Abstract
Anaerobic ammonium oxidation (anammox) and nitrite-dependent anaerobic methane oxidation (n-damo) are two new processes of recent discoveries linking the microbial nitrogen and carbon cycles. In this study, 16S ribosomal RNA (rRNA) gene of anammox bacteria and pmoA gene of n-damo bacteria were used to investigate their distribution and diversity in natural acidic and re-vegetated forest soils. The 16S rRNA gene sequences retrieved featured at least three species in two genera known anammox bacteria, namely Candidatus Brocadia anammoxidans, Candidatus Brocadia fulgida, and Candidatus Kuenenia stuttgartiensis while the pmoA gene amplified was affiliated with two species of known n-damo bacteria Candidatus Methylomirabilis oxyfera and a newly established Candidatus Methylomirabilis sp. According to the results, the diversity of anammox bacteria in natural forests was lower than in re-vegetated forests, but no significant difference was observed in n-damo community between them. Quantitative real-time PCR showed that both anammox and n-damo bacteria were more abundant in the lower layer (10–20 cm) than the surface layer (0–5 cm). The abundance of anammox bacteria varied from 2.21 × 105 to 3.90 × 106 gene copies per gram dry soil, and n-damo bacteria quantities were between 1.69 × 105 and 5.07 × 106 gene copies per gram dry soil in the two different layers. Both anammox and n-damo bacteria are reported for the first time to co-occur in acidic forest soil in this study, providing a more comprehensive information on more defined microbial processes contributing to C and N cycles in the ecosystems.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Anaerobic ammonium oxidation (anammox) process, the oxidation of ammonium to dinitrogen (N2) gas using nitrite as an electron acceptor under anoxic conditions, was first discovered in a denitrifying fluid reactor nearly 20 years ago (Mulder et al. 1995; Van de Graaf et al. 1995). The discovery changes the traditional knowledge that oxidation of ammonium occurs only under aerobic conditions. Currently, five genera, Candidatus Kuenenia (Chamchoi and Nitisoravut 2007; Egli et al. 2001; Schmid et al. 2000; Strous et al. 2006), Candidatus Brocadia (Jetten 2001; Kartal et al. 2008), Candidatus Scalindua (Kindaichi et al. 2007), Candidatus Anammoxglobus (Kartal et al. 2007), and Candidatus Jettenia (Quan et al. 2008) and a total of 13 species of anammox bacteria, have been established through culture independent approaches. All obtained anammox bacteria belong to the phylum of Planctomycetes (Strous et al. 1999).
Since 1995, anammox bacteria have been reported as distributed widely in various ecosystems, such as marine system (Dalsgaard et al. 2003; Dalsgaard et al. 2005); freshwater system, including lakes (Schubert et al. 2006), rivers (Zhang et al. 2007), wetland (Li et al. 2015; Shen et al. 2015), and groundwater (Moore et al. 2011); and terrestrial system (Long et al. 2013; Wang and Gu 2013; Zhu et al. 2011); even some extreme environments like polar regions (Rysgaard et al. 2008), high-temperature hot spring (Jaeschke et al. 2009), deep-sea hydrothermal vents (Byrne et al. 2009), oil reservoirs (Li et al. 2010), guts of invertebrates (Li and Gu 2016), and fishes (Chan et al. 2016). However, though anammox process has been identified to occur in various natural as well as artificial environments, the presence of anammox bacteria in forest soils has not been clearly studied and reported previously.
Nitrite-dependent anaerobic methane oxidation (n-damo) process was first discovered in 2006 and was a novel biochemical process contributing to microbial methane oxidation (Raghoebarsing et al. 2006). It has been shown that the bacterium Candidatus “Methylomirabilis oxyfera” can couple aerobic oxidation of methane (AOM) to nitrite reduction (NIR) [1] and suggested that it is an intra-aerobic methane oxidation pathway.
Currently, the n-damo process is the only known biological process providing a linkage between two major nutrient cycles—nitrogen and carbon cycles (Raghoebarsing et al. 2006). Through enrichment culture from freshwater canal sediments and then metagenomics analysis, it has been identified that the NIR process is mediated by the bacterium named Candidatus “Methylomirabilis oxyfera,” which is affiliated with the uncultured “NC10” phylum (Ettwig et al. 2009).
Up to date, M. oxyfera-like bacteria have been detected in various environments such as marine sediments of the South China Sea (Chen et al. 2014), freshwater sediments (Shen et al. 2014b), paddy soils (Shen et al. 2014a; Wang et al. 2012; Zhou et al. 2014), wetlands (Chen et al. 2015; Zhu et al. 2015), and waste water treatment facilities (Luesken et al. 2011a) through 16S ribosomal RNA (rRNA) gene and/or pmoA gene phylogenetic analysis. However, many other environments have not been investigated for its presence to provide a more comprehensive account of the n-damo process in ecosystems.
Forests drive the dynamics of the terrestrial C cycle, and forest soils contain approximately 73 % of C in the world soils (Sedjo 1993). More than 85 % of the total N in the forest ecosystem is stored in forest soils (Reichle 1981). Moreover, forests account for more than 75 % of gross primary production (GPP) in terrestrial ecosystem (Beer et al. 2010). Forest soils also act as an important sink of both carbon and nitrogen because these nutrients are processed from leaves and litter. Obviously, forest soils have a significant role in the carbon and nitrogen cycle, but both anammox and n-damo processes have not been studied in forest soils. Therefore, the objectives of this study were to (1) detect the presence of anammox bacteria and n-damo bacteria in acidic forest soils, (2) analyze the distribution and community structure of both natural and re-vegetated forest soils, and (3) quantify the abundance of anammox and n-damo bacteria within the different forest soils of tropical ecosystem.
Materials and methods
Sites description
The study sites are located at Nanling National Nature Reserve in Guangdong province of China (24° 37ʹ–24° 57′ N, 112° 30′–113° 04′ E) (Table 1), which covers an area of 58,368.4 hm2 and has a typical subtropical monsoon climate. The annual average precipitation is 2108.4 mm. The maximum average temperature is 26.2 and 7.1 °C in the hottest month (July) and the coldest month (January), respectively (Gan et al. 2015). Two types of forests are present within the study sites. Forests located at an altitude above 700 m from sea level are natural, undeveloped forests, while forests located at an altitude below 700 m were once deforested and then reforested artificially through re-vegetation. Soils in those natural forests are typical yellow containing granite, metamorphic rock, and sandstone, but red soils become dominant in re-vegetated forests (Gan et al. 2015).
In this study, soils from five representative forests at different levels of altitude were selected for investigation, including three from natural forests and two from re-vegetated forests. Briefly, soil samples were collected from (i) a natural bamboo forest at the top of a mountain (altitude of 1858 m, labeled as HLS and HLD, S and D represent soil of surface and lower layer, respectively); (ii) a natural coniferous forest, predominated by Pinus massoniana (Lambo) (1371 m, PS and PD); (iii) a natural evergreen broad-leaf forest, dominated by Schima superba and Machilus thunbergii (1009 m, EBLS and EBLD); (iv) a man-made young Cunninghamia lanceolata (Lamb.) forest (615 m, CLYS and CLYD); and (v) mature Cunninghamia lanceolata (Lamb.) forest (529 m, CLMS and CLMD), respectively.
Field sampling and physicochemical analysis
Soil samples were taken on January 8, 2015, from five locations along elevation from 1858 to 529 m for two layers (surface—A0 layer, 0–5 cm, and lower—A1 layer, 10–20 cm) (Table 1). Three replicates were sampled randomly at each site to form a composite sample. Soils were kept in an icebox immediately with coolant and separated into two parts. One was used for physicochemical analysis and the other was brought to The University of Hong Kong for molecular analysis.
Physical-chemical analysis was carried out at Guangdong Institute of Eco-environment and Soil Sciences. The methods used were described elsewhere (Gan et al. 2015). Soil moisture content was measured by drying in oven at 105 °C for 24 h to constant weight.
DNA extraction and PCR amplification
Genomic DNA was extracted from 0.25 g of soils using the PowerSoil® DNA isolation kit (MO BIO Laboratories, Inc., USA) according to the manual. DNA concentration was measured by NanoDrop™ ND-2000 (Thermo Scientific Inc., USA). The extracted DNA was stored at −20 °C for further analysis.
Nested PCR was adopted in this study to amplify gene fragments of the targeting microbial groups. For anammox bacteria, pla46F targeting Plantomycetes phylum and universal primer 630R were selected as outer primer, while Amx368f–Amx820r for anammox 16S rRNA gene were selected as inner primer. For n-damo bacteria, two sets of primers, A189_b-com682r and com182f-com568r (Luesken et al. 2011b), were used to amplify the characteristic pmoA gene in n-damo bacteria. Detailed information of primer and corresponding thermal profiles are listed in Table 2. After amplification, PCR products were examined with gel electrophoresis in 1 % agarose gels in TAE buffer.
Cloning and sequencing
Thirty individual PCRs (triplicates for each sample) were pooled and gel-purified by using GFX™ PCR DNA and Gel Band Purification Kit by following the manufacturer’s protocol (Amersham Biosciences). Amplified gene fragments were then ligated into pMD-18 vector (Takara, Japan) and transformed into DH5α competent cells for clone library construction. After blue-white screening, positive clones were selected randomly for sequencing.
Quantitative PCR assay
Primer sets of Amx 438f–Amx 684r and qP1F-qP1R were selected to conduct real-time quantitative PCR (qPCR) to estimate the abundance of 16S rRNA gene of anammox bacteria and n-damo bacteria in soil samples, respectively (Table 2).
The qPCR assays were performed on an ABI 7000 Sequence detection system (Applied Biosystems, Foster City, CA) using SYBR green method. Standard curves for qPCR were constructed by ten serial dilutions of a known copy number of plasmid through a sequenced clone. Triplication was conducted for each sample, and each reaction was performed in a volume of 20 μl containing 1 μl of DNA template (10 ~ 100 ng DNA templates), 1 μl of each primer (10 μM), and 10 μl of SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA). The reaction cycling conditions were in accordance with the PCR procedures described above. The R 2 values were 1.0, and amplification efficiency was 93.2 and 96.5 % of anammox and n-damo, respectively.
Sequences and phylogenetic analysis
Sequencing was conducted at Beijing Genomics Institute (Shenzhen, People’s Republic China). Clone sequences were processed by BioEdit (Hall 1999) and aligned by Clustal X (Thompson et al. 2002). DECIPHER’s Find Chimeras (Wright et al. 2012) was used to remove chimeras in anammox16S rRNA gene sequences to ensure the reliability of sequences obtained. Processed sequences were analyzed against those in GenBank with BLAST (Altschul et al. 1997). After the identity and validity of sequences confirmed through GeneBank, sequences of pmoA gene were translated into amino acids for phylogeny analysis. At last, all processed sequences were imported to MEGA version 6.0 to construct phylogenetic tree, using neighbor-joining algorithm with the best model of Kimura 2-parameter for anammox and JTT method amino acid substitution model for n-damo. The reliability of branching within the tree was tested with bootstrap of 1000 replicates, and the consensus tree was presented.
Statistical analysis
To examine the distribution of anammox bacteria and M. oxyfera-like bacteria, the operational taxonomic units (OTUs) cutoff values of 3 % was applied to determine the diversity of anammox bacterial 16S rRNA gene sequences and sequences of pmoA gene using the Mothur program (Schloss et al. 2009). The Chao1 estimator, richness, and Shannon-Wiener diversity index were also calculated with the Mothur software.
The data of physical-chemical parameters and abundance of anammox bacteria were imported to R to conduct independent sample t test and analysis of variance (ANOVA) to investigate any significant differences between samples from different layers or forests (Table S1). To investigate relationships between diversity of anammox communities, n-damo communities and soil physical-chemical parameters, the Pearson moment correlation was carried out using PAleontological STatistics version 3.0 (Hammer 2012). Detrended correspondence analysis (DCA) was conducted for anammox and n-damo analysis using Canoco for Windows 4.5 (Ter Braak and Smilauer 2012). A linear model was applied, and the constrained ordination method was redundancy analysis (RDA) using Canoco.
Finally, UniFrac function under Mothur software was used to perform the principal coordinate analysis (PCoA). Results generated by Mothur were imported into OriginPro8 (OriginLab, USA) to construct illustrations.
Nucleotide sequences
The anammox bacteria n-damo pmoA gene sequences were deposited in NCBI under accession number KU894711-KU894735 and KU894736-KU894777, respectively.
Results
Physical-chemical parameters
A range of soil properties were investigated including pH, organic matter, total N, total P, total K, NH4 +-N, NO3 −N, hydrolyzed nitrogen, available phosphorus, exchangeable Al3+, and water content for a total of these 11 parameters of 10 samples in triplicate. The pH values of soil samples ranged from 3.66 to 4.59, which were classified as extremely acidic to very strongly acidic (USDA 1999). The details of chemical properties of soil are listed in Table 1. According to the results of ANOVA (Table S1), the surface layer of soil was more acidic than the lower layer on average. Moreover, surface layer contained significantly higher concentration of organic matter, total N, NH4 +-N, NO3 −N, hydrolyzed N, and exchangeable Al3+ than lower layer. Total P and available P were higher in surface soil than the deep in natural forest, but this trend did not persist in re-vegetated forest. There was no significant difference of total K content between these two layers.
Comparison of soil properties between different forests was also made using ANOVA. The five forests were divided into natural and re-vegetated forests. Based on results obtained, soil samples from natural forest contained significantly higher organic matter, total N, NH4 +-N, NO3 −−N, total K, and available P than those of re-vegetated ones. The re-vegetated forest soils yielded higher total P than natural forest. There was no significant difference in pH values and exchangeable Al3+ between the two soil types. ANOVA was also conducted to compare the differences between the two layers of an individual forest. Regardless of the forest type, majority of physical-chemical parameters did not show a significant difference between the two forests. Significant differences were only observed for total K content in natural forest, and pH value and total P content in re-vegetated forest.
Phylogenetic analysis of anammox bacteria and n-damo bacteria
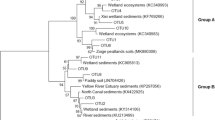
The coverage of clone library for anammox and n-damo bacteria reached 96 to 100 % (Table 3), indicating nearly complete obtainment of information of two communities. The phylogenetic tree constructed by 16S rRNA gene indicates that two known anammox bacterial genera, namely Brocadia and Kuenenia, were detected in the forest soils (Fig. 1). Majority of sequences (636 out of 702) were closely related to Candidatus Brocadia, and could be further divided into three subclusters according to the different environments that sequences affiliated with. Cluster I comprised mainly sequences from re-vegetated forest (20 out of 21). This clade was most closely related to Candidatus Brocadia anammoxidans and sequences from agriculture soil. Cluster II consisted of sequences from samples HLS, EBLS, and CLYS and were closely affiliated with uncultured anaerobic ammonium-oxidizing bacteria clones from sludge and reactor. In cluster III, all of the sequences in this cluster showed 98–99 % identity to the sequences of Candidatus Brocadia fulgida and grouped with references retrieved from freshwater ecosystem. The remaining 66 anammox 16S rRNA gene sequences were closely related to Candidatus Kuenenia stuttgartiensis 16S rRNA gene (AF375995) in GenBank (identities up to 97–99 % for nucleotide). But these sequences were only detected from sample EBLS and samples from re-vegetated forests. Overall, sequences from re-vegetated forest were more diverse and scattered, which could be detected in every cluster of Brocadia and Kuenenia. In natural forest, majority of sequences obtained were distributed in Brocadia in a more centralized manner, except sequences from EBLS.
Phylogenetic tree constructed by neighbor-joining method after an alignment of anammox bacterial 16S rRNA gene sequences along with some known species from GenBank. Numbers beside the sample ID refer to the number of clones are in each clone library. The numbers at the nodes are bootstrap calculation obtained based on 1000 times replicates
Primers targeting pmoA gene of n-damo bacteria were successfully applied on all samples in this study. Deduced amino acid sequences were grouped into six clusters with the pmoA sequence of M. oxyfera retrieved to show the phylogeny (Fig. 2). Cluster I, which contained 714 sequences in total, included sequences from all soil samples. Five hundred eighty-two sequences were 100 % identical to the sequence from sediment of Panjin swamp on nucleotide level; 93 sequences were closely affiliated with the sequences from freshwater sediments; interestingly, 11 sequences in this cluster having 95% identity to KC341442 (sediment of Panjin swamp) did not affiliate to any known cultured species. Cluster II was composed of 60 sequences, mainly from samples PS and PD, which were closely related to n-damo sequences from Qiantang River sediment. Cluster III contained only sequences from PS and PD and these sequences were most similar to M. oxyfera (FP565575) among all sequences retrieved. There were only two sequences in cluster IV, which were closely related to the sequence retrieved from aquifer environment. Cluster V contained four sequences closely related to samples from coastal wetland sediment with 93–96 % identity to the sequence of Candidatus Methylomirabilis sp. Rs1 (KT443986), which was a novel denitrifying methanotroph of NC10 phylum enriched (unpublished data). Cluster VI included nine sequences showing 100 % identity to the Candidatus Methylomirabilis sp. Rs1 in amino acid level (ALL28944). In short, clusters I to III were more related to known specie of M. oxyfera while clusters IV to VI were closely affiliated to novel specie Candidatus Methylomirabilis sp. Rs1.
Based on phylogenetic tree constructed, principal coordinate analysis (PCoA) was conducted. According to the results of PCoA, the first and second principles in total contributed about 66% of structure variations for anammox bacteria (Fig. 3). The community of samples retrieved from re-vegetated forests was more centralized while anammox community in natural forests was more scattered comparatively. For n-damo community structure, the first and second principles combined to contribute about 58% of variations in PCoA. Samples could be grouped together according to their corresponding forest types.
Genetic diversity analyzed of anammox bacteria and n-damo bacteria
A total of 708 sequences of anammox bacteria and 831 sequences of pmoA gene were analyzed. The diversity of anammox and n-damo bacteria was investigated via number of OTUs, Chao1 estimators and Shannon index calculations. The operational taxonomic unit (OTU) cutoff was examined by the 97 % sequence similarity to reflect phylogeny diversity in each sample (Table 3). A total of one to four and one to six OTUs of anammox 16S rRNA and pmoA genes were observed, respectively. The diversity of anammox bacteria in natural forest was lower than that in re-vegetated forest while no significant difference was detected in n-damo diversity between two types forests (Table 2). Compared to studies in other ecosystems, anammox bacteria showed a notably lower diversity while no difference was observed in n-damo community in this acidic forest ecosystem.
Abundance of anammox bacteria and n-damo bacteria
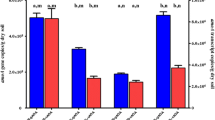
The primer pair of Amx438f and Amx684r was applied for the quantification of anammox bacterial abundance in the soil samples. Standard curves were generated by eight orders of magnitude of the standard plasmid containing cloned 16S rRNA gene fragment. A significantly linear relationship (R 2, 1.0) was obtained and an amplification efficiency of 93.2 % was yielded. The melting curve analyses revealed only one peak appearing at 84.5 °C, confirming that fluorescent signals were derived from specific PCR products in the process of the qPCR quantification, and thereby reliable results. The gene copy number measured in the surface sample (0–5 cm) was 2.21 × 105–3.91 × 105 gene copies per gram dry soil in natural forest and ranged from 4.44 × 105 to 1.20 × 106 gene copies per gram dry soil in the lower layer (Fig. 4a). Compared to natural forest, the abundances of anammox bacteria in re-vegetated forest were higher, ranging from 6.68 × 105 to 3.90 × 106 gene copies per gram dry soil.
The abundance of n-damo bacteria was estimated through qualifying 16S rRNA gene by qP1F and qP1R primers. With respect to its R 2 value (R 2, 1.0) and amplification efficiency (96.5%), the result was suggested to be reliable. The gene copy numbers ranged from 1.69 × 105 to 5.07 × 106 gene copies per gram dry soil. In natural forest, the abundance of surface samples was two to ten times lower than the deep layer samples, and the highest gene abundance was retrieved from pine forest sample (Fig. 4b), while samples from young forest resulted in fewer gene copies than matured forest under re-vegetation. The quantities of gene copies in the lower layer were higher than in the surface layers in two re-vegetated forests.
Discussion
Distribution and diversity of anammox bacteria and n-damo bacteria
In this study, anammox and n-damo bacteria were reported for the first time to be detected in acidic forest soils in tropic region. This contributes to better understanding on the global distribution of anammox and n-damo bacteria to fully account for C and N cycling. Two genera of anammox bacteria—Candidatus Brocadia and Candidatus Kuenenia—were found in this study. Both genera of anammox bacteria were reported as dominant types in terrestrial ecosystem (Humbert et al. 2010). Brocadia and Kuenenia were reported to be able to use ferrous iron and a variety of organic compounds, such like formate, acetate, propionate, and methylamines, as electron donors, and the versatile metabolism of Brocadia and Kuenenia organisms may result in better adaptions in various terrestrial environments (Kartal et al. 2008; Strous et al. 2006). The detection of these two genera was in agreement with our current understanding and information based on environmental conditions of the sampling sites.
Based on the number of OTU identified and diversity indices calculated, the results of phylogenetic study revealed that the diversity in these study sites were generally lower, compared to results reported in other ecosystems, such as rice paddy field or wetlands (Shen et al. 2015; Zhu et al. 2011). The lower diversity of anammox bacteria might be attributed to several factors. First, soil pH may be a critical one. According to a reported study on metabolism of anammox bacteria, and the performance of anammox bacteria in bioreactors, anammox bacteria grow within the pH range of 6.7 to 8.3, being optimum at slightly alkaline conditions (Kartal et al. 2011) whereas even slightly acidic condition would weaken the tolerance of resting anammox cells to NO2 − toxicity (Altschul et al. 1997). As mentioned previously, the pH of soil samples in this study ranged from 3.66 to 4.59. This highly acidic nature of soil may impede growth of anammox bacteria and lead to the result of low diversity.
Limited nitrogen sources may be another possible explanation for the low diversity observed. As anammox bacteria make use of ammonium and nitrite for metabolism, reactive nitrogen level would be an important factor for anammox bacteria to establish. Compared to environments in which anammox bacteria were frequently reported, such as rice paddy field (Zhu et al. 2011) and wastewater treatment facilities (Wu et al. 2015), forest soils are relatively infertile in terms of nitrogen. At the study sites, shed leave litter would probably be the sole nitrogen source to soil. On the other hand, agricultural soil and domestic wastewater generally contain a huge quantity of nitrogen available to microbes due to addition of fertilizers and decomposition of nitrogen-containing compounds, respectively. Thus, abundant nitrogen sources could support a more diverse, complex community of anammox bacteria in these environments.
Vegetation may also exert an adverse effect on the diversity of anammox bacteria. As the sampling sites were covered by extensive, various kinds of trees and herbaceous plants, a strong association between plants and soil properties could be expected in the rhizosphere. Plants release organic acids to the rhizosphere via the roots (Haoliang et al. 2007), and organic acids are generally unfavorable to anammox bacteria (Van de Graaf et al. 1996). Therefore, the growth and in turn diversity of anammox bacteria may be hindered by the extensive local vegetation.
Although anammox and n-damo bacteria seem to be competitive for nitrite which serves as the electron acceptor for each process (Haroon et al. 2013), co-occurrence was reported in paddy soil (Wang et al. 2012), saline lake (Yang et al. 2012), and wetlands (Shen et al. 2015) recently as well as in this study. According to the phylogeny analysis of n-damo bacteria (Fig. 2), all clones affiliated with the sequences from wetland and freshwater ecosystem, especially the cluster I accounting for 85.9% of all pmoA sequences closely related to the sediment of Panjin Swamp which is one of the best preserved wetlands in the world (Zhu et al. 2015).
This was indeed out of expectations because environmental conditions of forest soils were very different from that of wetland sediment. To explain this, in subtropical forest, moss and litter in surface layer may retain moisture for the soil from rainfall and, under anaerobic environments, litters would degrade to release substrates for n-damo bacteria. Degradation of organic carbon may provide similar conditions for n-damo bacteria in wetlands to establish and grow.
Another observation on n-damo bacteria was that the majority of sequences from different forests were highly concentrated in cluster I, except a sequence obtained from pine forest soil. Regarding sequences from samples PS and PD, an even distribution pattern was found in clusters I, II, and III, illustrating a more diverse community of n-damo bacteria in the soil samples concerned. Clear explanations and mechanisms of such difference are not available. It could be the consequences of complex interactions among physical-chemical parameters of soil, local vegetation, and microbial communities. Furthermore, less or minimum anthropogenic interruption may also contribute to the characteristic community of n-damo bacteria at this site for the pine forest is located at a steeper slope at relatively higher elevation.
Abundance of anammox bacteria and n-damo bacteria
The PCR primer Amx438f and Amx684r could recover a more refined community structure of anammox bacteria from a wider environmental samples (Han and Gu 2013; Humbert et al. 2012), they were used to detect the abundance of anammox bacteria in this study. Overall, the abundance of anammox bacteria observed in this study ranged from 2.21 × 105 to 3.90 × 106 gene copies g−1 dry soil, which is lower than that reported in paddy soil (105–107 copies g−1 dry soil) (Wang et al. 2012; Zhu et al. 2011). In fact, the surface layer of forest soil (A0 layer) is not exposed to air directly. Instead, it is generally covered by shed and moist leaves, in turn limiting the flux of air and producing a partially anaerobic environment at the surface soil. Thus, this may explain the existence of anammox bacteria in the surface layer of forest soil. However, growth and metabolism of anammox bacteria are highly sensitive and greatly inhibited by O2 (Jetten et al. 2009). In addition, unlike wetland sediment, forest soil in this study was porous in nature, and the anaerobic condition generated by shed leaves was not always guaranteed, e.g., drying season. Therefore, certain amount of oxygen may still frequently be present in the surface layer of soil and hamper the growth of anammox. This may explain the low detection frequency and lower abundance of anammox bacteria in the surface layer.
The abundance of n-damo bacteria (1.68 × 105–4.94 × 106) observed in this study was similar to those reported in flooded paddy field (105–106 copies per gram dry soil) (Shen et al. 2014a), freshwater lakes (104–106 copies per gram dry sediment) (Kojima et al. 2012; Liu et al. 2015), and wetland sediment (105 copies per gram dry sediment with qP1F-P2R) (Chen et al. 2015). Compared to anammox bacteria, n-damo bacteria yielded higher abundance and seemed to be more adaptive in acidic forest soil. However, as n-damo has been discovered only for a short period of time, current designed primers are not optimized and specific to n-damo bacteria. Instead, non-specific amplification of some other bacteria, such as Acidobacteria, has been detected in this acidic forest soil after sequencing. Thus, the value of abundance of n-damo bacteria obtained would be likely over-estimated, and it is hard to evaluate the real ratio of anammox and n-damo with the current available techniques.
Significant correlations were observed between the abundance of anammox bacteria and environmental factors (Table 4). Abundances of anammox and n-damo bacteria were observed to be negatively correlated to parameters including hydrolyzed N, NH4 +-N, and NO3 −−N and organic matter. Organic matter had a negative correlation with the abundance of anammox bacteria and this agrees with the general thought that organic matter (e.g., organic carbon) has an adverse effect on the growth of anammox bacteria because co-occurrence of ammonium, nitrite, and organic matter would encourage growth of heterotrophic denitrifiers which compete nitrite ions with anammox bacteria (Tang et al. 2010). Anammox bacteria, as a result, are less competitive because of their lithotrophic nature and slow growth (Tang et al. 2010). Also, organic alcohol, especially methanol, shows inhibitory effect on activities of anammox bacteria (Güven et al. 2005; Tang et al. 2010; Van de Graaf et al. 1996). Thus, negative correlation between organic matter and abundance of anammox bacteria was expected in forest soil.
An unusual phenomenon observed is that abundances of two bacteria gave negative correlation to nitrogen in soil, including ammonium, nitrite, or total nitrogen content. Since anammox bacteria utilize NH4 + and NO2 −, while n-damo bacteria utilize NO2 − as energy source, a positive correlation is expected between nitrogen content and abundance of these two bacteria (Fig. S1). However, the abundance of bacteria should be considered as the final outcome of all environmental conditions in an open and dynamic ecosystem. Indeed, both bacteria prefer anaerobic condition and oxygen level would be lower at deep layer and, meanwhile, the nitrogen content in soil usually decreases with depth. In case oxygen concentration is a determining factor than other parameters, the abundance of these two bacteria would be higher at lower layer than the surface. In other words, the negative correlation observed in our study would probably be a coincidence than a causation effect.
Anammox and n-damo bacteria co-occurred in the acidic forest soil of tropical forest ecosystem and they were more abundant in lower layers than the surface. Differences were observed between re-vegetated and natural forests on the diversity and abundance of them. Lower diversity of anammox community in the forest ecosystem than other ecosystems reported and n-damo community did not show much difference. These observations may be the results of interactions among various physical-chemical parameters, vegetation, and microbial communities, which warrants further investigations to reveal mechanisms involved in forest ecosystem to fully account the C and N cycling.
References
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi:10.1093/nar/25.17.3389
Beer C, Reichstein M, Tomelleri E, Ciais P, Jung M, Carvalhais N, Rödenbeck C, Arain M, Baldocchi D, Bonan GB, Bondeau A, Cescatti A, Lasslop G, Lindroth A, Lomas M, Luyssaert S, Margolis H, Oleson K, Roupsard O, Veenendaa E, Viovy N, Williams C, Woodward F, Papale D (2010) Terrestrial gross carbon dioxide uptake: global distribution and covariation with climate. Science 329:834–838. doi:10.1126/science
Byrne N, Strous M, Crépeau V, Kartal B, Birrien JL, Schmid M, Lesongeur F, Schouten S, Jaeschke A, Jetten M (2009) Presence and activity of anaerobic ammonium-oxidizing bacteria at deep-sea hydrothermal vents. ISME J 3:117–123. doi:10.1038/ismej.2008.72
Chamchoi N, Nitisoravut S (2007) Anammox enrichment from different conventional sludges. Chemosphere 66:2225–2232. doi:10.1016/j.chemosphere.2006.09.036
Chan HW, Meng H, Gu J-D (2016) Anamox bacteria detected in fish intestinal tract systems. Appl Environ Biotechnol 1(1):13–18. doi:10.18063/AEB.2016.01.010
Chen J, Zhou ZC, Gu J-D (2014) Occurrence and diversity of nitrite-dependent anaerobic methane oxidation bacteria in the sediments of the South China Sea revealed by amplification of both 16S rRNA and pmoA genes. Appl Microbiol Biotechnol 98:5685–5696. doi:10.1007/s00253-014-5733-4
Chen J, Zhou ZC, Gu J-D (2015) Complex community of nitrite-dependent anaerobic methane oxidation bacteria in coastal sediments of the Mai Po wetland by PCR amplification of both 16S rRNA and pmoA genes. Appl Microbiol Biotechnol 99:1463–1473. doi:10.1007/s00253-014-6051-6
Dalsgaard T, Canfield DE, Petersen J, Thamdrup B, Acuña-González J (2003) N2 production by the anammox reaction in the anoxic water column of Golfo Dulce, Costa Rica. Nature 422:606–608. doi:10.1038/nature01526
Dalsgaard T, Thamdrup B, Canfield DE (2005) Anaerobic ammonium oxidation (anammox) in the marine environment. Res Microbiol 156(4):457–464
Egli K, Fanger U, Alvarez PJJ, Siegrist H, van der Meer JR, Zehnder AJB (2001) Enrichment and characterization of an anammox bacterium from a rotating biological contactor treating ammonium-rich leachate. Arch Microbiol 175:198–207. doi:10.1007/s002030100255
Ettwig KF, Van Alen T, van de Pas-Schoonen KT, Jetten MS, Strous M (2009) Enrichment and molecular detection of denitrifying methanotrophic bacteria of the NC10 phylum. Appl Environ Microbiol 75:3656–3662. doi:10.1128/AEM.00067-09
Gan XH, Zhang FQ, Gu J-D, Guo YD, Li ZQ, Zhang WQ, Xu XY, Zhou Y, Wen XY, Xie GG, Wang YF (2015) Differential distribution patterns of ammonia-oxidizing archaea and bacteria in acidic soils of Nanling National Nature Reserve forests in subtropical China. Antonie van Leeuwenhoek 109(2):237–251. doi:10.1007/s10482-015-0627-8
Güven D, Dapena A, Kartal B, Schmid MC, Maas B, van de Pas-Schoonen K, Sozen S, Mendez R, den Camp HJO, Jetten MS (2005) Propionate oxidation by and methanol inhibition of anaerobic ammonium-oxidizing bacteria. Appl Environ Microbiol 71:1066–1071. doi:10.1128/AEM.71.2.1066-1071.2005
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic acids symposium series 41:95–98
Hammer Ø (2012) PAST: PAleontological STatistics. Version 3.0. University of Oslo, Norway
Han P, Gu J-D (2013) More refined diversity of anammox bacteria recovered and distribution in different ecosystems. Appl Microbiol Biotechnol 97:3653–3663. doi:10.1007/s00253-013-4756-6
Haroon MF, Hu S, Shi Y, Imelfort M, Keller J, Hugenholtz P, Yuan Z, Tyson GW (2013) Anaerobic oxidation of methane coupled to nitrate reduction in a novel archaeal lineage. Nature 500:567–570. doi:10.1038/nature12375
Humbert S, Tarnawski S, Fromin N, Mallet MP, Aragno M, Zopfi J (2010) Molecular detection of anammox bacteria in terrestrial ecosystems: distribution and diversity. ISME J 4:450–454. doi:10.1038/ismej.2009.125
Humbert S, Zopfi J, Tarnawski SE (2012) Abundance of anammox bacteria in different wetland soils. Environ Microbiol Rep 4:484–490. doi:10.1111/j.1758-2229.2012.00347.x
Jaeschke A, den Camp HJO, Harhangi H, Klimiuk A, Hopmans EC, Jetten MS, Schouten S, Damsté JSS (2009) 16S rRNA gene and lipid biomarker evidence for anaerobic ammonium-oxidizing bacteria (anammox) in California and Nevada hot springs. FEMS Microbiol Ecol 67:343–350. doi:10.1111/j.1574-6941.2008.00640.x
Jetten MS, Niftrik L, Strous M, Kartal B, Keltjens JT, Op den Camp HJ (2009) Biochemistry and molecular biology of anammox bacteria. Crit Rev Biochem Mol Biol 44:65–84. doi:10.1080/10409230902722783
Jetten MSM (2001) New pathways for ammonia conversion in soil and aquatic systems. Plant Soil 230:9–19. doi:10.1023/A:1004683807250
Kartal B, Geerts W, Jetten MSM (2011) Cultivation, detection, and ecophysiology of anaerobic ammonium-oxidizing bacteria. Method Enzymol 486:89–108. doi:10.1016/B978-0-12-381294-0.00004-3
Kartal B, Rattray J, van Niftrik LA, van de Vossenberg J, Schmid MC, Webb RI, Schouten S, Fuerst JA, Damste JSS, Jetten MSM, Strous M (2007) Candidatus “Anammoxoglobus propionicus” a new propionate oxidizing species of anaerobic ammonium oxidizing bacteria. Syst Appl Microbiol 30:39–49. doi:10.1016/j.syapm.2006.03.004
Kartal B, Van Niftrik L, Rattray J, Van De Vossenberg JL, Schmid MC, Damsté JS, Jetten MS, Strous M (2008) Candidatus ‘Brocadia fulgida’: an autofluorescent anaerobic ammonium oxidizing bacterium. FEMS Microbiol Ecol 63:46–55. doi:10.1111/j.1574-6941.2007.00408.x
Kindaichi T, Tsushima I, Ogasawara Y, Shimokawa M, Ozaki N, Satoh H, Okabe S (2007) In situ activity and spatial organization of anaerobic ammonium-oxidizing (anammox) bacteria in biofilms. Appl Environ Microb 73:4931–4939. doi:10.1128/Aem.00156-07
Kojima H, Tsutsumi M, Ishikawa K, Iwata T, Mußmann M, Fukui M (2012) Distribution of putative denitrifying methane oxidizing bacteria in sediment of a freshwater lake, Lake Biwa. Syst Appl Microbiol 35:233–238. doi:10.1016/j.syapm.2012.03.005
Li H, Chen S, Mu BZ, Gu J-D (2010) Molecular detection of anaerobic ammonium-oxidizing (anammox) bacteria in high-temperature petroleum reservoirs. Microb Ecol 60:771–783. doi:10.1007/s00248-010-9733-3
Li L, He C, Ji G, Zhi W, Sheng L (2015) Nitrogen removal pathways in a tidal flow constructed wetland under flooded time constraints. Ecol Eng 81:266–271. doi:10.1016/j.ecoleng.2015.04.073
Li M, Gu J-D (2016) Molecular evidence of the existence of anaerobic ammonia oxidation bacteria in the gut of polychaete (Neanthes glandicincta). Appl Environ Biotechnol 1(1):19–29. doi:10.18063/AEB.2016.01.011
Liu Y, Zhang J, Zhao L, Li Y, Yang Y, Xie S (2015) Aerobic and nitrite-dependent methane-oxidizing microorganisms in sediments of freshwater lakes on the Yunnan Plateau. Appl Microbiol Biotechnol 99:2371–2381. doi:10.1007/s00253-014-6141-5
Long A, Heitman J, Tobias C, Philips R, Song B (2013) Co-occurring anammox, denitrification, and codenitrification in agricultural soils. Appl Environmental Microbiol 79:168–176. doi:10.1128/AEM.02520-12
Luesken FA, Sánchez J, van Alen TA, Sanabria J, den Camp HJO, Jetten MS, Kartal B (2011a) Simultaneous nitrite-dependent anaerobic methane and ammonium oxidation processes. Appl Environmental Microbiol 77:6802–6807. doi:10.1128/AEM.05539-11
Luesken FA, Zhu BL, van Alen TA, Butler MK, Diaz MR, Song B, den Camp HJMO, Jetten MSM, Ettwig KF (2011b) pmoA primers for detection of anaerobic methanotrophs. Appl Environ Microbiol 77:3877–3880. doi:10.1128/AEM.02960-10
Moore TA, Xing Y, Lazenby B, Lynch MD, Schiff S, Robertson WD, Timlin R, Lanza S, Ryan MC, Aravena R (2011) Prevalence of anaerobic ammonium-oxidizing bacteria in contaminated groundwater. Environ Sci Technol 45:7217–7225. doi:10.1021/es201243t
Mulder A, Graaf A, Robertson L, Kuenen J (1995) Anaerobic ammonium oxidation discovered in a denitrifying fluidized bed reactor. FEMS Microbiol Ecol 16:177–184. doi:10.1016/0168-6496(94)00081-7
Quan ZX, Rhee SK, Zuo JE, Yang Y, Bae JW, Park JR, Lee ST, Park YH (2008) Diversity of ammonium-oxidizing bacteria in a granular sludge anaerobic ammonium-oxidizing (anammox) reactor. Environ Microbiol 10:3130–3139. doi:10.1111/j.1462-2920.2008.01642.x
Raghoebarsing AA, Pol A, Van de Pas-Schoonen KT, Smolders AJ, Ettwig KF, Rijpstra WIC, Schouten S, Damsté JSS, den Camp HJO, Jetten MS (2006) A microbial consortium couples anaerobic methane oxidation to denitrification. Nature 440:918–921. doi:10.1038/nature04617
Reichle DE (1981) Dynamic properties of forest ecosystems. Cambridge University Press, New York
Rysgaard S, Glud RN, Sejr MK, Blicher ME, Stahl HJ (2008) Denitrification activity and oxygen dynamics in Arctic Sea ice. Polar Biol 31:527–537. doi:10.1007/s00300-007-0384-x
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi:10.1128/AEM.01541-09
Schmid M, Twachtmann U, Klein M, Strous M, Juretschko S, Jetten M, Metzger JW, Schleifer K-H, Wagner M (2000) Molecular evidence for genus level diversity of bacteria capable of catalyzing anaerobic ammonium oxidation. Syst Appl Microbiol 23:93–106. doi:10.1016/S0723-2020(00)80050-8
Schmid M, Walsh K, Webb R, Rijpstra WI, van de Pas-Schoonen K, Verbruggen MJ, Hill T, Moffett B, Fuerst J, Schouten S (2003) Candidatus “Scalindua brodae”, sp. nov., Candidatus “Scalindua wagneri”, sp. nov., two new species of anaerobic ammonium oxidizing bacteria. Syst Appl Microbiol 26:529–538. doi:10.1078/072320203770865837
Schubert CJ, Durisch-Kaiser E, Wehrli B, Thamdrup B, Lam P, Kuypers MM (2006) Anaerobic ammonium oxidation in a tropical freshwater system (Lake Tanganyika). Environ Microbiol 8:1857–1863. doi:10.1111/j.1462-2920.2006.01074.x
Sedjo RA (1993) The carbon cycle and global forest ecosystem. Water Air Soil Poll 70:295–307. doi:10.1007/BF01105003
Shen LD, Liu S, He ZF, Lian X, Huang Q, He YF, Lou LP, Xu XY, Zheng P, Hu BL (2015) Depth-specific distribution and importance of nitrite-dependent anaerobic ammonium and methane-oxidising bacteria in an urban wetland. Soil Biol Biochem 83:43–51. doi:10.1016/j.soilbio.2015.01.010
Shen LD, Liu S, Huang Q, Lian X, He ZF, Geng S, Jin RC, He YF, Lou LP, Xu XY (2014a) Evidence for the cooccurrence of nitrite-dependent anaerobic ammonium and methane oxidation processes in a flooded paddy field. Appl Environmental Microbiol 80:7611–7619. doi:10.1128/AEM.02379-14
Shen LD, Liu S, Zhu Q, Li XY, Cai C, Cheng DQ, Lou LP, Xu XY, Zheng P, Hu BL (2014b) Distribution and diversity of nitrite-dependent anaerobic methane-oxidizing bacteria in the sediments of the Qiantang River. Microb Ecol 67:341–349. doi:10.1007/s00248-013-0330-0
Strous M, Fuerst JA, Kramer EHM, Logemann S, Muyzer G, van de Pas-Schoonen KT, Webb R, Kuenen JG, Jetten MSM (1999) Missing lithotroph identified as new planctomycete. Nature 400:446–449. doi:10.1038/22749
Strous M, Pelletier E, Mangenot S, Rattei T, Lehner A, Taylor MW, Horn M, Daims H, Bartol-Mavel D, Wincker P, Barbe V, Fonknechten N, Vallenet D, Segurens B, Schenowitz-Truong C, Medigue C, Collingro A, Snel B, Dutilh BE, Op den Camp HJM, van der Drift C, Cirpus I, van de Pas-Schoonen KT, Harhangi HR, van Niftrik L, Schmid M, Keltjens J, van de Vossenberg J, Kartal B, Meier H, Frishman D, Huynen MA, Mewes HW, Weissenbach J, Jetten MSM, Wagner M, Le Paslier D (2006) Deciphering the evolution and metabolism of an anammox bacterium from a community genome. Nature 440:790–794. doi:10.1038/Nature04647
Tang CJ, Zheng P, Zhang L, Chen JW, Mahmood Q, Chen XG, Hu BL, Wang CH, Yu Y (2010) Enrichment features of anammox consortia from methanogenic granules loaded with high organic and methanol contents. Chemosphere 79:613–619. doi:10.1016/j.chemosphere.2010.02.045
Ter Braak CJ, Smilauer P (2012) Canoco reference manual and user’s guide: software for ordination, version 5.0. Microcomputer Power, Ithaca
Thompson JD, Gibson T, Higgins DG (2002) Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics. 2.3. 1–2.3. 22. Wiley, New York
US Department of Agriculture (1999) Soil taxonomy—a basic system of soil classification for making and interpreting soil surveys. USDA, Washington
Van de Graaf AA, de Bruijn P, Robertson LA, Jetten MS, Kuenen JG (1996) Autotrophic growth of anaerobic ammonium-oxidizing micro-organisms in a fluidized bed reactor. Microbiol 142:2187–2196. doi:10.1099/13500872-142-8-2187
Van de Graaf AA, Mulder A, de Bruijn P, Jetten M, Robertson LA, Kuenen JG (1995) Anaerobic oxidation of ammonium is a biologically mediated process. Appl Environmental Microbiol 61:1246–1251
Wang J, Gu J-D (2013) Dominance of Candidatus Scalindua species in anammox community revealed in soils with different duration of rice paddy cultivation in Northeast China. Appl Microbiol Biotechnol 97:1785–1798. doi:10.1007/s00253-012-4036-x
Wang Y, Zhu G, Harhangi HR, Zhu B, Jetten MS, Yin C, den Camp HJO (2012) Co-occurrence and distribution of nitrite-dependent anaerobic ammonium and methane-oxidizing bacteria in a paddy soil. FEMS Microbiol Lett 336:79–88. doi:10.1111/j.1574-6968.2012.02654.x
Wright ES, Yilmaz LS, Noguera DR (2012) DECIPHER, a search-based approach to chimera identification for 16S rRNA sequences. Appl Environmental Microbiol 78:717–725. doi:10.1128/AEM.06516-11
Wu L, Zhang L, Shi X, Liu T, Peng Y, Zheng J (2015) Analysis of the impact of reflux ratio on coupled partial nitrification-anammox for co-treatment of mature landfill leachate and domestic wastewater. Bioresource Technol 198:207–214. doi:10.1016/j.biortech.2015.08.072
Yang J, Jiang H, Wu G, Hou W, Sun Y, Lai Z, Dong H (2012) Co-occurrence of nitrite-dependent anaerobic methane oxidizing and anaerobic ammonia oxidizing bacteria in two Qinghai-Tibetan saline lakes. Front Earth Sci 6:383–391. doi:10.1007/s11707-012-0336-9
Zhang Y, Ruan XH, Op den Camp HJ, Smits TJ, Jetten MS, Schmid MC (2007) Diversity and abundance of aerobic and anaerobic ammonium-oxidizing bacteria in freshwater sediments of the Xinyi River (China). Environ Microbiol 9:2375–2382. doi:10.1111/j.1462-2920.2007.01357.x
Zhou LL, Wang Y, Long XE, Guo JH, Zhu GB (2014) High abundance and diversity of nitrite-dependent anaerobic methane-oxidizing bacteria in a paddy field profile. FEMS Microbiol Lett 360:33–41. doi:10.1111/1574-6968.12567
Zhu G, Wang S, Wang Y, Wang C, Risgaard-Petersen N, Jetten MS, Yin C (2011) Anaerobic ammonia oxidation in a fertilized paddy soil. ISME J 5:1905–1912. doi:10.1038/ismej.2011.63
Zhu GB, Zhou LL, Wang Y, Wang SY, Guo JH, Long XE, Sun XB, Jiang B, Hou QY, Jetten MSM, Yin CQ (2015) Biogeographical distribution of denitrifying anaerobic methane oxidizing bacteria in Chinese wetland ecosystems. Env Microbiol Rep 7:128–138. doi:10.1111/1758-2229.12214
Acknowledgments
This project was supported by the National Natural Science Foundation of China (grant No. 31470562 to YFW), Hong Kong PhD Fellowship (HM), and RGC GRF Grant No. 701913 (J-DG). Additional financial support for this research project was from the laboratory fund.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM
(PDF 395 kb)
Rights and permissions
About this article
Cite this article
Meng, H., Wang, YF., Chan, HW. et al. Co-occurrence of nitrite-dependent anaerobic ammonium and methane oxidation processes in subtropical acidic forest soils. Appl Microbiol Biotechnol 100, 7727–7739 (2016). https://doi.org/10.1007/s00253-016-7585-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-016-7585-6