suggested, we could not confirm it because authentic 2-Cl-2′,6-DHDE was not available. Thus, we also tested DD degradation in the same manner to find 2,2′-DHDE as the metabolite. Fig. 4 shows the TICs of TMS-derivatized DD metabolites of strain YA of OD660 = 10 after 18 hours incubation. Along with catechol, pyrogallol, hydroxylated DD, THDE, and di-hydroxylated DD, metabolite E was observed. This metabolite was identified as DHDE with the same fragmentation pattern and the retention time as those of the TMS derivative of the authentic DHDE. This result supports the formation of 2-Cl-2′,6-DHDE from 1-CDD.
Discussion
Strain YA, isolated from the sediment of the Ayase River in Japan, can grow on DF. It was identified as Janibacter sp. with respect to 16S rDNA sequences. Janibacter sp. strain YY-1 has also been reported to utilize DF [20], and some Terrabacter strains, which are phylogenetically close to Janibacter, have been reported to degrade dioxins [3, 9, 15]. As for the genus of Janibacter, strain YA is first reported to degrade chlorinated DDs.
Strain YA produced Cl-THDE (Fig. 3C) and 3-CC (Fig. 3A) in 1-CDD degradation. Chlorinated THDE and 3-CC were also formed in degradation of 1-CDD by S. wittichii RW1 [16]. Chlorinated THDE was also produced in 2-CDD degradation by Pseudomonas resinovorans strain CA10, Terrabacter sp. strain DBF63 [3] and Burkholderia sp. strain JB1 [13]. The formation of THDE and 3-CC has also been also reported in degradation of DD by many bacteria [5, 18]. These metabolites were formed by way of the angular dioxygenation pathway. Angular dioxygenation specifically attacks the angular position on the two carbon atoms adjacent to the ether bridge of DD or DF. After this reaction, an unstable hemiacetal is formed, which decays spontaneously to 2,2′,3-THDE [10, 17]. These results indicated that the degradation of 1-CDD by strain YA follows the angular dioxygenation pathway (Fig. 5). The production of dihydroxylated 1-CDD (Fig. 3D) might be the result of lateral dioxygenation of 1-CDD (Fig. 5). This type of dioxygenation was reported in DF degradation by Ralstonia sp. strain SBUG [1]. Furthermore, strain YA formed 2-Cl-2′,6-DHDE (Fig. 3B) from 1-CDD. 2-Cl-2′,6-DHDE is a new metabolite of 1-CDD degradation. This compound has not been reported as the metabolite of 1-CDD through lateral and angular dioxygenation. In the lateral dioxygenation of DD, di-hydroxylated DD is formed (Fig. 5), and it can not be dehydrated to DHDE. In the angular dioxygenation of DD, THDE is formed spontaneously from unstable hemiacetal (Fig. 5). When this hemiacetal is dehydrated, it rearomatizes to yield hydroxylated DD but not DHDE [5]. Chlorinated DHDE might be formed by mono-oxygenation, which leads to cleavage of the ether bridge (Fig. 5). This reaction is similar to that found in the dibenzothiophene (DBT) desulfurization pathway by Rhodococcus erythropolis D-1 [12]. Dibenzothiophene sulfone mono-oxygenase catalyzes the conversion of DBT sulfone to form 2′-hydroxybiphenyl 2-sulfinic acid. In this reaction, mono-oxygenation occurs and one of the two C-S bonds of the DBT molecule is cleaved.
In 2-CDD degradation, TICs of the extract containing 2-CDD metabolites showed many peaks of the products (data not shown). Among them, only a small peak of dihydroxylated 2-CDD was identified by its mass spectra. This result suggests that strain YA degraded 2-CDD by lateral dioxygenation. However, we could not find any products formed by angular dioxygenation or mono-oxygenation by comparing their mass spectrum. These results suggested that the degradation pattern of 2-CDD was different from that of 1-CDD.
Strain YA, classified as Janibacter, degraded chlorinated dioxins. It was suggested that both types of reported degradation pathways were present in 1-CDD degradation. In addition, 2-Cl-2′,6-DHDE was observed as a new metabolite. This metabolite might be the result of mono-oxygenation of 1-CDD. It is necessary to elucidate these oxygenation mechanisms to further understand CDDs degradation.
Literature Cited
Becher D, Specht M, Hammer E, Francke W, Schauer F (2000) Cometabolic degradation of dibenzofuran by biphenyl-cultivated Ralstonia sp. strain SBUG 290. Appl Environ Microbiol 66:4528–4531
Fukuda K, Nagata S, Taniguchi H (2002) Isolation and characterization of dibenzofuran-degrading bacteria. FEMS Microbiol Lett 208:179–185
Habe H, Chung JS, Lee JH, Kasuga K, Yoshida T, Nojiri H, et al. (2001) Degradation of chlorinated dibenzofurans and dibenzo-p-dioxins by two types of bacteria having angular dioxygenases with different features. Appl Environ Microbiol 67:3610–3617
Harms H, Wilkes H, Sinnwell V, Wittich RM, Figge K, Francke W, et al. (1991) Transformation of 3-chlorodibenzofuran by Pseudomonas sp. HH69. FEMS Microbiol Lett 81:25–29
Harms H, Wittich RM, Sinnwell V, Meyer H, Fortnagel P, Francke W (1990) Transformation of dibenzo-p-dioxin by Pseudomonas sp. strain HH69. Appl Environ Microbiol 56:1157–1159
Hong HB, Chang YS, Choi SD, Park YH (2000) Degradation of dibenzofuran by Pseudomonas putida PH-01. Water Res 34:2404–2407
Kimura N, Urushigawa Y (2001) Metabolism of dibenzo-p-dioxin and chlorinated dibenzo-p-dioxin by a gram-positive bacterium, Rhodococcus opacus SAO101. J Biosci Bioeng 92:138–143
Lang E, Kroppenstedt RM, Swiderski J, Schumann P, Ludwig W, Schmid A, et al. (2003) Emended description of Janibacter terrae, including ten dibenzofuran-degrading strains and Janibacter brevis as its later heterotypic synonym. Int J Syst Evol Microbiol 53:1999–2005
Monna L, Omori T, Kodama T (1993) Microbial-degradation of dibenzofuran, fluorene, and dibenzo-p-dioxin by Staphylococcus-auriculans DBF63. Appl Environ Microbiol 59:285–289
Nojiri H, Habe H, Omori T (2001) Bacterial degradation of aromatic compounds via angular dioxygenation. J Gen Appl Microbiol 47:279–305
Nojiri H, Nam JW, Kosaka M, Morii K, Takemura T, Furihata K, et al. (1999) Diverse oxygenations catalyzed by carbazole 1,9a-dioxygenase from Pseudomonas sp. strain CA10. J Bacteriol 181:3105–3113
Ohshiro T, Kojima T, Torii K, Kawasoe H, Izumi Y (1999) Purification and characterization of dibenzothiophene (DBT) sulfone monooxygenase, an enzyme involved in DBT desulfurization from Rhodococcus erythropolis D-1. J Biosci Bioeng 88:610–616
Parsons JR, De Brujine JA, Weiland AR (1998) Biodegradation pathway of 2-chlorodibenzo-p-dioxin and 2-chlorodibenzofuran in the biphenyl-utilising strain JB1. Chemosphere 137:1915–1922
Resnick SM, Gibson DT (1996) Regio- and stereospecific oxidation of fluorene, dibenzofuran, and dibenzothiophene by naphthalene dioxygenase from Pseudomonas sp. strain NCBI 9816-4. Appl Environ Microbiol 62:4073–4080
Strubel V, Engesser KH, Fischer P, Knackmuss HJ (1991) 3-(2-Hydroxyphenyl)catechol as substrate for proximal meta ring cleavage in dibenzofuran degradation by Brevibacterium sp. strain DPO1361. J Bacteriol 173:1932–1937
Wilkes H, Wittich RM, Timmis KN, Fortnagel P, Franche W (1996) Degradation of chlorinated dibenzofurans and dibenzo-p-dioxins by Sphingomonas sp. strain RW1. Appl Environ Microbiol 62:367–371
Wittich RM (1998) Degradation of dioxin-like compounds by microorganisms. Appl Microbiol Biotechnol 49:489–499
Wittich RM, Wilkes H, Sinnwell V, Francke W, Fortnagel P (1992) Metabolism of dibenzo-p-dioxin by Sphingomonas sp. strain RW1. Appl Environ Microbiol 58:1005–1010
Yamazoe A, Yagi O, Oyaizu H (2004) Biotransformation of fluorine, diphenyl ether, dibenzo-p-dioxin and carbazole by Janibacter sp. Biotechnol Lett 26:479–486
Yamazoe A, Yagi O, Oyaizu H (2004) Degradation of polycyclic aromatic hydrocarbons by a newly isolated dibenzofuran-utilizing Janibacter sp. strain YY-1. Appl Microbiol Biotechnol 65:211–218
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Iwai, S., Yamazoe, A., Takahashi, R. et al. Degradation of Mono-chlorinated Dibenzo-p-Dioxins by Janibacter sp. Strain YA Isolated from River Sediment. Curr Microbiol 51, 353–358 (2005). https://doi.org/10.1007/s00284-005-0099-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-005-0099-6