Abstract
Some members of the Bacillus velezensis (Bv) group (e.g., Bv FZB42T and AS3.43) were previously assigned grouping with B. subtilis and B. amyloliquefaciens, based on the fact that they shared a 99% DNA–DNA percentage phylogenetic similarity. However, hinging on current assessments of the pan-genomic reassignments, the differing phylogenomic characteristics of Bv from B. subtilis and B. amyloliquefaciens are now better understood. Within this re-grouping/reassignment, the various strains within the Bv share a close phylogenomic resemblance, and a number of these strains have received a lot of attention in recent years, due to their genomic robustness, and the growing evidence for their possible utilization in the agricultural industry for managing plant diseases. Only a few applications for their use medicinally/pharmaceutically, environmentally, and in the food industry have been reported, and this may be due to the fact that the majority of those strains investigated are those typically occurring in soil. Although the intracellular unique biomolecules of Bv strains have been revealed via in silico genome modeling and investigated using transcriptomics and proteomics, a further inquisition into the Bv metabolome using newer technologies such as metabolomics could elucidate additional applications of this economically relevant Bacillus species, beyond that of primarily the agricultural sector.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Over the past six decades, Gram-positive endospores have received a great deal of attention due to the various discoveries made pertaining to their use in various sectors of industry, especially agriculturally. Additional advantages when using these spore-forming bacteria are that they can be easily cultured, stored, and manipulated for biotechnological purposes (Cao et al. 2018). To date, B. licheniformis, B. subtilis, and B. amyloliquefaciens are among the most exploited species from the genus; however, over the last decade, a vast amount of research has been done on B. velezensis (Bv). Although Bv is categorized as a heterotypic synonym of B. amyloliquefaciens subsp. plantarum FZB42T, B. methylotrophicus KACC 13015T, and B. oryzicola KACC 18228, based on DNA–DNA hybridization values greater than 84% (Dunlap et al. 2016; Fan et al. 2017), strains within the species still show distinct genomic characteristics (Adeniji and Babalola 2018). Bv was originally cultured from the river bank of Vélez in Málaga in Southern Spain, was reported to grow at a pH of between 5.0 and 10.0 and at temperatures of between 15 and 45 °C (Ruiz-García et al. 2005), and produce diverse metabolic intermediates, which include antibiotics, enzymes, phytohormones, iron chelators, antioxidants, growth promoters, and antitumor agents (Gao et al. 2017b; Liu et al. 2010b; Meena et al. 2018).
Genome sequence reports for many of the Bv subspecies are now readily available (Baptista et al. 2018; Chen 2017; Kim et al. 2017a; Kim et al. 2017b; Lee et al. 2015, 2017; Li et al. 2018; Liu et al. 2017) which is also considered extremely advantageous from a biotechnological exploitation perspective. To date, Bv has been extensively utilized in the area of biocontrol and plant growth promotion; however, studies involving their use in other industries or various other environmental applications are scarce and hence a topic of further research. Better described Bacillus sp. such as B. licheniformis, for instance, has been comparatively well exploited for various commercial applications ranging from the manufacture of enzymes (proteases, aminopeptidases, pectinolytic enzymes) to the production of antibiotics, biochemicals, and also applied in the textile industry. Bv, on the other hand, although considered extremely robust to work with, and despite the numerous studies on its possible biotech applications, has as yet not been extensively applied commercially, nor if these applications have reduced reliance on the use of synthetic products for industrial, agricultural, environmental, and medical purposes.
Comparative genomic, mutagenic, transcriptomic, histology, and proteomic analyses have all contributed to characterizing these beneficial Bacillus strains and in the production of improved bioformulations for various biotech applications (Wu et al. 2015b). A limitation to using these technologies, however, is that it appears that only those compounds previously known to be present in other Bacillus spp., such as difficidin, surfactin, bacilysin, macrolactin, bacillaene, bacillibactin, and fengycin, are being re-identified in many of the Bacillus strains and species reported lately in literature. Considering this, there is a desperate need for using other research approaches for identifying and characterization new Bv secondary biomolecules in vitro. Metabolomics, one of the newest additions to “omics” research, is likely to identify new microbial biomarkers and, in so doing, expand our knowledge and subsequently the applications and commercial value of Bv and other species of Bacillus. Here, we expound on the genomic relatedness existing among the Bv species, their rapid deployment over the years, and we suggest future approaches for exploiting underutilized beneficial microbes such as Bv.
Phylogenomic interrelation between the B. velezensis strains
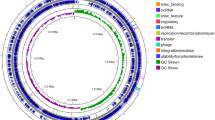
Dunlap et al. (2016) report that members of the Bv sp. share phenotypic and genotypic coherence based on their morphology, physiology, chemotaxonomy, and phylogeny and since then confirmed by others (Ye et al. 2018). In order to fully understand the genetic relationship and biochemical diversity which exists within the species, we collected 17 Bv genome sequences from the National Center for Biotechnological Information (NCBI), with identity similarity ≥ 98% and phylogenomically surveyed them using the Kbase bioinformatics platform (Arkin et al. 2016). Subsets (10) of the genomes were also compared using the online web server of EDGAR 2.3 https://edgar.computational.bio.uni-giessen.de/. From the in silico analysis, the conserved genes located in all of the collected strains (representing the core genome), and the supersets of all the genes (the pan-genome) (Dunlap et al. 2013) were identified and described (Fig. 1a, b).
a Pan-genomic atlas of 17 Bv strains computed on the Kbase platform (Arkin et al. 2016). Core and non-core genome similarities and dissimilarities are indicated on the left key of the figure. Peculiar genes in the genome 0 (Bv NWUMFkBS10.5) are indicated in its ring as light blue arcs; genome 1: VCC genbank genome (gbg) genome; genome 2: ATCC19217.gbg; genome 3: BS10.5.gbg; genome 4: SX.gbg; genome 5: AS43.3.gbg; genome 6: LABIM40.gbg; genome 7: J-5.gbg; genome 8: DR08.gbg; genome 9: GYL4.gbg; genome 10: UCMB5033.gbg; genome 11: M7.gbg; genome 12: SQR9.gbg; genome 13: TrigoCor1448.gbg; genome 14: CBMB205.gbg; genome 15: WS8.gbg; Genome 16: LS69.gbg). b Circular map showing pairwise alignment of Bv strain_LS69 (NZ_CP015911) with other 9 Bv genome subsets
In Fig. 2a, b, the values overlapping are the gene coding proteins (GCPs) common within the genomes and values outside the overlaps signify the GCPs in each genome without orthologs in the other genomes. The Venn diagram was computed as described by Bardou et al. (2014) using the BIOiPLUG Comparative Genomics Database (https://www.bioiplug.com/). Also, using the default parameters of the web server BIOiPLUG Apps (https://www.bioiplug.com/apps), the Pan-genome Orthologous Groups (POGs) and Gene Content (GC) of a subset of the 17 strains (5 highly referenced and 5 somewhat less popular B. velezensis strains) were determined. The Pan-genome Orthologous Groups (POGs) which represent the basic unit of core genes/pan-genome and gene presence/absence representing total genetic content were analyzed using BIOiPLUG Comparative Genomics Database (https://www.bioiplug.com/) (Fig. 3a, b).
Both the core genes and dispensable genes are crucial for determining bacterial species diversity. The core genes are responsible for indicating fundamental functions of the organism’s biology which includes replication, translation, and maintenance of cellular homeostasis, and in Fig. 3a, b, these core genes indicate the genetic diversity among the strains compared. The dispensable genes are associated with survivability, antimicrobial resistance, virulence traits, and development of novel gene functions. Some of these genes, when present within the strains, also confer an adaptive superiority over others lacking them (Carlos Guimaraes et al. 2015). Average nucleotide identity (ANI) was also computed on the online web server of EDGAR 2.3 https://edgar.computational.bio.uni-giessen.de/. Majority of the core genes irrespective of their functions and location are necessary for better phylogenomic reassignments among the Bv strains. The distance matrices heatmap (Fig. 4) indicates the genomic similarities and differences among 10 of the 17 Bv strains. Based on this ANI analyses, we reported that the phylogeny of these closely related bacteria corresponds to previous reports. A total of 154 core functions and 784 core protein families were peculiar to all 17 the strains probed. Additionally, the 17 strains have in combination 60,538 protein-coding genes of which 59,263 are present in homolog families and 1275 in singleton families. The total numbers of families identified in the 17 strains are 5538, of which 4207 are in the homolog families and 1331 in the singleton families (Table 1). Figure 5 indicates the collinearity of genes among 10 of the Bv subset. The conserved genomic regions of the Bv strains were probably due to their common ancestry, either by duplication or by speciation of the genes. Several industries have benefited from the versatile genomic resource of Bv, and this will subsequently be discussed in greater detail in the latter sections of this review.
Agricultural applications of B. velezensis
As previously mentioned, the majority of research supports the use of B. velezensis for its possible utilization in the agricultural industry, as alternatives to the current synthetic fertilizers and chemical pesticides (as summarized in Table 2). In an attempt to reduce the incidence of wheat powdery mildew (Cai et al. 2017), Cai et al. (2016) performed a field trial showing the capacity of a bioactive metabolite extract of Bv CC09 to reduce the severity of mildew disease by 86.12% when compared to a commercial fungicide triazolone which only showed a 50.39% reduction when applied as pretreatment samples. Similarly, Chen et al. (2018c) indicated that Bv extract, LM2303, significantly reduced the incidence and severity of wheat FHB (caused by F. graminearum) under in planta conditions and similarly to CC09, it also had a higher biocontrol efficacy when compared with that of a chemical fungicide. The authors further proposed that the biocontrol potential of Bv LM2303 was most likely via four major mechanisms: (i) antibiosis mediated by Bacillus antibacterial metabolites and lipopeptides, (ii) activation of induced systemic resistance (ISR) in wheat by the volatiles and the lipopeptide surfactin, (iii) enhanced growth of wheat due to an elevated production of growth hormone and nutrient uptake, and (iv) competitive exclusion of other microflora. Gilardi et al. (2015) investigated a commercially obtained Bv (Cilus Plus IT45) towards the biocontrol of Phytophthora capsici, which results in root and crown rot of zucchini in parts in Italy, however, in their investigation the biocontrol effect of the strain was determined to be non-significant. Using an HPLC-MS, in combination with genomic analyses, Jin et al. (2017) identified the metabolic intermediates that were responsible for the plant protection and growth promoting attributes of Bv S3-1 during an anti-Botrytis cinerea experiment. The bioactivity of the Bv was attributed to the production of various lipopeptide antibiotics (iturin, fengycin, and surfactin). It is important to note, however, that many of the aforementioned compounds ascribed to Bv functionality are not necessarily novel compounds.
Streptomyces scabies is a pathogen that affects diverse crop types, ranging from, but not limited to the following: beet, carrot, cucumber, pepper, potato, radish, squash, tomato, and turnip. Bv BAC03 was identified as an effective antagonist of S. scabies; however, antagonism and plant growth promotion of the strain was dependent on its prior application before disease onset. Additionally, strain BAC03 increased the biomass of the radish roots and leave harvest, in spite of S. scabies infection (Chen et al. 2015; Meng and Hao 2017; Meng et al. 2016). Likewise, Bv YC7010, a multi-functional endophyte with the capacity to induce systemic resistance against several rice pathogens, was also found to induce systemic resistance in Arabidopsis against Myzus persicae (Rashid et al. 2017). Green peach aphid (GPA), Myzus persicae, is a destructive phloem sap insect–pest that causes significant agricultural losses. Root drenching of Arabidopsis with the aforementioned Bv strain resulted in a significant reduction in the settling, feeding, and reproduction of the pest on the Arabidopsis leaves. The authors attributed the ISR to the expression of the senescence-promoting gene, phytoalexin deficient 4 (PAD4), and the subsequent suppression of botrytis-induced kinase 1 (BIK1) of M. persicae, which warrants further experiments on the use of Bv YC7010 as a pesticide.
Lastly, Nam et al. (2009) proved the efficacy of various formulations made up of Bv BS87 and RK1 to be highly effective for the bioprotection of strawberries against Fusarium oxysporum f. sp. fragariae, by performing in vitro and in planta studies. Although the bioprotective efficacy of the RK1 formulation was higher than its counterpart BS87, it exhibited similar antagonism in planta in comparison to a conventional fungicide (copper hydroxide). Application of Bv along with a Glomus intraradices (mycorrhizal fungi) as a potential biofertilizer for strawberry crops was later reported by Palencia et al. (2015). These studies show the capacity of Bv for use as a very effective alternative for conventional biofertilizers and chemical pesticides, with subsequently comparatively even better crop yields with less harm to the environment.
Industrial and environmental applications of B. velezensis
Various Bv strains have been shown to have applications for degrading various toxic and harmful industrial byproducts. The azo dyes, for instance, found in textile industry effluents, are toxic to aquatic environments, due to their mutagenic and recalcitrant properties. A preliminary evaluation by Bafana et al. (2008) indicated the capacity of Bv strain AB to decolorize and detoxify these azo dyes and, subsequently, its applications for treating and preventing azo dye effluent pollution. Similarly, Lan (2016) indicated a total reduction in nitrogen and COD of 83.5% and 93.2% respectively, in a study testing the capacity and possible applications of Bv strain M2 for application for slaughterhouse wastewater bioremediation. An in silico study by Chen et al. (2018a) indicated that a number of Bv strains, employed for in vivo pretreatments of lignocellulosic material, had the capacity to efficiently degrade cellulose and hemicelluloses due to the presence of genes coding for various lignocellulolytic enzymes including endo-glucanases, endo-glucosidases, phospho-glucosidases, phospho-galactosidase, sucrose-phosphate hydrolase, and levanases.
In a follow-up study, Chen et al. (2018b) isolated a Bv strain (157) with the capacity for degrading various agro-industrial byproducts including soybean meal, wheat bran, sugarcane bagasse, wheat straw, rice husk, maize flour and maize straw, and the subsequent utilization of these degraded agro-materials in biofuel production. The authors additionally indicated that during solid-state fermentation, several important industrial enzymes are produced by this Bv strain 157. In an explorative study, searching for Bv strains with the capacity to efficiently depolymerize various types of lignocelluloses (cellulose, hemicellulose, and lignin) into fermentable sugars (Nair et al. 2018) isolated Bv ASN1 which had the capacity to synthesize cellulase, which has a broad range of applications in food, textile, animal feed, petroleum, waste management, biosurfactant, and pulp/paper industries. Cellulase from Bv ASN1 is a candidate for depolymerizing recalcitrant lignocellulosic biomass from waste office paper.
There is currently a high demand for biosurfactants, for application as commercial bioemulsifiers due to their improved biodegradability as compared to their synthetic surfactant counterparts. A comprehensive analysis by Liu et al. (2010b), using rapid molecular PCR and chemical elucidation of various Bv H3 metabolomic intermediates, indicated that the investigated strain has excellent bioemulsifying properties. Due to the fact that primer detection efficiency for Bv PCR analysis is suboptimal at present, a species–subspecies in silico exploration of Bv and B. subtilis genomes was carried out by Cho et al. (2018) in order to identify and produce Bacillus-specific primers. The study selected various Bacillus-specific genes, and the designed primers were for the purpose of improved detection efficiency during quantitative and real-time PCR procedures. The improved PCR assay was subsequently further applied towards a targeted detection of the Bacillus strains in a fermented food sample.
Microbial inoculants containing a number of different Bv strains have also been previously investigated for their capacity for reducing N2O and CO2 emissions occurring due to various anthropogenic activities when manufacturing and using fertilizers, which is considered an environmental hazard (Calvo et al. 2013; Calvo et al. 2016). The data provided preliminary proof for the ex-situ application of the Bv bio-inoculants for these applications since the Bv strains tested show excellent nitrogen use capacities. Lastly, an inoculant containing Bv strain NRRL B-23189 was indicated to be an antagonist of Penicillium roqueforti sensu stricto (s.s.) (toxigenic mold) which can have a profound effect on improving processes related to corn silage (Wambacq et al. 2018). Although the initial inhibitory tests showed promise, the latter in vitro and in vivo results from the study were less encouraging.
Applications in biomedicine
The increased prevalence of drug-resistant pathogens and subsequent shortage of possible drugs for eliminating these organisms and curing the associated diseases have led to an upsurge in research on possible new drug compounds, in particular, those from natural origins. In recent years, microbial produced exopolysaccharide (EPS) biopolymers have been shown to be of particular value for various applications relating to medical health, bio-nanotechnology, food and cosmetic industry gelling agents, and bio-flocculation (Moghannem et al. 2018). Moghannem et al. (2018) applied a Plackett–Burman statistical experimental design, in order to screen for microbes producing EPS and identify the most optimal culture and environmental conditions conducive for EPS production by these species, using a response surface methodology (RSM). The Bv KY498625 strain tested and produced comparatively the highest EPS, which was easily extracted and purified using diethylaminoethyl (DEAE) cellulose. Further identification of the EPS was carried out using gel permeation chromatography GPC), Fourier transform infrared (FTIR), and gas chromatography-mass spectrometry (GC–MS). Since this microbial produced EPS is biodegradable, it is considered a considerably substantial breakthrough in terms of environmental sustainability.
Microbial produced compounds with possible anticancer/tumor properties (e.g., B. subtilis surfactins) are also now being investigated due to their high pharmacological potency and medicinal value for treating these conditions (Wu et al. 2017). An exopolysaccharide (EPS) compound, isolated from a marine Bv MHM3 strain designated as MHM3EPS, showed an extremely high anticancer (MCF-7 cells) capacity at very low concentrations, with no apparent cytotoxic effects against health host cells (Mahgoub et al. 2018). In a similar study, Meena et al. (2018) indicated a lipopeptide antibiotic extract from a novel Bv strain (KLP2016), inhibited both Aspergillus niger and a Mucor sp., and additionally exhibited high (~ 90%) cytotoxicity against human cervical carcinoma cells Hep2-C. Furthermore, Rehman et al. (2018) recently reported the antiproliferative activity of five compounds extracted from Bv RA5401 against breast cancer cell lines, two of which function via inhibition of the intracellular cancer proteases and the remaining three via inhibition of the G protein-coupled receptors of the cancer cells (Meena et al. 2018).
A Bv V4 strain, isolated from an aquaculture system, with biocontrol activity against Aeromonas salmonicida subsp. salmonicida, the causative agent of furunculosis in fish (Gao et al. 2017a), was also investigated for possible probiotic activity on Oncorhynchus mykiss (rainbow trout). In the study, Bv V4 caused a significant reduction in the fish’s mortality while also improving its growth. The authors used metabolic profiling (genomics and metabolomics (LC-MS/MS and NMR)) to identify the active metabolites produced by Bv V4, which were lipopeptides from the iturin, difficidin, and macrolactin group, with known mechanisms of action. Bv V4 has the potential for use as an aquaculture probiotic and as an antifurunculosis agent. With similar objectives, Van Doan et al. (2018) reported an improved immunity (via significantly elevated serum and skin mucus lysozyme and peroxidase activities, alternative complement activity, phagocytic activity, and respiratory burst activities) of Nile Tilapia when treated with a probiotic mixture of Lactobacillus plantarum N11 and Bv H3.1., as a feed additive. However, adequate information on the specific identity of the active metabolites (from the Bv strain) responsible for the immune boosts and mechanism by which the metabolite worked synergistically with other components of the probiotic is currently lacking.
And lastly, Yoo et al. (2018) isolated Bv K68 from traditional Korean-fermented foods and indicated its functionality and possible use in the prevention of dental caries, caused by Streptococcus mutans by inhibiting S. mutans biofilm formation, adhesion, and GTF gene expression, through deoxynojirimycin (DNJ) production. This study and all of the aforementioned literature substantiate the use of Bv, various Bv extracts and purified compounds from Bv for various medicinal/clinical applications in both humans and animals.
Possible future use of metabolic information in combination with other omics derived data
Microbial genome-scale metabolic models (GEMs) have been previously developed but to date rarely used as systems’ metabolic engineering strategies for strain design and development (Covert et al. 2001; Lee et al. 2010; Oh et al. 2007). Applying microbial metabolic models or predictive pathway analyses could greatly assist researchers in predicting structural and functional properties of various microorganisms, including that of the Bv strains discussed in this review. Microbial metabolic models can additionally be used as a tool for generating hypotheses and engineering the metabolism of several organisms (Liu et al. 2010a; Patil et al. 2004). For example, a recombinant B. subtilis strain (BBG100), obtained from a wild strain ATCC 6633, was engineered to over secrete mycosubtilin (15-fold increase), via manipulation of its internal mycosubtilin operon (Leclère et al. 2005). A similar approach was used to generate various microbial strains for the production of novel bio-based compounds, medical antibiotics, plant/crop inoculants, and industrial chemicals via fermentation (Mienda 2017; Zachow et al. 2015).
The discovery of the non-ribosomal peptide synthetase (NRPS) siderophores (cupriachelin and taiwachelin) via in silico genome mining of the nitrogen-fixing bacteria Cupriavidus necator H16 (syn. Ralstonia eutropha H16) and C. taiwanensis LMG19424 by Kreutzer et al. (2012); Kreutzer and Nett (2012) also follow a genome-scale metabolic modeling. The predicted metabolic map (from the genome sequence data) by the same researchers subsequently assisted in the reconstruction of the putative pathways for the biosynthesis of the aforementioned lipopeptides. The reconstructed pathway was then used in the formulation of the fermentation growth media for the synthesis of these aforementioned compounds, which were subsequently purified, extracted, and identified.
Biosynthetic modeling of B. velezensis NWUMFkBS10.5 genome
Using the KEGG platform and the RAST server genome annotation of the Fusarium-biosuppressor Bv NWUMFkBS10.5 Adeniji et al. (2018), a genome-scale metabolic model of the Bv NWUMFkBS10.5 was mapped and or constructed. The final model includes 1558 chemical reactions, employing 1559 compounds, associated with 1000 genes. Twenty-eight possible significant compounds were predicted from the modeled metabolic pathway. In Fig. 6, the biosynthetic uniqueness of this genome is illustrated and compared to the genomic map of three other referenced Bv strains (using the default parameters of the web server BIOiPLUG Apps (https://www.bioiplug.com/apps). One of the major novelties of the NWUMFkBS10.5 genome is the presence of the betalain and novobiocin metabolic pathways, responsible for the synthesis of interesting compounds including gomphrenin-1, lampranthin-2, celosianin-2, 2-decarboxybetanidin, betalamic acid, miraxanthin V, albamycin, and maltol. These compounds have been previously documented to be useful antioxidants, food additives, and flavor enhancers. A summary of the possible metabolic pathways for the synthesis of some of these beneficial compounds is given in Table 3.
Exploiting the Bacillus velezensis metabolome using practicable metabolomic insights
Interpretation of microbial intracellular information derived from all the other omics data, in conjunction with metabolomics data, would potentially revolutionize the quest for identifying and synthesizing various novel and useful natural microbial compounds (Adeniji and Babalola 2018), thereby further enhancing natural products discovery (Fig. 7). The benefits of using the multi-omics platform approach with deliberate convergence to metabolomics, for microbial bioengineering applications, are clear. Various studies (as discussed in this review) have attempted to use a multi-dimensional omics approach to determine the role of several candidate biocontrol compounds synthesized by various Bv strains (Chen et al. 2018c). Other practicable insights are further described below. For instance, during a host–microbe interaction (within a clinical, industrial, environmental, and agricultural setting), various metabolic processes may be disrupted within a treated pathogen or living host, after the application of a beneficial microbe, or its purified beneficial compounds. Metabolomics can provide valuable information on the bio-transformation that typically occurs in the metabolome of these beneficial microbes when they are utilized directly as a treatment alternative (Xu et al. 2014).
Furthermore, metabolomics can be used to monitor systemic acquired resistance/induced systemic resistance in plants induced by exogenous microbial treatment. Its application during microbial industrial fermentation studies may additionally elucidate changes to the metabolic flux, enzyme functions, and gene function, which in turn can be applied to optimizing fermentation protocols (Gao and Xu 2015). Zhao et al. (2016) investigated the inhibitory effect of a phage protein (gp70.1) on Pseudomonas aeruginosa using transcriptomics and NMR-based metabolomics. The phage protein caused a significant reduction in the rate of amino acid consumption by P. aeruginosa. The up-production of alanine and pyroglutamate and downregulation of ornithine which was revealed by the NMR spectroscopy confirmed the phenotypic disruptions observed in the extracellular components of the organism.
Lastly, Wu et al. (2015a) elucidated the mechanism by which the biocontrol bacteria Penicillium citrinum W1 could improve its production of the antifungal protein, PcPAF. Using a GC-MS metabolomics approach, they determined an upregulation in the fatty acid synthesis and TCA cycle intermediates when cultured in media supplemented with glycine, serine, and threonine, correlating to the phenotypic expression of increased bacterial growth and improved antagonism (Wu et al. 2015a). Similarly, Koen et al. (2018a) and Koen et al. (2018b) used a GCXGC-TOFMS metabolomics approach to determine the intracellular response of the organism Mycobacterium tuberculosis (Mtb) to the antimicrobials colistin sulfate and colistin methanesulfonate respectively, in order to determine possible mechanisms of action of the antimicrobial. Increased uptake of various metabolic precursors (e.g., glucose) and a subsequent elevation of other underutilized precursors within the cell, all of which were related to fatty acid synthesis and cell wall repair, were reported by the authors. The metabolomics study corroborated previously proposed genomics and histological generated hypotheses, regarding colistin’s mechanism of action against Mtb. Negative alterations in the metabolome of beneficial microbes (like Bv) could be monitored and controlled borrowing from the perspectives above.
Final remarks and conclusion
It is becoming paramount that the biosynthetic capacity of microbes identified through other omics strategies be validated via metabolomics, and the presupposed or predicted significant microbial biomolecule (or the products of microbial fermentation) is subsequently detected or synthesized in vitro (Palazzotto and Weber 2018). Apparently at inception, research into microbial metabolomics was limited by the capacity of analytical techniques available at the time; however, the recent advancement of the platform (metabolomics) has resulted in an exponential increase in new and exciting discoveries (Xu et al. 2014). We anticipate an exponential increase in the use of systems biology research approaches towards the discovery of new compounds from microbial sources over the course of the next 10 years. A number of challenges however still exist, and these include the difficulty in extracting or purifying novel compounds from fermentation broths produced by these beneficial microbes (e.g., Bv). The latter limitation is largely attributed to suboptimized growth conditions, improper selection of extraction solvents, and a lack of availability for novel compounds commercially for absolute identification of novel compounds, especially those synthesized in lower concentrations (Machado et al. 2017). The reported non-selectiveness of many microbial-derived beneficial agents and their apparent inability to competently function in vivo as detected in vitro, in comparison to synthetic antimicrobials, is a challenge. A further question is whether the location or sources of isolation of these beneficial bacteria (e.g., Bv) influence the diversity of their applications.
This study expounds on the phylogeny of Bv (using current omics tools) to actually corroborate previous claims on the close knitting of the species, while comprehensively revealing the omics techniques employed in the past investigations relating to Bv. Using metabolic modeling, we identified possible novel biomolecules from our Bv strain (NWUMfkBS10.5). We have also shared interesting perspectives on how the Bv metabolome could be further exploited, using current metabolomic trends capable of detecting biomarkers pertinent to novel biosynthetic pathways. Despite the inexhaustible diversity of microbial metabolites and the beneficial compounds produced by beneficial bacteria, there are only a few databases which currently exist, documenting these compounds and their biotechnological applications. Metabolomics would serve a useful approach to generating such info quickly, which deserves attention considering the need for sustainable production of novel biotechnological products.
References
Adeniji AA, Babalola OO (2018) Tackling maize fusariosis: in search of Fusarium graminearum biosuppressors. Arch Microbiol 200:1239–1255. https://doi.org/10.1007/s00203-018-1542-y
Adeniji AA, Aremu OS, Babalola OO (2018) Selecting lipopeptide-producing, Fusarium-suppressing Bacillus spp.: metabolomic and genomic probing of Bacillus velezensis NWUMFkBS10. Microbiologyopen 25:e742
Arkin AP, Stevens RL, Cottingham RW, Maslov S, Henry CS, Dehal P, Ware D, Perez F, Harris NL, Canon S (2016) The DOE systems biology knowledgebase (KBase) bioRxiv 096354
Bafana A, Chakrabarti T, Devi SS (2008) Azoreductase and dye detoxification activities of Bacillus velezensis strain AB. Appl Microbiol Biotechnol 77:1139–1144. https://doi.org/10.1007/s00253-007-1212-5
Baptista JP, Sanches PP, Teixeira GM, Morey AT, Tavares ER, Yamada-Ogatta SF, da Rocha SPD, Hungria M, Ribeiro RA, Balbi-Peña MI (2018) Complete genome sequence of Bacillus velezensis LABIM40, an effective antagonist of fungal plant pathogens. Genome Announc 6:e00595–e00518
Bardou P, Mariette J, Escudié F, Djemiel C, Klopp C (2014) jvenn: an interactive Venn diagram viewer. BMC Bioinformatics 15:293
Cai X, Kang X, Xi H, Liu C, Xue Y (2016) Complete genome sequence of the endophytic biocontrol strain Bacillus velezensis CC09. Genome Announc 4:e01048–e01016
Cai X-C, Liu C-H, Wang B-T, Xue Y-R (2017) Genomic and metabolic traits endow Bacillus velezensis CC09 with a potential biocontrol agent in control of wheat powdery mildew disease. Microbiol Res 196:89–94. https://doi.org/10.1016/j.micres.2016.12.007
Calvo P, Watts DB, Ames RN, Kloepper JW, Torbert HA (2013) Microbial-based inoculants impact nitrous oxide emissions from an incubated soil medium containing urea fertilizers. J Environ Qual 42:704–712. https://doi.org/10.2134/jeq2012.0300
Calvo P, Watts DB, Kloepper JW, Torbert HA (2016) The influence of microbial-based inoculants on N2O emissions from soil planted with corn (Zea mays L.) under greenhouse conditions with different nitrogen fertilizer regimens. Can J Microbiol 62:1041–1056
Cao Y, Hualiang P, Chandrangsu P, Yongtao L, Wang Y, Zhou H, Xiong H, Helmann JD, Cai Y (2018) Antagonism of two plant-growth promoting Bacillus velezensis isolates against Ralstonia solanacearum and Fusarium oxysporum. Sci Rep 8:4360. https://doi.org/10.1038/s41598-018-22782-z
Carlos Guimaraes L, Benevides de Jesus L, Vinicius Canario Viana M, Silva A, Thiago Juca Ramos R, de Castro Soares S, Azevedo V (2015) Inside the pan-genome—methods and software overview. Curr Genomics 16:245–252
Chen L (2017) Complete genome sequence of Bacillus velezensis LM2303, a biocontrol strain isolated from the dung of wild yak inhabited Qinghai-Tibet plateau. J Biotechnol 251:124–127. https://doi.org/10.1016/j.jbiotec.2017.04.034
Chen Y, Gao Q, Huang M, Liu Y, Liu Z, Liu X, Ma Z (2015) Characterization of RNA silencing components in the plant pathogenic fungus Fusarium graminearum. Sci Rep 5:12500
Chen L, Gu W, Xu H-Y, Yang G-L, Shan X-F, Chen G, Kang Y-H, Wang C-F, Qian A-D (2018a) Comparative genome analysis of Bacillus velezensis reveals a potential for degrading lignocellulosic biomass. 3 Biotech 8:253. https://doi.org/10.1007/s13205-018-1270-7
Chen L, Gu W, Xu H-Y, Yang G-L, Shan X-F, Chen G, Wang C-F, Qian A-D (2018b) Complete genome sequence of Bacillus velezensis 157 isolated from Eucommia ulmoides with pathogenic bacteria inhibiting and lignocellulolytic enzymes production by SSF. 3 Biotech 8:114. https://doi.org/10.1007/s13205-018-1125-2
Chen L, Heng J, Qin S, Bian K (2018c) A comprehensive understanding of the biocontrol potential of Bacillus velezensis LM2303 against Fusarium head blight. PLoS One 13:e0198560
Cho MS, Jin YJ, Kang BK, Park YK, Kim C, Park DS (2018) Understanding the ontogeny and succession of Bacillus velezensis and B. subtilis subsp. subtilis by focusing on kimchi fermentation. Sci Rep 8:7045. https://doi.org/10.1038/s41598-018-25514-5
Covert MW, Schilling CH, Famili I, Edwards JS, Goryanin II, Selkov E, Palsson BO (2001) Metabolic modeling of microbial strains in silico. Trends Biochem Sci 26:179–186. https://doi.org/10.1016/S0968-0004(00)01754-0
Disi JO, Kloepper JW, Fadamiro HY (2018a) Seed treatment of maize with Bacillus pumilus strain INR- 7 affects host location and feeding by Western corn rootworm, Diabrotica virgifera virgifera. J Pest Sci 91:515–522. https://doi.org/10.1007/s10340-017-0927-z
Disi JO, Zebelo S, Kloepper JW, Fadamiro H (2018b) Seed inoculation with beneficial rhizobacteria affects European corn borer (Lepidoptera: Pyralidae) oviposition on maize plants. Entomol Sci 21:48–58. https://doi.org/10.1111/ens.12280
Dunlap CA, Bowman MJ, Schisler DA (2013) Genomic analysis and secondary metabolite production in Bacillus amyloliquefaciens AS 43.3: a biocontrol antagonist of Fusarium head blight. Biol Control 64:166–175
Dunlap CA, Kim S-J, Kwon S-W, Rooney AP (2016) Bacillus velezensis is not a later heterotypic synonym of Bacillus amyloliquefaciens; Bacillus methylotrophicus, Bacillus amyloliquefaciens subsp. plantarum and ‘Bacillus oryzicola’ are later heterotypic synonyms of Bacillus velezensis based on phylogenomics. Int J Syst Evol Microbiol 66:1212–1217. https://doi.org/10.1099/ijsem.0.000858
Fan B, Blom J, Klenk HP, Borriss R (2017) Bacillus amyloliquefaciens, Bacillus velezensis, and Bacillus siamensis form an “operational group B. amyloliquefaciens” within the B. subtilis species complex. Front Microbiol 8:22. https://doi.org/10.3389/fmicb.2017.00022
Gao P, Xu G (2015) Mass-spectrometry-based microbial metabolomics: recent developments and applications. Anal Bioanal Chem 407:669–680
Gao X-Y, Liu Y, Miao L-L, Li E-W, Sun G-X, Liu Y, Liu Z-P (2017a) Characterization and mechanism of anti-Aeromonas salmonicida activity of a marine probiotic strain, Bacillus velezensis V4. Appl Microbiol Biotechnol 101:3759–3768. https://doi.org/10.1007/s00253-017-8095-x
Gao Z, Zhang B, Liu H, Han J, Zhang Y (2017b) Identification of endophytic Bacillus velezensis ZSY-1 strain and antifungal activity of its volatile compounds against Alternaria solani and Botrytis cinerea. Biol Control 105:27–39. https://doi.org/10.1016/j.biocontrol.2016.11.007
Gerst MM, Dudley EG, Xiaoli L, Yousef AE (2017) Draft genome sequence of Bacillus velezensis GF610, a producer of potent Anti-Listeria agents. Genome Announc 5:e01046-01017
Gilardi G, Demarchi S, Gullino ML, Garibaldi A (2015) Nursery treatments with non-conventional products against crown and root rot, caused by Phytophthora capsici, on zucchini. Phytoparasitica 43:501–508. https://doi.org/10.1007/s12600-015-0461-6
Huang L, Li Q-C, Hou Y, Li G-Q, Yang J-Y, Li D-W, Ye J-R (2017a) Bacillus velezensis strain HYEB5-6 as a potential biocontrol agent against anthracnose on Euonymus japonicus. Biocontrol Sci Technol 27:636–653. https://doi.org/10.1080/09583157.2017.1319910
Huang MH, Zhang SQ, Di GL, Xu LK, Zhao TX, Pan HY, Li YG (2017b) Determination of a Bacillus velezensis strain for controlling soybean root rot. Biocontrol Sci Technol 27:696–701. https://doi.org/10.1080/09583157.2017.1328483
Hwangbo K, Um Y, Kim KY, Madhaiyan M, Sa TM, Lee Y (2016) Complete genome sequence of Bacillus velezensis CBMB205, a phosphate-solubilizing bacterium Isolated from the rhizoplane of rice in the Republic of Korea. Genome Announc 4. https://doi.org/10.1128/genomeA.00654-16
Jiang C-H, Liao M-J, Wang H-K, Zheng M-Z, Xu J-J, Guo J-H (2018) Bacillus velezensis, a potential and efficient biocontrol agent in control of pepper gray mold caused by Botrytis cinerea. Biol Control 126:147–157. https://doi.org/10.1016/j.biocontrol.2018.07.017
Jin Q, Jiang Q, Zhao L, Su C, Li S, Si F, Li S, Zhou C, Mu Y, Xiao M (2017) Complete genome sequence of Bacillus velezensis S3-1, a potential biological pesticide with plant pathogen inhibiting and plant promoting capabilities. J Biotechnol 259:199–203. https://doi.org/10.1016/j.jbiotec.2017.07.011
Kang X, Zhang W, Cai X, Zhu T, Xue Y, Liu C (2018) Bacillus velezensis CC09: a potential ‘vaccine’ for controlling wheat diseases. Mol Plant-Microbe Interact 31:623–632. https://doi.org/10.1094/MPMI-09-17-0227-R
Kim SY, Lee SY, Weon H-Y, Sang MK, Song J (2017a) Complete genome sequence of Bacillus velezensis M75, a biocontrol agent against fungal plant pathogens, isolated from cotton waste. J Biotechnol 241:112–115. https://doi.org/10.1016/j.jbiotec.2016.11.023
Kim SY, Song H, Sang MK, Weon H-Y, Song J (2017b) The complete genome sequence of Bacillus velezensis strain GH1-13 reveals agriculturally beneficial properties and a unique plasmid. J Biotechnol 259:221–227. https://doi.org/10.1016/j.jbiotec.2017.06.1206
Koen N, van Breda SV, Loots DT (2018a) Elucidating the antimicrobial mechanisms of colistin sulfate on Mycobacterium tuberculosis using metabolomics Tuberculosis 111:14-19
Koen N, van Breda SV, Loots DT (2018b) Metabolomics of colistin methanesulfonate treated Mycobacterium tuberculosis. Tuberculosis 111:154–160
Kreutzer MF, Nett M (2012) Genomics-driven discovery of taiwachelin, a lipopeptide siderophore from Cupriavidus taiwanensis. Org Biomol Chem 10:9338–9343. https://doi.org/10.1039/C2OB26296G
Kreutzer MF, Kage H, Nett M (2012) Structure and biosynthetic assembly of cupriachelin, a photoreactive siderophore from the bioplastic producer Cupriavidus necator H16. J Am Chem Soc 134:5415–5422. https://doi.org/10.1021/ja300620z
Lan TTT (2016) Study on the growth of Bacillus velezensis M2 and applying it for treatment of the cattle slaughterhouse wastewater. Vietnam J Sci Technol 54:213
Leclère V, Béchet M, Adam A, Guez J-S, Wathelet B, Ongena M, Thonart P, Gancel F, Chollet-Imbert M, Jacques P (2005) Mycosubtilin overproduction by Bacillus subtilis BBG100 enhances the organism’s antagonistic and biocontrol activities. Appl Environ Microbiol 71:4577–4584
Lee KY, Park JM, Kim TY, Yun H, Lee SY (2010) The genome-scale metabolic network analysis of Zymomonas mobilis ZM4 explains physiological features and suggests ethanol and succinic acid production strategies. Microb Cell Factories 9:94–94. https://doi.org/10.1186/1475-2859-9-94
Lee HH, Park J, Lim JY, Kim H, Choi GJ, Kim J-C, Seo Y-S (2015) Complete genome sequence of Bacillus velezensis G341, a strain with a broad inhibitory spectrum against plant pathogens. J Biotechnol 211:97–98. https://doi.org/10.1016/j.jbiotec.2015.07.005
Lee HJ, Chun B-H, Jeon HH, Kim YB, Lee SH (2017) Complete genome sequence of Bacillus velezensis YJ11-1-4, a strain with broad-spectrum antimicrobial activity, isolated from traditional Korean fermented soybean paste. Genome Announc 5:e01352–e01317
Li Z, Chen M, Ran K, Wang J, Zeng Q, Song F (2018) Draft genome sequence of Bacillus velezensis Lzh-a42, a plant growth-promoting rhizobacterium isolated from tomato rhizosphere. Genome Announc 6:e00161–e00118
Lim SM, Yoon M-Y, Choi GJ, Choi YH, Jang KS, Shin TS, Park HW, Yu NH, Kim YH, Kim J-C (2017) Diffusible and volatile antifungal compounds produced by an antagonistic Bacillus velezensis G341 against various phytopathogenic fungi. Plant Pathol J 33:488–498. https://doi.org/10.5423/PPJ.OA.04.2017.0073
Liu S, Lin L, Jiang P, Wang D, Xing Y (2010a) A comparison of RNA-Seq and high-density exon array for detecting differential gene expression between closely related species. Nucleic Acids Res 39:39–588. https://doi.org/10.1093/nar/gkq817
Liu X, Ren B, Chen M, Wang H, Kokare CR, Zhou X, Wang J, Dai H, Song F, Liu M, Wang J, Wang S, Zhang L (2010b) Production and characterization of a group of bioemulsifiers from the marine Bacillus velezensis strain H3. Appl Microbiol Biotechnol 87:1881–1893. https://doi.org/10.1007/s00253-010-2653-9
Liu G, Kong Y, Fan Y, Geng C, Peng D, Sun M (2017) Whole-genome sequencing of Bacillus velezensis LS69, a strain with a broad inhibitory spectrum against pathogenic bacteria. J Biotechnol 249:20–24. https://doi.org/10.1016/j.jbiotec.2017.03.018
Machado H, Tuttle RN, Jensen PR (2017) Omics-based natural product discovery and the lexicon of genome mining. Curr Opin Microbiol 39:136–142. https://doi.org/10.1016/j.mib.2017.10.025
Mahgoub AM, Mahmoud MG, Selim MS, Awady E, Mohamed E (2018) Exopolysaccharide from marine Bacillus velezensis MHM3 induces apoptosis of human breast cancer MCF-7 cells through a mitochondrial pathway. Asian Pac J Cancer Prev 19:1957–1963
Martínez-Raudales I, De La Cruz-Rodríguez Y, Alvarado-Gutiérrez A, Vega-Arreguín J, Fraire-Mayorga A, Alvarado-Rodríguez M, Balderas-Hernández V, Fraire-Velázquez S (2017) Draft genome sequence of Bacillus velezensis 2A-2B strain: a rhizospheric inhabitant of Sporobolus airoides (Torr.) Torr., with antifungal activity against root rot causing phytopathogens. Stand Genomic Sci 12:73. https://doi.org/10.1186/s40793-017-0289-4
Meena KR, Tandon T, Sharma A, Kanwar SS (2018) Lipopeptide antibiotic production by Bacillus velezensis KLP2016. J Appl Pharm Sci 8:091–098
Meng Q, Hao JJ (2017) Optimizing the application of Bacillus velezensis BAC03 in controlling the disease caused by Streptomyces scabies. BioControl 62:535–544. https://doi.org/10.1007/s10526-017-9799-7
Meng Q, Jiang H, Hao JJ (2016) Effects of Bacillus velezensis strain BAC03 in promoting plant growth. Biol Control 98:18–26. https://doi.org/10.1016/j.biocontrol.2016.03.010
Mienda BS (2017) Genome-scale metabolic models as platforms for strain design and biological discovery. J Biomol Struct Dyn 35:1863–1873. https://doi.org/10.1080/07391102.2016.1197153
Moghannem SAM, Farag MMS, Shehab AM, Azab MS (2018) Exopolysaccharide production from Bacillus velezensis KY471306 using statistical experimental design. Braz J Microbiol 49:452–462. https://doi.org/10.1016/j.bjm.2017.05.012
Nair AS, Al-Battashi H, Al-Akzawi A, Annamalai N, Gujarathi A, Al-Bahry S, Dhillon GS, Sivakumar N (2018) Waste office paper: a potential feedstock for cellulase production by a novel strain Bacillus velezensis ASN1. Waste Manage 79:491–500. https://doi.org/10.1016/j.wasman.2018.08.014
Nam MH, Park MS, Kim HG, Yoo SJ (2009) Biological control of strawberry Fusarium wilt caused by Fusarium oxysporum f. sp. fragariae using Bacillus velezensis BS87 and RK1 formulation. J Microbiol Biotechnol [En línea] 19:520–524
Nannan C, Gillis A, Caulier S, Mahillon J (2018) Complete genome sequence of Bacillus velezensis CN026 exhibiting antagonistic activity against gram-negative foodborne pathogens. Genome Announc 6:e01543-01517
Oh Y-K, Palsson BO, Park SM, Schilling CH, Mahadevan R (2007) Genome-scale reconstruction of metabolic network in Bacillus subtilis based on high-throughput phenotyping and gene essentiality data. J Biol Chem 282:28791–28799. https://doi.org/10.1074/jbc.M703759200
Palazzotto E, Weber T (2018) Omics and multi-omics approaches to study the biosynthesis of secondary metabolites in microorganisms. Curr Opin Microbiol 45:109–116. https://doi.org/10.1016/j.mib.2018.03.004
Palazzini JM, Dunlap CA, Bowman MJ, Chulze SN (2016) Bacillus velezensis RC 218 as a biocontrol agent to reduce Fusarium head blight and deoxynivalenol accumulation: Genome sequencing and secondary metabolite cluster profiles. Microbiol Res 192:30–36. https://doi.org/10.1016/j.micres.2016.06.002
Palencia P, Martínez F, Pestana M, Oliveira JA, Correia PJ (2015) Effect of Bacillus velezensis and Glomus intraradices on fruit quality and growth parameters in strawberry soilless growing system. Hort J 84:122–130
Pan HQ, Li QL, Hu JC (2017) The complete genome sequence of Bacillus velezensis 9912D reveals its biocontrol mechanism as a novel commercial biological fungicide agent. J Biotechnol 247:25–28. https://doi.org/10.1016/j.jbiotec.2017.02.022
Pandin C, Le Coq D, Deschamps J, Vedie R, Rousseau T, Aymerich S, Briandet R (2018) Complete genome sequence of Bacillus velezensis QST713: a biocontrol agent that protects Agaricus bisporus crops against the green mould disease. J Biotechnol 278:10–19. https://doi.org/10.1016/j.jbiotec.2018.04.014
Patil KR, Åkesson M, Nielsen J (2004) Use of genome-scale microbial models for metabolic engineering. Curr Opin Biotechnol 15:64–69. https://doi.org/10.1016/j.copbio.2003.11.003
Rashid MH-O, Khan A, Hossain MT, Chung YR (2017) Induction of systemic resistance against aphids by endophytic Bacillus velezensis YC7010 via expressing phytoalexin deficient4 in Arabidopsis. Front Plant Sci 8:211. https://doi.org/10.3389/fpls.2017.00211
Rehman NU, Abed RMM, Hussain H, Khan HY, Khan A, Khan AL, Ali M, Al-Nasri A, Al-Harrasi K, Al-Rawahi AN, Wadood A, Al-Rawahi A, Al-Harrasi A (2018) Anti-proliferative potential of cyclotetrapeptides from Bacillus velezensis RA5401 and their molecular docking on G-protein-coupled receptors. Microb Pathog 123:419–425. https://doi.org/10.1016/j.micpath.2018.07.043
Ruiz-García C, Béjar V, Martínez-Checa F, Llamas I, Quesada E (2005) Bacillus velezensis sp. nov., a surfactant-producing bacterium isolated from the river Vélez in Málaga, southern Spain. Int J Syst Evol Microbiol 55:191–195. https://doi.org/10.1099/ijs.0.63310-0
Van Doan H, Hoseinifar SH, Khanongnuch C, Kanpiengjai A, Unban K, Van Kim V, Srichaiyo S (2018) Host-associated probiotics boosted mucosal and serum immunity, disease resistance and growth performance of Nile tilapia (Oreochromis niloticus). Aquaculture 491:94–100. https://doi.org/10.1016/j.aquaculture.2018.03.019
Wambacq E, Audenaert K, Hofte M, De Saeger S, Haesaert G (2018) Bacillus velezensis as antagonist towards Penicillium roqueforti s.l. in silage: in vitro and in vivo evaluation. J Appl Microbiol 125:1–11. https://doi.org/10.1111/jam.13944
Wu C-W, Zhao X, Wu X-J, Wen C, Li H, Chen X-H, Peng X-X (2015a) Exogenous glycine and serine promote growth and antifungal activity of Penicillium citrinum W1 from the south-west Indian Ocean FEMS. Microbiol Lett 362:fnv040
Wu L, Wu H-J, Qiao J, Gao X, Borriss R (2015b) Novel routes for improving biocontrol activity of Bacillus based bioinoculants. Front Microbiol 6:1395. https://doi.org/10.3389/fmicb.2015.01395
Wu Y-S, Ngai S-C, Goh B-H, Chan K-G, Lee L-H, Chuah L-H (2017) Anticancer activities of surfactin and potential application of nanotechnology assisted surfactin delivery. Front Pharmacol 8:761. https://doi.org/10.3389/fphar.2017.00761
Xu Y-J, Wang C, Ho WE, Ong CN (2014) Recent developments and applications of metabolomics in microbiological investigations TrAC. Trends Anal Chem 56:37–48. https://doi.org/10.1016/j.trac.2013.12.009
Ye M, Tang X, Yang R, Zhang H, Li F, Tao F, Li F, Wang Z (2018) Characteristics and application of a novel species of Bacillus: Bacillus velezensis. ACS Chem Biol 13:500–505. https://doi.org/10.1021/acschembio.7b00874
Yoo Y, Seo D-H, Lee H, Nam Y-D, Seo M-J (2018) Inhibitory effect of Bacillus velezensis on biofilm formation by Streptococcus mutans. bioRxiv:313965
Zachow C, Jahanshah G, de Bruijn I, Song C, Ianni F, Pataj Z, Gerhardt H, Pianet I, Lämmerhofer M, Berg G, Gross H, Raaijmakers JM (2015) The novel lipopeptide poaeamide of the endophyte Pseudomonas poae RE*1-1-14 is involved in pathogen suppression and root colonization. Mol Plant-Microbe Interact 28:800–810. https://doi.org/10.1094/MPMI-12-14-0406-R
Zaki SA, Elkady MF, Farag S, Abd-El-Haleem D (2013) Characterization and flocculation properties of a carbohydrate bioflocculant from a newly isolated Bacillus velezensis 40B. J Environ Biol 34:51
Zhao X, Chen C, Jiang X, Shen W, Huang G, Le S, Lu S, Zou L, Ni Q, Li M, Zhao Y, Wang J, Rao X, Hu F, Tan Y (2016) Transcriptomic and metabolomic analysis revealed multifaceted effects of phage protein Gp70.1 on Pseudomonas aeruginosa. Front Microbiol 7:1519. https://doi.org/10.3389/fmicb.2016.01519
Acknowledgements
The first author appreciates North-West University for post-doctoral fellowship. This work is based on the research supported by National Research Foundation, South Africa (UID95111) France/SA bilateral.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies involving human participants or animals.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Adeniji, A.A., Loots, D.T. & Babalola, O.O. Bacillus velezensis: phylogeny, useful applications, and avenues for exploitation. Appl Microbiol Biotechnol 103, 3669–3682 (2019). https://doi.org/10.1007/s00253-019-09710-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-019-09710-5