Abstract
Subcellular compartmentalization of the biosynthetic enzymes is one of the limiting factors for isobutanol production in Saccharomyces cerevisiae. Previously, it has been shown that mitochondrial compartmentalization of the biosynthetic pathway through re-locating cytosolic Ehrlich pathway enzymes into the mitochondria can increase isobutanol production. In this study, we improved mitochondrial isobutanol production by increasing mitochondrial pool of pyruvate, a key substrate for isobutanol production. Mitochondrial isobutanol biosynthetic pathway was introduced into bat1Δald6Δlpd1Δ strain, where genes involved in competing pathways were deleted, and MPC1, MPC2, and MPC3 genes encoding the subunits of mitochondrial pyruvate carrier (MPC) hetero-oligomeric complex were overexpressed with different combinations. Overexpression of Mpc1 and Mpc3 forming high-affinity MPCOX was more effective in improving isobutanol production than overexpression of Mpc1 and Mpc2 forming low-affinity MPCFERM. The final engineered strain overexpressing MPCOX produced 330.9 mg/L isobutanol from 20 g/L glucose, exhibiting about 22-fold increase in production compared to wild type.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Microbial production of higher alcohols has attracted great attention as renewable next-generation biofuels because of fluctuating oil prices and increasing environmental concerns (Connor and Liao 2009; Weber et al. 2010). Especially, isobutanol, a kind of branched-chain higher alcohol, has desirable properties as transport fuels, such as lower moisture absorption and higher energy density than ethanol, and higher octane value than its linear-chain counterpart (Atsumi et al. 2008; Blombach and Eikmanns 2011; Weber et al. 2010). Isobutanol has been successfully produced with high titer in several bacteria including Escherichia coli, Corynebacterium glutamicum, and Bacillus subtilis by introducing the last two enzymes of Ehrlich pathway, 2-ketoacid decarboxylase (KDC) and alcohol dehydrogenase (ADH), which catalyze the conversion of 2-ketoacid into corresponding alcohols (Blombach and Eikmanns 2011). In these engineered bacterial strains, 2-ketoisovalerate (2-KIV) is produced from two molecules of pyruvate by sequential actions of α-acetolactate synthase (ALS), ketol acid reductoisomerase (KARI), and dihydroxyacid dehydratase (DHAD), and then 2-KIV is further converted into isobutanol by KDC and ADH (Atsumi et al. 2008; Hazelwood et al. 2008). However, the low tolerance to butanol can be a limiting factor for industrial-scale isobutanol production by using non-native producers (Knoshaug and Zhang 2009; Weber et al. 2010).
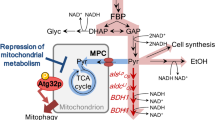
Saccharomyces cerevisiae has been believed as a promising host for isobutanol production because it is recognized as safe and has high tolerance to isobutanol and harsh industrial conditions (Hohmann 2002; Nevoigt 2008; Porro et al. 2011). S. cerevisiae can naturally produce small amounts of isobutanol from glucose (Hazelwood et al. 2008). Although several metabolic engineering efforts have been made to produce isobutanol in S. cerevisiae, the reported production levels are still low compared with those produced in bacteria (Avalos et al. 2013; Brat et al. 2012; Buijs et al. 2013; Chen et al. 2011; Ida et al. 2015; Kondo et al. 2012; Lee et al. 2012; Milne et al. 2015; Park et al. 2014). Because isobutanol production pathway is subdivided by subcellular compartmentalization in S. cerevisiae, it is essential to transport intermediates across the membranes, which could be one of the bottlenecks in isobutanol production (Avalos et al. 2013). In S. cerevisiae, cytosolic pyruvate generated by glycolysis is imported into the mitochondrial matrix by mitochondrial pyruvate carrier (MPC) (Fig. 1) (Bricker et al. 2012; Herzig et al. 2012). Two molecules of pyruvate are converted into a 2-KIV by consecutive actions of Ilv2 (ALS), Ilv5 (KARI), and Ilv3 (DHAD) in the mitochondria, and 2-KIV is further converted into isobutanol in the cytoplasm by endogenous KDCs and ADHs in yeast (Fig. 1) (Hazelwood et al. 2008). 2-KIV is also used for the production of branched-chain amino acids including valine and leucine. Valine is generated from 2-KIV by mitochondrial branched-chain amino acid transferase (Bat1) (Fig. 1) (Kispal et al. 1996; Lilly et al. 2006). On the other hand, 2-KIV is first converted to 2-isopropylmalate (2-IPM) by 2-isopropylmalate synthase (Leu4), followed by three enzymatic steps mediated by Leu1, Leu2, and Bat2 to produce leucine (Fig. 1) (Baichwal et al. 1983). To overcome the limitations resulting from subcellular compartmentalization of isobutanol pathway enzymes in yeast, the whole biosynthetic pathway was relocated either to the cytoplasm by expressing mitochondrial enzymes (Ilv2, Ilv3, and Ilv5) in the cytoplasm (Brat et al. 2012; Lee et al. 2012) or to the mitochondria by expressing cytosolic enzymes (KDCs and ADHs) in the mitochondria (Avalos et al. 2013). In a study comparing the effects of the two approaches, the mitochondrial pathway led to higher isobutanol production level than the cytosolic pathway (Avalos et al. 2013).
Biosynthetic pathways for isobutanol production from glucose in S. cerevisiae. Cytosolic pyruvate, produced by glycolysis, is imported into the mitochondria through mitochondrial pyruvate carrier (MPC) complex. Two molecules of mitochondrial pyruvate are converted to one molecule of isobutanol via 2-ketoisovalerate (2-KIV) and isobutyraldehyde by sequential actions of Ilv2 (acetolactate synthase), Ilv5 (ketol acid reductoisomerase), Ilv3 (dihydroxyacid dehydratase), 2-ketoacid decarboxylases (KDCs), and alcohol dehydrogenases (ADHs). Leu3 is a transcriptional activator of ILV2, ILV5, and BAT1 genes. Dashed line indicates multiple enzymatic reactions
Previously, we improved isobutanol production in S. cerevisiae by deleting competing pathway enzymes involved in isobutyrate production from isobutyraldehyde (Ald6) and valine biosynthesis from 2-KIV (Bat1), and overexpressing enzymes in the isobutanol production pathway (Ilv2, Ilv3, Ilv5, Aro10, and Adh2) in their natural compartments (Park et al. 2014). In this study, we improved our isobutanol-production strain by deleting LPD1, encoding a subunit of pyruvate dehydrogenase (PDH) and by mitochondrial compartmentalization of the biosynthetic pathway based on previous studies (Matsuda et al. 2013). The mitochondrial isobutanol production was further improved by increasing the transport of pyruvate from cytoplasm to the mitochondrial matrix by overexpressing MPC.
Materials and methods
Strains and media
All strains used in this study are described in Table 1. S. cerevisiae CEN.PK2-1C was used as a parental strain of all engineered strains. Gene deletion strains were generated by polymerase chain reaction (PCR)-based homologous recombination using Cre/loxP recombination system (Gueldener et al. 2002). Gene-specific deletion cassette, generated by PCR amplification from pUG72 plasmid, was integrated into S. cerevisiae chromosome, and then the selection marker gene was removed by introducing Cre recombinase expression vector, pSH63 (Gueldener et al. 2002). Yeast cells were cultured in YPD medium (10 g/L yeast extract, 20 g/L bacto-peptone, and 20 g/L glucose) or in synthetic complete (SC) medium (6.7 g/L yeast nitrogen base without amino acids, 20 g/L glucose, and 1.67 g/L amino acids dropout mixture lacking His, Trp, Leu, and Ura (0.083 g/L of each amino acid) supplemented with auxotrophic amino acids as needed.
Construction of plasmids
Plasmids used in this study are described in Table 2. To construct p426-MLS plasmid, mitochondrial localization signal (MLS) fragment was generated by annealing complementary oligomers corresponding to the N-terminal 25 residues of COX4 gene and was inserted between SpeI and BamHI sites of p426GPD plasmid (Mumberg et al. 1995). All genes were amplified from S. cerevisiae CEN.PK2-1C genomic DNA using target-specific primer pairs. Construction of single-gene-expression plasmid p413TEF-LEU3Δ601, containing LEU3Δ601 under the control of TEF1 promoter and CYC1 terminator, was described previously (Park et al. 2014). To generate single-gene-expression plasmids for other genes, PCR-amplified ORFs were cloned into p413GPD, p414GPD, p416GPD, p423GPD, p425GPD, or p426GPD plasmid under the control of TDH3 promoter and CYC1 terminator (Mumberg et al. 1995), resulting in p413GPD-ILV2, p414GPD-ILV5, p416GPD-ILV3, p425GPD-ARO10, p426GPD-ADH2, p426-MLS-ARO10, p426-MLS-ADH2, p423GPD-MPC1, p423GPD-MPC2, and p423GPD-MPC3.
To construct multigene-expression plasmids for isobutanol biosynthetic pathway, multiple cloning system was used as previously described with minor modifications (Kim and Hahn 2015). To generate p413-L, LEU3Δ601 expression cassette (P TEF1 -LEU3Δ601-T CYC1 ) flanked by SacI and KpnI was amplified by PCR from p413TEF-LEU3Δ601 and cloned between SacI and KpnI sites of p413-D plasmid, replacing alsD expression cassette (Kim and Hahn 2015). ILV2 expression cassette (P TDH3 -ILV2-T CYC1 ) flanked by MauBI and NotI sites was amplified by PCR using a universal primer pair, Univ F2 (5′-GACTCGCGCGCGGGAACAAAAGCTGGAGCTC-3′, MauBI site is underlined) and Univ R (5′-GACTACGCGTGCGGCCGCTAATGGCGCGCCATAGGGCGAATTGGGTACC-3′, NotI and AscI sites are underlined) and sequentially cloned into AscI and NotI sites of the p413-L plasmid. Other gene expression cassettes flanked by MauBI and NotI sites were also obtained from the single-gene-expression vectors using the same universal primers and sequentially cloned into AscI and NotI sites, resulting in pJIB1, pJIB2, pJIB3, pJIB4, and pJIB5.
Culture conditions and analytical methods
Yeast cells harboring proper plasmid were pre-cultured in selective SC-His medium containing 20 g/L glucose and then inoculated to A600 of 0.2 in the same medium with 6.5 mL culture volume in a 50-mL conical tube. Cells were grown at 30 °C with constant shaking at 170 rpm. To determine isobutanol concentration, 1.5 mL of culture supernatants was filtered through a 0.22-μm syringe filter and then analyzed by gas chromatography (GC) equipped with a DB-WAX capillary column (30 m × 0.32 mm, 0.25 μm, Agilent, Waldbronn, Germany), an auto-sampler (CP-8410, Varian, Netherlands), and a flame ionization detector (FID) following a previously described procedure (Park et al. 2014). The concentrations of glucose, ethanol, and glycerol were detected by high-performance liquid chromatography (HPLC) as described previously (Park et al. 2014).
Results
Disruption of competing pathways to increase isobutanol production
In our previous study, we improved isobutanol production by deleting ALD6 encoding aldehyde dehydrogenase and BAT1 encoding branched-chain amino acid aminotransferase, which are involved in oxidation of isobutyraldehyde and valine synthesis, respectively, competing with isobutanol production (Park et al. 2014). To further improve isobutanol production in bat1Δald6Δ (JHY43) strain, we additionally deleted genes involved in other competing pathways. Pyruvate imported into the mitochondria is converted to acetyl-CoA by PDH complex and then used for production of ATP via TCA cycle (Fig. 1) (Pronk et al. 1996). In S. cerevisiae, PDH complex is encoded by five genes, PDB1, PDA1, LAT1, LPD1, and PDX1 (Pronk et al. 1996), among which deletion of LPD1 was shown to be most effective in increasing isobutanol production (Matsuda et al. 2013). Therefore, we additionally deleted LPD1 in JHY43, generating bat1Δald6Δlpd1Δ (JHY46) strain. In agreement with previous studies (Park et al. 2014), JHY43 showed an increase in isobutanol production compared with wild type, and isobutanol production increased further in JHY46 even with reduced growth rate (Fig. 2). JHY46 produced 112.6 mg/L isobutanol after 48 h, indicating a 7.1- and 3.0- fold increase compared with WT (15.9 mg/L) and JHY43 (37.3 mg/L), respectively (Fig. 2).
Improvement of isobutanol production by deleting genes in competing pathways. WT (CEN.PK2-1C), JHY43 (ald6Δbat1Δ), JHY46 (ald6Δbat1Δlpd1Δ), JHY47 (ald6Δbat1Δleu4Δ), and JHY48 (ald6Δbat1Δlpd1Δleu4Δ) cells were grown in SC medium containing 2 % glucose for 48 h. Each value indicates the average ± SD of triplicate experiments
In addition, we tested the effect of deleting LEU4 encoding 2-isopropylmalate (2-IPM) synthase, which is the first enzyme in the leucine biosynthetic branch from 2-KIV (Baichwal et al. 1983). The resulting bat1Δald6Δleu4Δ (JHY47) strain showed increased isobutanol production up to 53.5 mg/L compared with JHY43, indicating a positive effect of LEU4 deletion in bat1Δald6Δ strain background (Fig. 2). Therefore, we next combined LEU4 and LPD1 deletion, generating bat1Δald6Δlpd1Δleu4Δ (JHY48) strain. Although JHY48 produced 1.8-fold higher level of isobutanol than did JHY47, the final production level (95.3 mg/L) was lower than that of JHY46. Consequently, JHY46 showing the highest level of isobutanol production was chosen as a platform strain for the following metabolic engineering.
Overexpression of mitochondrially re-localized isobutanol biosynthetic pathways
To further increase isobutanol production, we overexpressed genes involved in isobutanol production in JHY46 strain. Isobutanol biosynthesis from pyruvate requires six enzymes, Ilv2, Ilv5, Ilv3, KDC, and ADH (Fig. 1) (Dickinson et al. 1998; Hazelwood et al. 2008). Previously, we overexpressed these enzymes in their natural compartments: Ilv2, Ilv5, and Ilv3 in the mitochondria and Aro10 (KDC) and Adh2 (ADH) in the cytoplasm (Park et al. 2014). In addition, LEU3Δ601, encoding a constitutively active form of Leu3 transcription factor for ILV2 and ILV5 (Friden et al. 1989), was also expressed to increase endogenous expression levels of Leu3 target genes (Park et al. 2014). By overexpressing these 6 genes in bat1Δald6Δ by co-transformation of three plasmids including two high copy number plasmids, 146.9 mg/L isobutanol was produced form 2 % glucose (Park et al. 2014). In this study, to ensure more stable gene expression, we constructed a single multigene-expression plasmid containing all the required genes by using multiple cloning system (Kim and Hahn 2015). The CEN/ARS-based low copy number plasmid pJIB1 consists of ILV2, ILV3, ILV5, ARO10, ADH2, and LEU3Δ601 controlled by the strong constitutive promoter, P TDH3 or P TEF1 and terminator, T CYC1 (Table 2). The JHY46 strain harboring empty p413TEF vector (JHY46–1) produced 127.4 mg/L isobutanol, while JHY46 harboring pJIB1 plasmid (JHY461) produced up to 155.0 mg/L isobutanol in SC-His medium containing 2 % glucose after 48 h (Fig. 3), showing mild effect of gene overexpression. Although overexpressing isobutanol pathway enzymes in their natural compartments improved isobutanol titer, this native pathway requires the transport of an intermediate, 2-KIV, from the mitochondria to the cytoplasm. It has been shown previously that construction of mitochondrial pathway by re-localization of cytosolic enzymes KDC and ADH into the mitochondria could improve isobutanol production by overcoming such a limitation (Avalos et al. 2013). Therefore, we also developed mitochondrial pathway by expressing Aro10 and Adh2 in the mitochondria by tagging the proteins with N-terminal mitochondrial localization signal (MLS) of COX4 (Maarse et al. 1984), as described previously (Avalos et al. 2013). In pJIB2 plasmid, ARO10 and ADH2 of pJIB1 were replaced with MLS-ARO10 and MLS-ADH2. JHY46 harboring pJIB2 (JHY462) produced up to 185.7 mg/L isobutanol after 48 h, showing a 1.2-fold higher isobutanol titer than that produced in JHY461. These results suggest that confining Ehrlich pathway in the mitochondria might lead to spatial concentration of intermediates and enzymes involved in isobutanol production, resulting in more efficient isobutanol production as demonstrated previously (Avalos et al. 2013).
Improvement of isobutanol production by overexpressing genes involved in isobutanol biosynthetic pathway. JHY46–1 (ald6Δbat1Δlpd1Δ [EV]), JHY461 (ald6Δbat1Δlpd1Δ [JIB1]), and JHY462 (ald6Δbat1Δlpd1Δ [JIB2]) cells were grown in SC-His medium containing 2 % glucose for 48 h. Each value indicates the average ± SD of triplicate experiments
Enhancing mitochondrial pyruvate uptake by overexpressing mitochondrial pyruvate carrier (MPC)
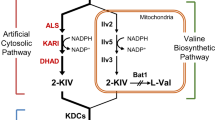
Pyruvate is a critical intermediate for the isobutanol production. Especially, the size of mitochondrial pyruvate pool might be directly related to isobutanol titer produced via mitochondrial isobutanol biosynthetic pathway. In yeast, cytosolic pyruvate is considered to cross the mitochondrial outer membrane through a kind of porins, the voltage-dependent anion channel (VDAC) (Lee et al. 1998). The mitochondrial pyruvate carrier (MPC) in the mitochondrial inner membrane is responsible for the transport of pyruvate into the mitochondrial matrix (Fig. 4a) (Bricker et al. 2012; Herzig et al. 2012). S. cerevisiae has three homologous MPC subunit proteins, Mpc1, Mpc2, and Mpc3, which generate two types of hetero-oligomeric complex (Bricker et al. 2012; Herzig et al. 2012). The low-affinity MPCFERM is composed of Mpc1 and Mpc2, whereas the high-affinity MPCOX is composed Mpc1 and Mpc3 (Fig. 4a) (Bender et al. 2015). The common subunit Mpc1 is expressed regardless of the carbon source, but Mpc2 and Mpc3 are expressed under fermentative and respiratory conditions, respectively, leading to differential expression levels of MPCFERM and MPCOX, and subsequent differential pyruvate flux to the mitochondria, depending on the carbon metabolic conditions (Bender et al. 2015).
Effects of overexpressing different forms of MPC complex on isobutanol production. a Roles of MPC in mitochondrial pyruvate transport. Under fermentative conditions, the low-activity MPCFERM, composed of Mpc1 and Mpc2, is expressed and competing for cytosolic pyruvate with pyruvate decarboxylase (PDC). Most of the pyruvate is left in the cytosol and is finally converted to ethanol by PDC. On the other hand, under respiratory conditions, the high-activity MPCOX, composed of Mpc1 and Mpc3, is expressed, whereas PDC is downregulated. Consequently, pyruvate is efficiently transported to the mitochondrial matrix and then enters the TCA cycle via pyruvate dehydrogenase (PDH). EtOH ethanol, PYR pyruvate, BCAA branched chain amino acid. b JHY462 ( ald6Δbat1Δlpd1Δ [JIB2]), JHY463 (ald6Δbat1Δlpd1Δ [JIB3]), JHY464 (ald6Δbat1Δlpd1Δ [JIB4]), and JHY465 (ald6Δbat1Δlpd1Δ [JIB5]) were grown in SC-His medium containing 2 % glucose for 48 h. Each value indicates the average ± SD of triplicate experiments
To further improve isobutanol production in the mitochondria, we investigated the effects of overexpressing MPCFERM or MPCOX in JHY462 strain. MPC1, MPCFERM (MPC1 and MPC2), or MPCOX (MPC1 and MPC3) genes were additionally cloned into pJIB2, generating plasmids pJIB3, pJIB4, and pJIB5, respectively (Table 2). JHY46 strain harboring pJIB3 (JHY463) showed improved isobutanol production up to 265.5 mg/L compared with JHY462, indicating that overexpression of MPC1 alone can contribute to isobutanol production in the mitochondria (Fig. 4b). Overexpression of MPCFERM (JHY464) also improved isobutanol production, but to a lesser extent than the overexpression of MPC1. Cells overexpressing MPCOX (JHY465) showed the highest level of isobutanol production, reaching 330.9 mg/L, which is a 1.9-fold higher level than that produced in JHY462. Taken together, mitochondrial isobutanol production could be successfully improved by increasing mitochondrial pyruvate pool via overexpressing MPC, especially in the form of MPCOX.
Compared to wild-type control strain harboring empty vector (WT-1), the engineered strain JHY462 showed reduced glucose uptake rate and lower final cell density, whereas producing about 22-fold higher level of isobutanol (Fig. 5). Glycerol production also increased up to 1.63 g/L in JHY462 compared to WT-1 (0.38 g/L). Inhibition of TCA cycle pathway by LPD1 deletion might be mainly responsible for the growth phenotype of JHY465. The consumed glucose was mostly used to produce ethanol in both WT-1 and JHY465, producing similar levels of ethanol, suggesting that the overexpressed MPCOX might redirect only minor part of cytosolic pyruvate to the mitochondria (Fig. 4a).
Discussion
S. cerevisiae is a promising host for isobutanol production, but subcellular compartmentalization of biosynthetic pathway is one of the bottlenecks preventing efficient production. The natural isobutanol biosynthetic pathway in S. cerevisiae requires import of pyruvate into the mitochondria and export of 2-KIV into the cytoplasm. Previously, it has been shown that construction of mitochondrial pathway by re-localizing the last two cytosolic enzymes, PDC and ADH, into the mitochondria could improve isobutanol production (Avalos et al. 2013). Mitochondrial pathway might not only solve the problem of 2-KIV transport across the membrane but also provide additional advantages such as increasing reaction rates through concentration of substrates. In addition, iron-sulfur cluster is synthesized exclusively in the mitochondria, making it more available for Ilv3, an iron-sulfur protein (Muhlenhoff and Lill 2000). However, since S. cerevisiae has reduced mitochondrial function and pyruvate transport into the mitochondria at high glucose concentrations or during anaerobic conditions (Bender et al. 2015; Brat et al. 2012), it is essential to increase mitochondrial pyruvate pool to improve mitochondrial isobutanol synthesis.
In this study, we improved isobutanol production via mitochondrial pathway by increasing mitochondrial uptake of pyruvate through MPC. We first generated S. cerevisiae strain having mitochondrial isobutanol biosynthetic pathway based on previous studies including ours (Avalos et al. 2013; Matsuda et al. 2013; Park et al. 2014). Isobutanol production was improved by deleting competing pathway enzymes (Bat1, Ald6, and Lpd1), overexpressing all biosynthetic enzymes (Ilv2, Ilv5, Ilv3, Aro10, and Adh2) in the mitochondria by re-locating Aro10 and Adh2 into the mitochondria, and overexpressing a constitutively active transcription factor, Leu3Δ601. Although LEU4 deletion was also effective in increasing isobutanol production in the bat1Δald6Δ background, it exerted a negative effect in combination with lpd1Δ. The reasons for these results are not clear yet, but since 2-IPM synthesized by Leu4 activates endogenous transcription factor Leu3, deleting LEU4 gene might cause reduced expression levels of Leu3 target genes including ILV2 and ILV5 (Baichwal et al. 1983; Boer et al. 2005; Friden and Schimmel 1988), affecting isobutanol synthesis.
MPC in the mitochondrial inner membrane is necessary for pyruvate uptake into the mitochondrial matrix (Bricker et al. 2012; Herzig et al. 2012). S. cerevisiae has three homologous proteins, Mpc1, Mpc2, and Mpc3, forming two types of hetero-oligomeric complex, MPCFERM and MPCOX (Bender et al. 2015). Mpc1, a common subunit of MPC, is constitutively expressed, but Mpc2 and Mpc3 are expressed under fermentative and respiratory conditions, respectively, forming MPCFERM and MPCOX (Bender et al. 2015). Previously, overexpression of MPC1 and MPC2 from S. cerevisiae was shown to improve acetoin production via reconstructed mitochondrial pathway in Candida glabrata (Li et al. 2015). In this study, we compared the effects of overexpressing Mpc1, MPCFERM (Mpc1 and Mpc2), or MPCOX (Mpc1 and Mpc3) on mitochondrial isobutanol production. Overexpression of MPCOX was most effective in increasing isobutanol production, followed by overexpression of Mpc1 alone. Overexpression of MPCFERM was least effective. These results agree with a recent finding showing that MPCOX has about twofold higher rate of pyruvate import than MPCFERM (Bender et al. 2015). Since fermentation pathway is dominant during the growth of S. cerevisiae on glucose, pyruvate flux to the mitochondria through the low-affinity MPCFERM might not be sufficient for isobutanol production. Overexpression of the high-affinity MPCOX using a strong constitutive promoter might increase pyruvate pool in the mitochondria even under fermentative conditions, leading to an improved isobutanol production. Overexpression of MPCOX might also be a useful strategy for the production of other pyruvate-derived chemicals in the mitochondria.
Taken together, our final engineered strain JHY465 with improved mitochondrial pyruvate transport showed a significant increase in isobutanol titer (330.9 mg/L) compared to the wild-type strain and previous reports (Avalos et al. 2013; Matsuda et al. 2013; Park et al. 2014). However, even in cells overexpressing MPCOX, pyruvate is mainly used for ethanol production. Therefore, further studies might be necessary for more efficient redirection of pyruvate flux into the mitochondria.
References
Atsumi S, Hanai T, Liao JC (2008) Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature 451:86–89
Avalos JL, Fink GR, Stephanopoulos G (2013) Compartmentalization of metabolic pathways in yeast mitochondria improves the production of branched-chain alcohols. Nat Biotechnol 31:335–341
Baichwal VR, Cunningham TS, Gatzek PR, Kohlhaw GB (1983) Leucine biosynthesis in yeast: identification of two genes (LEU4, LEU5) that affect alpha-isopropylmalate synthase activity and evidence that LEU1 and LEU2 gene expression is controlled by alpha-isopropylmalate and the product of a regulatory gene. Curr Genet 7:369–377
Bender T, Pena G, Martinou JC (2015) Regulation of mitochondrial pyruvate uptake by alternative pyruvate carrier complexes. EMBO J 34:911–924
Blombach B, Eikmanns BJ (2011) Current knowledge on isobutanol production with Escherichia coli, Bacillus subtilis and Corynebacterium glutamicum. Bioeng Bugs 2:346–350
Boer VM, Daran JM, Almering MJ, de Winde JH, Pronk JT (2005) Contribution of the Saccharomyces cerevisiae transcriptional regulator Leu3p to physiology and gene expression in nitrogen- and carbon-limited chemostat cultures. FEMS Yeast Res 5:885–897
Brat D, Weber C, Lorenzen W, Bode HB, Boles E (2012) Cytosolic re-localization and optimization of valine synthesis and catabolism enables inseased isobutanol production with the yeast Saccharomyces cerevisiae. Biotechnol Biofuels 5:65
Bricker DK, Taylor EB, Schell JC, Orsak T, Boutron A, Chen YC, Cox JE, Cardon CM, Van Vranken JG, Dephoure N, Redin C, Boudina S, Gygi SP, Brivet M, Thummel CS, Rutter J (2012) A mitochondrial pyruvate carrier required for pyruvate uptake in yeast, drosophila, and humans. Science 337:96–100
Buijs NA, Siewers V, Nielsen J (2013) Advanced biofuel production by the yeast Saccharomyces cerevisiae. Curr Opin Chem Biol 17:480–488
Chen X, Nielsen KF, Borodina I, Kielland-Brandt MC, Karhumaa K (2011) Increased isobutanol production in Saccharomyces cerevisiae by overexpression of genes in valine metabolism. Biotechnol Biofuels 4:21
Connor MR, Liao JC (2009) Microbial production of advanced transportation fuels in non-natural hosts. Curr Opin Biotechnol 20:307–315
Dickinson JR, Harrison SJ, Hewlins MJ (1998) An investigation of the metabolism of valine to isobutyl alcohol in Saccharomyces cerevisiae. J Biol Chem 273:25751–25756
Friden P, Reynolds C, Schimmel P (1989) A large internal deletion converts yeast LEU3 to a constitutive transcriptional activator. Mol Cell Biol 9:4056–4060
Friden P, Schimmel P (1988) LEU3 of Saccharomyces cerevisiae activates multiple genes for branched-chain amino acid biosynthesis by binding to a common decanucleotide core sequence. Mol Cell Biol 8:2690–2697
Gueldener U, Heinisch J, Koehler GJ, Voss D, Hegemann JH (2002) A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res 30:e23
Hazelwood LA, Daran JM, van Maris AJ, Pronk JT, Dickinson JR (2008) The Ehrlich pathway for fusel alcohol production: a century of research on Saccharomyces cerevisiae metabolism. Appl Environ Microbiol 74:2259–2266
Herzig S, Raemy E, Montessuit S, Veuthey JL, Zamboni N, Westermann B, Kunji ERS, Martinou JC (2012) Identification and functional expression of the mitochondrial pyruvate carrier. Science 337:93–96
Hohmann S (2002) Osmotic adaptation in yeast-control of the yeast osmolyte system. Int Rev Cytol 215:149–187
Ida K, Ishii J, Matsuda F, Kondo T, Kondo A (2015) Eliminating the isoleucine biosynthetic pathway to reduce competitive carbon outflow during isobutanol production by Saccharomyces cerevisiae. Microb Cell Factories 14
Kim S, Hahn JS (2015) Efficient production of 2,3-butanediol in Saccharomyces cerevisiae by eliminating ethanol and glycerol production and redox rebalancing. Metab Eng 31:94–101
Kispal G, Steiner H, Court DA, Rolinski B, Lill R (1996) Mitochondrial and cytosolic branched-chain amino acid transaminases from yeast, homologs of the myc oncogene-regulated Eca39 protein. J Biol Chem 271:24458–24464
Knoshaug EP, Zhang M (2009) Butanol tolerance in a selection of microorganisms. Appl Biochem Biotechnol 153:13–20
Kondo T, Tezuka H, Ishii J, Matsuda F, Ogino C, Kondo A (2012) Genetic engineering to enhance the Ehrlich pathway and alter carbon flux for increased isobutanol production from glucose by Saccharomyces cerevisiae. J Biotechnol 159:32–37
Lee AC, Xu X, Blachly-Dyson E, Forte M, Colombini M (1998) The role of yeast VDAC genes on the permeability of the mitochondrial outer membrane. J Membr Biol 161:173–181
Lee WH, Seo SO, Bae YH, Nan H, Jin YS, Seo JH (2012) Isobutanol production in engineered Saccharomyces cerevisiae by overexpression of 2-ketoisovalerate decarboxylase and valine biosynthetic enzymes. Bioprocess Biosyst Eng 35:1467–1475
Li SB, Liu LM, Chen J (2015) Compartmentalizing metabolic pathway in Candida glabrata for acetoin production. Metab Eng 28:1–7
Lilly M, Bauer FF, Styger G, Lambrechts MG, Pretorius IS (2006) The effect of increased branched-chain amino acid transaminase activity in yeast on the production of higher alcohols and on the flavour profiles of wine and distillates. FEMS Yeast Res 6:726–743
Maarse AC, Van Loon AP, Riezman H, Gregor I, Schatz G, Grivell LA (1984) Subunit IV of yeast cytochrome c oxidase: cloning and nucleotide sequencing of the gene and partial amino acid sequencing of the mature protein. EMBO J 3:2831–2837
Matsuda F, Ishii J, Kondo T, Ida K, Tezuka H, Kondo A (2013) Increased isobutanol production in Saccharomyces cerevisiae by eliminating competing pathways and resolving cofactor imbalance. Microb Cell Factories 12:119
Milne N, van Maris AJA, Pronk JT, Daran JM (2015) Comparative assessment of native and heterologous 2-oxo acid decarboxylases for application in isobutanol production by Saccharomyces cerevisiae. Biotechnol Biofuels 8
Muhlenhoff U, Lill R (2000) Biogenesis of iron-sulfur proteins in eukaryotes: a novel task of mitochondria that is inherited from bacteria. Bba-Bioenergetics 1459:370–382
Mumberg D, Muller R, Funk M (1995) Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156:119–122
Nevoigt E (2008) Progress in metabolic engineering of Saccharomyces cerevisiae. Microbiol Mol Biol Rev 72:379–412
Park SH, Kim S, Hahn JS (2014) Metabolic engineering of Saccharomyces cerevisiae for the production of isobutanol and 3-methyl-1-butanol. Appl Microbiol Biotechnol 98:9139–9147
Porro D, Gasser B, Fossati T, Maurer M, Branduardi P, Sauer M, Mattanovich D (2011) Production of recombinant proteins and metabolites in yeasts: when are these systems better than bacterial production systems? Appl Microbiol Biotechnol 89:939–948
Pronk JT, Steensma HY, van Dijken JP (1996) Pyruvate metabolism in Saccharomyces cerevisiae. Yeast 12:1607–1633
Weber C, Farwick A, Benisch F, Brat D, Dietz H, Subtil T, Boles E (2010) Trends and challenges in the microbial production of lignocellulosic bioalcohol fuels. Appl Microbiol Biotechnol 87:1303–1315
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
This work was supported by the National Research Foundation of Korea (NRF) Grant funded by the Korean Government (NRF-2015R1A2A2A01005429).
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Rights and permissions
About this article
Cite this article
Park, SH., Kim, S. & Hahn, JS. Improvement of isobutanol production in Saccharomyces cerevisiae by increasing mitochondrial import of pyruvate through mitochondrial pyruvate carrier. Appl Microbiol Biotechnol 100, 7591–7598 (2016). https://doi.org/10.1007/s00253-016-7636-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-016-7636-z