Abstract
A newly reported 16S rRNA gene-based PCR primer set was successfully applied to detect anammox bacteria from four ecosystem samples, including sediments from marine, reservoir, mangrove wetland, and wastewater treatment plant sludge. This primer set showed ability to amplify a much wider coverage of all reported anammox bacterial genera. Based on the phylogenetic analyses of 16S rRNA gene of anammox bacteria, two new clusters were obtained, one closely related to Candidatus Scalindua, and the other in a previously reported novel genus related to Candidatus Brocadia. In the Scalindua cluster, four new subclusters were also found in this study, mainly by sequences of the South China Sea sediments, presenting a higher diversity of Candidatus Scalindua in marine environment. Community structure analyses indicated that samples were grouped together based on ecosystems, showing a niche-specific distribution. Phylogenetic analyses of anammox bacteria in samples from the South China Sea also indicated distinguished community structure along the depth. Pearson correlation analysis showed that the amount of anammox bacteria in the detected samples was positively correlated with the nitrate concentration. According to Canonical Correspondence Analysis, pH, temperature, nitrite, and nitrate concentration strongly affected the diversity and distribution of anammox bacteria in South China Sea sediments. Results collectively indicated a promising application of this new primer set and higher anammox bacteria diversity in the marine environment.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Anaerobic ammonium-oxidizing (anammox) bacteria are an important addition to the comprehensive understanding of nitrogen cycle. The existence of anaerobic ammonium oxidation with nitrite as an electron acceptor was predicted based on thermodynamics calculations in 1977 (Broda 1977). The organisms involved were discovered in bioreactors of wastewater treatment plants in the 1990s (Mulder et al. 1995; van de Graaf et al. 1995). Currently, five anammox genera have been described, including the genera of Candidatus Brocadia (Kartal et al. 2008; Oshiki et al. 2011), C. Anammoxoglobus (Kartal et al. 2007), C. Kuenenia (Schmid et al. 2000; Strous et al. 2006), C. Scalindua (Kuypers et al. 2003; Schmid et al. 2003; van de Vossenberg et al. 2008), and C. Jettenia (Quan et al. 2008), all belong to the same monophyletic branch named the Brocadiacaea and are related to the phylum Planctomycetes. Since their discovery, anammox bacteria have been detected in various environments, such as terrestrial environments (Zopfi et al. 2009), mangrove (Li et al. 2011b; Li et al. 2011d; Meyer et al. 2005; Wang et al. 2012b; Wang and Gu 2012b), oil field (Li et al. 2010a), wastewater treatment systems (Wang et al. 2010), and also fresh water and marine ecosystems (Hirsch et al. 2011; Hong et al. 2011; Schmid et al. 2007).
Even though anammox bacteria are distributed widely in environments, there is no pure culture available. Because of this, many culture-independent methods have been developed to detect the diversity and distribution of anammox bacteria in samples from various ecosystems. PCR amplification of anammox bacterial 16S rRNA and functional genes are commonly used for detection and community analysis (Chouari et al. 2003; Li and Gu 2011; Li et al. 2011d; Penton et al. 2006; Quan et al. 2008; Schmid et al. 2005; Wang and Gu 2012a; Wang et al. 2012b). Fluorescence in situ hybridization and stable isotope labeling also serve as useful tools to confirm the presence and activity of anammox bacteria in natural environments (Schmid et al. 2007). Functional gene encoding hydrazine dehydrogenase/oxidoreductase have been reported to amplify anammox bacteria from different environmental samples (Schmid et al. 2008) including coastal and deep-ocean sediments (Dang et al. 2010; Hirsch et al. 2011; Li et al. 2011d), rice paddy soils (Wang and Gu 2012a), oilfields (Li et al. 2010a), or wastewater treatment plants (Quan et al. 2008). The cytochrome cd1-containing nitrite reductase encoding gene (nirS) was used for anammox detection in the ocean and coastal sediment samples (Lam et al. 2009; Li et al. 2011c). The recently reported hydrazine synthase encoding gene (hzsA and hzsB) markers were also applied to detect all known anammox genera (Harhangi et al. 2012; Wang et al. 2012a), yet the efficiency and specificity need to be further studied with more samples from different ecosystems.
In this study, the efficiency of a recently reported 16S rRNA gene-based primer (Humbert et al. 2012) set in detecting anammox bacteria was estimated with several samples. In order to get results with smallest bias, samples from a variety of ecosystems were involved. The primer set was successfully applied to detect anammox bacteria in eleven samples from four ecosystems, including sediments from marine ecosystem, reservoir, mangrove wetland, and wastewater treatment plant sludge. Sequences obtained in this study were compared with sequences from other similar environment like ocean ecosystems (Jensen et al. 2011; Kuypers et al. 2005; Lam and Kuypers 2011; Lam et al. 2009) and mangrove wetland (Li et al. 2011b; Li et al. 2011d), etc. The primer set was successfully applied with our samples and a more refined diversity of anammox bacteria was highlighted in this study.
Materials and methods
Sample collection and chemical analysis
Marine surface sediment samples E702, E704, E707, E708, E709, and E525S were collected from the South China Sea in 2008, and information of sampling sites were reported before (Cao et al. 2012). A2 was collected from a full-scale wastewater treatment plant in July of 2012. R2 was collected from Tai Lam Chung Water Reservoir of Hong Kong SAR in October 2011. Both 1S and 3S were collected from surface layer (0–10 cm) of wetland sediment in Mai Po Nature Reserve in Hong Kong in April 2012. 3Sm was collected in the mangrove surface layer (0–10 cm) near sample 3S (50 m away). The locations and detailed information of Mai Po Nature Reserve are available elsewhere (Li et al. 2011a; Li et al. 2011b; Wang et al. 2012b). Temperature, pH, and redox potential of the sediment samples were measured in situ using IQ180G Bluetooth Multi-Parameter System (Hach Company, Loveland, CO). Concentration of ammonia, nitrite, and nitrate in pore water of sediment samples were measured with an auto-analyzer (QuickChem, Milwaukee, WI) according to the standard methods for the examination of water and wastewater (Association APH 1995).
DNA extraction and PCR amplification
Thawed and homogenized soil or sludge (approximately 0.5 g, wet weight) were transferred into sterile 1.5 mL centrifuge tubes for DNA extraction. The total genomic DNA was extracted using the Power Soil Isolation Kit (Mo Bio, Carlsbad, California) according to the manufacturer's instructions. All extracted DNAs were separately stored at −20 °C for further analysis. For the gene amplification, in a final volume of 25 μl, the PCR reaction mixture contained: 1 μL of extracted template DNA (1–10 ng), 5 μL of 5 × GoTaq Flexi buffer (Promega, Hong Kong) and 2.5 μL of MgCl2 (25 mM, Promega), 0.5 μL of dNTPs (10 mM, Invitrogen, Hong Kong), 1 μL BSA (stock 10 mg mL−1), 0.5 μL of each forward (A438f: 5′-GTCRGGAGTTADGAAATG-3′) and reverse primer (A684r: 5′- ACCAGAAGTTCCACTCTC-3′ ) (20 μM) and 0.2 μL of GoTaq Flexi polymerase (5 U ml−1, Promega, Hong Kong). The PCR program consisted of 95 °C of initial denaturation for 5 min, 33 cycles of 95 °C for 45 s, 54 °C for 30 s, 72 °C for 50 s, followed by 10 min of final elongation at 72 °C. PCR products were analyzed by electrophoresis in 1 % agarose gels in TAE (20 mM Tris-acetate pH 8.0; 0.5 mM EDTA) at 125 V, 400 mA for 25 min (Amersham Biosciences, Electrophoresis Power Supply 301). The gels were stained by addition of GelRed nucleic acid stain (Biotium) and photographed using a Bio-Rad® GelDoc™ station.
Cloning, sequencing, and phylogenetic analyses
Clone libraries were constructed from the PCR products according to the method described elsewhere (Weidner et al. 1996). Briefly, the amplified PCR products were checked in gel as described above and then purified using a Qiaex II Gel Extraction Kit (QIAGen, Hilden, Germany). Ligation with a pMD® 18-T vector (Takara, Japan) was performed at 16 °C for 10 h in a 11.4-μL reaction volume consisting of 4 μL of Solution I (containing T4 DNA Ligase and ligation buffer), 7 μL of purified PCR products, and 0.4 μL of pMD® 18-T vector. Transformation of the vectors to E. coli competent cells was performed (Chung et al. 1989). The cloned single colonies were then randomly picked for PCR amplification using the primer sets M13 (M13F: 5′-TGTAAAACGACGGCCAGT-3′; M13R: 5′-CAGGAAACAGCTATGACC-3′) in a 25-μL PCR reaction volume consisting of the same components except primers as described above. Positive PCR products were sent for DNA sequencing performed with the Big Dye Terminator Kit (Applied Sciences, Foster City, CA) on an ABI Prism 3730 DNA analyzer.
Clone sequences obtained were first checked using Black Box Chimera Check tool (Gontcharova et al. 2010), and their sequence similarity with known references was analyzed with reference sequences retrieved from the GenBank using BLAST tool. Sequences were aligned and phylogenetic trees were constructed using MEGA version 5.05 and subjected to phylogenetic inference using the neighbor-joining algorithm followed by 1,000 times of bootstrap.
Quantitative real-time PCR
The 16S rRNA gene abundance of anammox bacteria including all 11 samples was determined in triplicate using an Applied Biosystems StepOnePlus™ Real-Time PCR System. The quantification was based on the fluorescent dye SYBR-Green I. Each reaction was performed in a 20-μL volume containing 10 μL of Power SYBR Green PCR Master Mix (Applied BioSystems), 1 μL of DNA template (1–10 ng), 0.2 μL of each primer (20 μM; A438f, A684r), and 8.8 μL of autoclaved DD water. The PCR profile was 10 min at 95 °C, followed by a total of 40 cycles of 15 s at 95 °C, 1 min at 54 °C, 15 s at 72 °C. A standard plasmid carrying 16S rRNA gene was generated by amplifying the gene from DNA extracted from samples described above and cloning into the pMD 18-T Vector (Takara). The plasmid DNA concentration was determined, and the gene abundance in terms of copy number of 16S rRNA gene was calculated directly from the concentration of the extracted plasmid DNA. Tenfold serial dilutions of a known copy number of the plasmid DNA were subjected to a quantitative PCR assay in triplicate to generate an external standard curve. The correlation coefficient (r 2) of the standard curve was 0.96.
Anammox bacterial community and statistical analyses
All sequences related to anammox bacteria 16S rRNA gene sequences were analyzed using the DOTUR (Distance-Based OTU and Richness) program to compare their diversity and richness (Schloss and Handelsman 2005). Operational taxonomic units (OTUs) for community analysis were defined at 3 % variation in nucleic acid sequences. In order to examine the distribution of anammox bacteria within the different environmental samples, all clone libraries of 16S rRNA gene were analyzed with UniFrac (http://bmf.colorado.edu/unifrac/) using the principal coordinates analysis tool (PCoA) (Lozupone and Knight 2005). Pearson correlation analyses were used to test correlations between the diversity of anammox bacterial, abundance and different physiochemical characteristics. Correlations of the Scalindua anammox subclusters with environmental factors of the South China Sea surface sediments were explored with canonical correspondence analysis (CCA) using the software Canoco (version 4.5; Microcomputer Power).
Nucleotide sequence accession numbers
Partial sequences of 16S rRNA gene amplified products from the South China Sea, reservoir, mangrove wetland sediments, and wastewater treatment plant have been deposited in GenBank under accession numbers: JX898303-JX898432 and KC539017-KC539100, JX898289-JX898302 and KC539006-KC539016, JX898254-JX898288 and KC538948-KC539005, and JX898221-JX898236, respectively.
Results
Biogeochemical characteristics of sampling sites
The physiochemical characteristics of the five samples are shown in Table 1. The physiochemical characteristics of six South China Sea sediments were reported in previous studies (Cao et al. 2011). There was no significant pH difference among them. The NH+ concentration of samples collected from Mai Po Nature Reserve ranged from 2.94 to 8.20 μg/g. The concentration of NO2 − and NO3 − in these samples ranged from 0.49 to 1.05 μg/g and from 1.24 to 8.82 μg/g, respectively. In addition, NO3 − concentration in Mai Po samples was higher than others, but NO2 − concentration showed a reverse trend in other samples except R2 and E525S. Considering data of the South China Sea sediment samples, it could be referred to previous studies (Cao et al. 2011).
PCR amplification and statistical analyses
This PCR primer set used in this study worked well with almost all included samples. Based on the 11 clone libraries constructed, diversity and richness indices of anammox bacteria with 3 % cutoff was analyzed using DOTUR program (Table 2). Rarefaction curves of 16S rRNA gene sequences retrieved from all samples in this study are available in Fig. S1. A total of 368 gene sequences 16S rRNA genes were obtained from 11 sediment samples in this study. Sequence analyses confirmed that 310 (84.2 %) of the amplified sequences were related to known anammox genera. Overall, a total of 73 OUTs were obtained based on 3 % cutoff. The numbers of OTUs varied among samples ranging from 2 in the waste water treatment plant sample A2 to 13 in the South China Sea sediment sample E707. Generally, the South China Sea and reservoir samples contained more OTUs than other samples. Both the Chao1 diversity estimator and the Shannon index showed a similar variability difference for all samples. Among all the 11 samples, the primer efficiency for anammox detection ranges from 59.3 % for E525S to 100 % for E704, 3Sm, R2, and A2.
Phylogenetic analyses
According to the sequences obtained from this study and the relevant sequences from NCBI, one phylogenetic tree was constructed (Fig. 1). A total of 16 clusters can be visualized based on all obtained and reference sequences. For anammox bacteria, besides the large Scalindua cluster, clusters of Jettenia, Anammoxoglobus, Kuenenia, and Brocadia were established and at the same time, two novel clusters (B.R2-cluster, unknown cluster) were also revealed in this study. One sequence from R2 fell into B.R2 cluster which was a novel cluster described previously (Humbert et al. 2010; Humbert et al. 2012; Quan et al. 2008; Tal et al. 2005). One sequence from 3Sm fell into an unknown cluster which also include sequences retrieved from Jiaojiang estuary (Hu et al. 2012). The similarity of these two clusters to closest typical species was 94.7–97.3 % and 91.4–94.1 %, respectively (Table S1). There are also two R2 groups closely related to Jettenia and Anammoxoglobus which indicated potential independent clusters. They have 98 % nucleotides similarity with sequences retrieved from a eutrophic freshwater lake in Japan (Yoshinaga et al. 2011). Besides all the cluster described above, 10.5 % (2/19) sequences from 1S, 31.5 % (6/19) from 3S and 27.9 % (12/43) from 3Sm contributed to cluster Kuenenia stuttgartiensis, most (15/16) A2 sequences formed Brocadia anammoxidans cluster except one sequence for cluster Anammoxoglobus propionicus, 20.8 % (5/24) sequences from R2 were included in Brocadia fulgida cluster.
Phylogenetic tree constructed with distance and neighbor-joining method from an alignment of PCR amplified 16S rRNA gene sequences along with their some standard species from GenBank. Numbers in racket refer to number of clones. The numbers at the nodes are percentages that indicate the levels of bootstrap support based on 1,000 resampled data sets. Branch lengths correspond to sequence differences as indicated by the scale bar. Phylogenetic tree including all sequences obtained in this study and their related sequences
Sequences belonging to the Scalindua cluster formed another big group, which is in agreement with previous studies considering microdiversity of marine anammox bacteria, namely wagneri, arabica, brodae, zhenghei-I, zhenghei-II, and zhenghei-III subclusters (Hong et al. 2011; Woebken et al. 2008). In this study, no sequence from the South China Sea sediment samples fell into wagneri or zhenghei-III subcluster, 52.6 %(10/19) sequences from 1S and 6.9 % (3/42) sequences from 3Sm contribute to the wagneri subcluster, while zhenghei-III includes 2.3 % (1/42) sequence from 3Sm. The big arabica subcluster contained sequences from all South China Sea samples except E525S; it includes 14.3 % (6/42) sequences from E702, 30.2 % (13/43) for E704, 22.9 % (8/35) for E707, 10 % (3/30) for E708 and 9.5 % (2/21) for E709. The brodae subcluster has 4.8 % (2/42) sequences from E702, 11.6 % (5/43) for E704, 28.6 % (10/35) for E707, 23.3 % (7/30) for E708 and 28.6 % (6/21) for E709. The zhenghei-I subcluster contains 66.7 % (28/42) of sequences from E702, 9.3 % (4/43) for E704, 2.9 % (1/35) for E707, 3.3 % (1/30) for E708 and 6.3 % (1/16) for E525S. For the zhenghei-II subcluster, there are 23.3 % (7/30) sequences from E708, 28.6 % (6/21) for E709, 87.5 % (14/16) for E525S, 42.1 % (8/19) for 3S, 60.5 % (26/43) for 3Sm and 12.5 % (3/24) for R2.
In addition to the above six Scalindua subclusters, four more new subclusters have also been discovered in this study, namely S1, S2, S3, and S4 subclusters. They are specific to the South China Sea except S1 which also include 26.3 % (5/19) sequences from 1S. Besides, in S1 subcluster, there are 14.3 % (6/42), 13.9 % (6/43), 28.6 % (10/35), 23.3 % (7/30), 23.8 % (5/21), and 6.3 % (1/16) of sequences from E702, E704, E707, E708, E709, and E525S respectively. Approximately, 34.9 % (15/43), 5.7 % (2/35), and 3.3 % (1/30) sequences are from E704, E707, and E708 respectively, contribute to S2 subcluster. S3 subcluster contains 6.7 % (2/30) of sequences from E708 only. S4 subcluster contains 11.4 % (4/35), 6.7 % (2/30), and 9.5 % (2/21) of sequences from E707, E708, and E709 respectively. The similarity of above four clusters (S1, S2, S3, and S4) to closest typical species was 95.2–96.8 %, 95.0–95.2 %, 95.7–96.8 %, and 95.2–96.8 %, respectively (Table S1).
Quantity of anammox bacteria
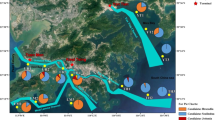
The abundance of anammox bacteria in the 11 samples was estimated by the presence of the anammox bacterial 16S rRNA gene with the primer sets A438f and A684r. The amplified anammox 16S rRNA gene abundance ranged from 2.24 × 103 copies per gram sediments (dry weight) in 1S to 1.02 × 106 copies per gram granule (dry weight) in the A2 sample (Fig. 2). For South China Sea sediment samples, E702 had the highest amount (13.18 × 103 copies/g), while the other five samples did not show large differences (3.45–4.46 × 103 copies/g). The water reservoir sample R2 contained more anammox than other Mai Po samples from the mangrove and mudflat.
Anammox bacterial community structure comparison and correlation with environmental factors
All retrieved 16S rRNA gene sequences were analyzed with UniFrac principal coordinates analysis (PCoA) and Jackknife Environment Clusters analyses (Fig. 3a). Sequences from the South China Sea sediment samples were analyzed separately (Fig. 3b). It was apparently that the South China Sea 16S rRNA clone libraries were significantly different from samples of Mai Po, reservoir and wastewater treatment plant. Besides, it was needed to point that their clone libraries from Mai Po shared similar community structures and sample 1S is separated with the other two samples (3S and 3Sm). Reservoir sample R2 and wastewater treatment plant sample A2 represent their different and close community structure which was indicated by PCoA and Jackknife Environment Clusters analyses. The anammox 16S rRNA gene clone libraries from the six South China Sea samples were also analyzed and compared (Fig. 3b). We tentatively define samples with more than 100 m depth as deep-sea sediments and less than 100 m depth as shallow sediments. It was revealed that these six samples grouped separately according to their depth. However, what's needed to be mentioned is that in the deep sea group, sample E702(2370 m) is closer to E704(175 m) than to E525S (1100 m), indicating that the influence of depth on anammox bacteria community in the South China Sea sediments is more complicated.
Unweighted Unifrac Jackknife Environmental Clusters including all sequences obtained (3a-1) and Scalindua related sequences obtained from South China Sea sediments (3b-1); PCoA analysis including all sequences obtained (3a-2) and Scalindua related sequences obtained from the South China Sea sediments (3b-2). Solid circles in trees represent Jackknife support for the monophyletic at that node. Pink circles, >99.9 %; yellow circles, 90–99.9 %; green, 70–90 %; blue circles, 50–70 %, and gray, <50 %
We also tried to find out the correlation of environmental factors to anammox community structure. Pearson correlation analysis showed that the nitrate concentration significantly influenced the diversity and amount of anammox bacteria within all the 11 samples (Table 3). Besides, the South China Sea samples and the ten described Scalindua subclusters based on 16S rRNA phylogenetic analyses with environmental parameters were analyzed via CCA (Fig. 4). The correlation of samples with environmental variables was shown in the first two CCA dimensions (CCA1 and CCA2), which explained 45.2 % of the total variance in samples and 78.3 % of the cumulative variance of the anammox environment relationship (Fig.4a). The correlation of Scalindua subclusters with environmental variables was shown in the first two CCA dimensions (CCA1 and CCA2), which explained 41.4 % of the total variance in samples and 68.9 % of the cumulative variance of the anammox environment relationship (Fig. 4b).
CCA ordination plots for the first two principal dimensions of the relationship between the distributions of the South China Sea surface sediment samples (a) and anammox Scalindua subclusters (b) with environmental parameters of the used in this study. Correlations between environmental variables and CCA axes are represented by the length and angle of arrows (environmental factor vectors)
The temperature, pH, and concentration of nitrite and ammonia were strikingly correlated with the diversity of four sea samples (E704, E707, E708, E709, depth <200 m). For ten Scalindua subclusters, pH was strongly correlated with subcluster S1, S2, S3, S4, arabica, and brodae. Concentration of nitrite and nitrate were profoundly correlated with subcluster zhenghei-II, zhenghei-III, and wagneri. In addition, correlations of the anammox bacterial assemblages with environmental parameters analyzed by CCA indicated that depth is significantly correlated with zhenghei-I subcluster and E702 sample.
Discussion
Diversity of anammox bacteria in different ecosystems
In this study, all the five known anammox genera were detected from the 11 samples used in this study with the newly reported PCR primer set. For samples from Mai Po Nature Reserve, a wide coverage of anammox diversity including Kuenenia, Brocadia, Jettenia, and Scalindua had been observed showing a good agreement with previous studies considering anammox bacteria in this area (Li et al. 2011b; Li et al. 2011d). For the reservoir sample R2, a higher diversity and wider coverage of anammox bacteria including Brocadia, Scalindua, one sequence which may fall into a novel cluster were evident. To our knowledge, few studies on anammox bacteria diversity in water reservoir are available and our study provides a basis to investigate its microbial diversity including ammonia-oxidizing bacteria and archaea in this ecosystem. In the wastewater treatment plant (WWTP) sample A2, Brocadia dominated in the anammox community with only one individual Anammoxoglobus sequence, showing differences from other WWTP samples in which the dominant anammox bacteria are Kuenenia (Kumar and Lin 2010). So far, anammox bacteria were reported in marine oxygen minimum zones (OMZs) from Peruvian, Namibian, and the Arabian Sea (Woebken et al. 2008), OMZs off northern Chile (Galan et al. 2009) and OMZs located in the Arabian Sea, the eastern tropical North Pacific (ETNP) and the eastern tropical South Pacific (ETSP) (Jayakumar et al. 2009). By comparing sequences from the above OMZs and also sequences retrieved from anoxic wetland sediments (Wang et al. 2012b), mangrove sediment (Amano et al. 2011), sponge (Hoffmann et al. 2009; Mohamed et al. 2010), bioreactors (Isaka et al. 2006; Tal et al. 2006), freshwater (Zhang et al. 2007) and estuarial sediment (Hu et al. 2012; Li et al. 2011b), four new Scalindua subclusters were obtained from the South China Sea sediments, which further highlight the microdiversity of marine Scalindua in marine and subsurface ecosystem from existing information (Hong et al. 2011; Woebken et al. 2008).
Anammox community structure comparison
All retrieved 16S rRNA gene sequences were analyzed with UniFrac principal coordinates analysis (PCoA). According to the results, all samples from the South China Sea were grouped together and samples from Mai Po Nature Reserve grouped sparsely. This generally indicated a niche-specific distribution pattern of anammox bacteria and also in agreement with previous studies (Li et al. 2010b). Sequences from the South China Sea sediment samples were analyzed separately due to the requirements for a more detailed Scalindua analyses with higher resolution. Samples were grouped along a depth-related distribution pattern which may be defined as a coastal-ocean pattern. Four sea samples (E704, E707, E708 and E709) were distributed separately with the other two deep-sea samples (E702 and E525S). The observed pattern may not only be caused along depth pattern, but also by the anthropogenic pollution gradient from coastal Pearl River estuary to the South China Sea as discussed in previous study considering diversity and abundance of ammonia-oxidizing prokaryotes (Cao et al. 2012).
The influence of environmental factors on the anammox community structure
Pearson correlation analysis showed that the nitrate concentration significantly influenced the diversity and amount of anammox bacteria (Table 3), which is also reported in the influence of intense chemical pollution on the community and abundance of anammox bacteria in the Jiaojiang Estuary (China) (Hu et al. 2012). That also adds evidence to the statement that the nitrate concentration can be identified as an important factor influencing the distribution of anammox communities in estuarial mudflat sediment of the Mai Po Nature Reserve (Li et al. 2011b). Base on canonical correspondence analysis, pH, temperature, concentration of nitrite, nitrate, and ammonia have significant influences on certain sea samples or Scalindua subclusters, depth of samples also has a strong correlation with deep-sea subcluster, zhenghei-I.
The new 16S rRNA gene-based primer set (A438f, A684r) was successfully applied in detecting anammox bacteria from widely different ecosystems. Two new clusters and four new Scalindua subclusters were recovered, showing the robustness of this primer set and also the hidden diversity of anammox bacteria in environment, especially in marine ecosystems. In addition, this primer set could also be applied in quantitative real-time PCR. Results collectively indicate a much higher diversity of anammox bacteria in environment, and more analyses should be conducted on samples from ocean sediments in the future to unravel the anammox diversity.
References
Amano T, Yoshinaga I, Yamagishi T, Thuoc CV, Thu PT, Ueda S, Kato K, Sako Y, Suwa Y (2011) Contribution of anammox bacteria to benthic nitrogen cycling in a mangrove forest and shrimp ponds, haiphong, Vietnam. Microbes Environ 26(1):1–6
Association APH (1995) Standard methods for the examination of water and wastewater. APHA, Washington, DC
Broda E (1977) Two kinds of lithotrophs missing in nature. Z Allg Mikrobiol 17(6):491–493
Cao H, Hong Y, Li M, Gu J-D (2011) Diversity and abundance of ammonia-oxidizing prokaryotes in sediments from the coastal Pearl River estuary to the South China Sea. Antonie Leeuwenhoek 100(4):545–556
Cao H, Hong Y, Li M, Gu J-D (2012) Community shift of ammonia-oxidizing bacteria along an anthropogenic pollution gradient from the Pearl River Delta to the South China Sea. Appl Microbiol Biotechnol 94(1):247–259
Chouari R, Le Paslier D, Daegelen P, Ginestet P, Weissenbach J, Sghir A (2003) Molecular evidence for novel planctomycete diversity in a municipal wastewater treatment plant. Appl Environ Microbiol 69(12):7354–7363
Chung CT, Niemela SL, Miller RH (1989) One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc Natl Acad Sci U S A 86(7):2172–2175
Dang H, Li J, Chen R, Wang L, Guo L, Zhang Z, Klotz MG (2010) Diversity, abundance, and spatial distribution of sediment ammonia-oxidizing Betaproteobacteria in response to environmental gradients and coastal eutrophication in Jiaozhou Bay, China. Appl Environ Microbiol 76(14):4691–4702
Galan A, Molina V, Thamdrup B, Woebken D, Lavik G, Kuypers MMM, Ulloa O (2009) Anammox bacteria and the anaerobic oxidation of ammonium in the oxygen minimum zone off northern Chile. Deep-Sea Res II 56(16):1125–1135
Gontcharova V, Youn E, Wolcott RD, Hollister EB, Gentry TJ, Dowd SE (2010) Black Box Chimera Check (B2C2): a Windows-Based Software for Batch Depletion of Chimeras from Bacterial 16S rRNA Gene Datasets. Open Microbiol J 4:47–52
Harhangi HR, Le Roy M, van Alen T, Hu BL, Groen J, Kartal B, Tringe SG, Quan ZX, Jetten MS, Op den Camp HJ (2012) Hydrazine synthase, a unique phylomarker with which to study the presence and biodiversity of anammox bacteria. Appl Environ Microbiol 78(3):752–758
Hirsch MD, Long ZT, Song B (2011) Anammox bacterial diversity in various aquatic ecosystems based on the detection of hydrazine oxidase genes (hzoA/hzoB). Microb Ecol 61(2):264–276
Hoffmann F, Radax R, Woebken D, Holtappels M, Lavik G, Rapp HT, Schlappy ML, Schleper C, Kuypers MM (2009) Complex nitrogen cycling in the sponge Geodia barretti. Environ Microbiol 11(9):2228–2243
Hong YG, Li M, Cao H, Gu J-D (2011) Residence of habitat-specific anammox bacteria in the deep-sea subsurface sediments of the South China Sea: analyses of marker gene abundance with physical chemical parameters. Microb Ecol 62(1):36–47
Hu B, Shen L, Du P, Zheng P, Xu X, Zeng J (2012) The influence of intense chemical pollution on the community composition, diversity and abundance of anammox bacteria in the Jiaojiang Estuary (China). PLoS One 7(3):e33826
Humbert S, Tarnawski S, Fromin N, Mallet MP, Aragno M, Zopfi J (2010) Molecular detection of anammox bacteria in terrestrial ecosystems: distribution and diversity. ISME J 4(3):450–454
Humbert S, Zopfi J, Tarnawski SE (2012) Abundance of anammox bacteria in different wetland soils. Env Microbiol Rep 4(5):484–490
Isaka K, Date Y, Sumino T, Yoshie S, Tsuneda S (2006) Growth characteristic of anaerobic ammonium-oxidizing bacteria in an anaerobic biological filtrated reactor. Appl Microbiol Biotechnol 70(1):47–52
Jayakumar A, O’Mullan GD, Naqvi SW, Ward BB (2009) Denitrifying bacterial community composition changes associated with stages of denitrification in oxygen minimum zones. Microb Ecol 58(2):350–362
Jensen MM, Lam P, Revsbech NP, Nagel B, Gaye B, Jetten MS, Kuypers MM (2011) Intensive nitrogen loss over the Omani Shelf due to anammox coupled with dissimilatory nitrite reduction to ammonium. ISME J 5(10):1660–1670
Kartal B, Rattray J, van Niftrik LA, van de Vossenberg J, Schmid MC, Webb RI, Schouten S, Fuerst JA, Damste JS, Jetten MS, Strous M (2007) Candidatus “Anammoxoglobus propionicus” a new propionate oxidizing species of anaerobic ammonium oxidizing bacteria. Syst Appl Microbiol 30(1):39–49
Kartal B, van Niftrik L, Rattray J, van de Vossenberg JL, Schmid MC, Sinninghe Damste J, Jetten MS, Strous M (2008) Candidatus ‘Brocadia fulgida’: an autofluorescent anaerobic ammonium oxidizing bacterium. FEMS Microbiol Ecol 63(1):46–55
Kumar M, Lin JG (2010) Co-existence of anammox and denitrification for simultaneous nitrogen and carbon removal—strategies and issues. J Hazard Mater 178(1–3):1–9
Kuypers MM, Lavik G, Woebken D, Schmid M, Fuchs BM, Amann R, Jorgensen BB, Jetten MS (2005) Massive nitrogen loss from the Benguela upwelling system through anaerobic ammonium oxidation. Proc Natl Acad Sci U S A 102(18):6478–6483
Kuypers MM, Sliekers AO, Lavik G, Schmid M, Jorgensen BB, Kuenen JG, Sinninghe Damste JS, Strous M, Jetten MS (2003) Anaerobic ammonium oxidation by anammox bacteria in the Black Sea. Nature 422(6932):608–611
Lam P, Kuypers MM (2011) Microbial nitrogen cycling processes in oxygen minimum zones. Ann Rev Mar Sci 3:317–345
Lam P, Lavik G, Jensen MM, van de Vossenberg J, Schmid M, Woebken D, Gutierrez D, Amann R, Jetten MS, Kuypers MM (2009) Revising the nitrogen cycle in the Peruvian oxygen minimum zone. Proc Natl Acad Sci U S A 106(12):4752–4757
Li H, Chen S, Mu BZ, Gu J-D (2010a) Molecular detection of anaerobic ammonium-oxidizing (anammox) bacteria in high-temperature petroleum reservoirs. Microb Ecol 60(4):771–783
Li M, Cao H, Hong Y, Gu J-D (2011a) Spatial distribution and abundances of ammonia-oxidizing archaea (AOA) and ammonia-oxidizing bacteria (AOB) in mangrove sediments. Appl Microbiol Biotechnol 89(4):1243–1254
Li M, Cao H, Hong YG, Gu J-D (2011b) Seasonal dynamics of anammox bacteria in estuarial sediment of the Mai Po Nature Reserve revealed by analyzing the 16S rRNA and hydrazine oxidoreductase (hzo) genes. Microbes Environ 26(1):15–22
Li M, Ford T, Li X, Gu J-D (2011c) Cytochrome cd1-containing nitrite reductase encoding gene nirS as a new functional biomarker for detection of anaerobic ammonium oxidizing (Anammox) bacteria. Environ Sci Technol 45(8):3547–3553
Li M, Gu J-D (2011) Advances in methods for detection of anaerobic ammonium oxidizing (anammox) bacteria. Appl Microbiol Biotechnol 90(4):1241–1252
Li M, Hong Y, Klotz MG, Gu J-D (2010b) A comparison of primer sets for detecting 16S rRNA and hydrazine oxidoreductase genes of anaerobic ammonium-oxidizing bacteria in marine sediments. Appl Microbiol Biotechnol 86(2):781–790
Li M, Hong YG, Cao HL, Gu J-D (2011d) Mangrove trees affect the community structure and distribution of anammox bacteria at an anthropogenic-polluted mangrove in the Pearl River Delta reflected by 16S rRNA and hydrazine oxidoreductase (HZO) encoding gene analyses. Ecotoxicology 20(8):1780–1790
Lozupone C, Knight R (2005) UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71(12):8228–8235
Meyer RL, Risgaard-Petersen N, Allen DE (2005) Correlation between anammox activity and microscale distribution of nitrite in a subtropical mangrove sediment. Appl Environ Microbiol 71(10):6142–6149
Mohamed NM, Saito K, Tal Y, Hill RT (2010) Diversity of aerobic and anaerobic ammonia-oxidizing bacteria in marine sponges. ISME J 4(1):38–48
Mulder A, Vandegraaf AA, Robertson LA, Kuenen JG (1995) Anaerobic ammonium oxidation discovered in a denitrifying fluidized-bed reactor. FEMS Microbiol Ecol 16(3):177–183
Oshiki M, Shimokawa M, Fujii N, Satoh H, Okabe S (2011) Physiological characteristics of the anaerobic ammonium-oxidizing bacterium ‘Candidatus Brocadia sinica’. Microbiology 157(Pt 6):1706–1713
Penton CR, Devol AH, Tiedje JM (2006) Molecular evidence for the broad distribution of anaerobic ammonium-oxidizing bacteria in freshwater and marine sediments. Appl Environ Microbiol 72(10):6829–6832
Quan ZX, Rhee SK, Zuo JE, Yang Y, Bae JW, Park JR, Lee ST, Park YH (2008) Diversity of ammonium-oxidizing bacteria in a granular sludge anaerobic ammonium-oxidizing (anammox) reactor. Environ Microbiol 10(11):3130–3139
Schloss PD, Handelsman J (2005) Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl Environ Microbiol 71(3):1501–1506
Schmid M, Twachtmann U, Klein M, Strous M, Juretschko S, Jetten M, Metzger JW, Schleifer KH, Wagner M (2000) Molecular evidence for genus level diversity of bacteria capable of catalyzing anaerobic ammonium oxidation. Syst Appl Microbiol 23(1):93–106
Schmid M, Walsh K, Webb R, Rijpstra WI, van de Pas-Schoonen K, Verbruggen MJ, Hill T, Moffett B, Fuerst J, Schouten S, Damste JS, Harris J, Shaw P, Jetten M, Strous M (2003) Candidatus “Scalindua brodae”, sp. nov., Candidatus “Scalindua wagneri”, sp. nov., two new species of anaerobic ammonium oxidizing bacteria. Syst Appl Microbiol 26(4):529–538
Schmid MC, Hooper AB, Klotz MG, Woebken D, Lam P, Kuypers MM, Pommerening-Roeser A, Op den Camp HJ, Jetten MS (2008) Environmental detection of octahaem cytochrome c hydroxylamine/hydrazine oxidoreductase genes of aerobic and anaerobic ammonium-oxidizing bacteria. Environ Microbiol 10(11):3140–3149
Schmid MC, Maas B, Dapena A, van de Pas-Schoonen K, van de Vossenberg J, Kartal B, van Niftrik L, Schmidt I, Cirpus I, Kuenen JG, Wagner M, Sinninghe Damste JS, Kuypers M, Revsbech NP, Mendez R, Jetten MS, Strous M (2005) Biomarkers for in situ detection of anaerobic ammonium-oxidizing (anammox) bacteria. Appl Environ Microbiol 71(4):1677–1684
Schmid MC, Risgaard-Petersen N, van de Vossenberg J, Kuypers MM, Lavik G, Petersen J, Hulth S, Thamdrup B, Canfield D, Dalsgaard T, Rysgaard S, Sejr MK, Strous M, den Camp HJ, Jetten MS (2007) Anaerobic ammonium-oxidizing bacteria in marine environments: widespread occurrence but low diversity. Environ Microbiol 9(6):1476–1484
Strous M, Pelletier E, Mangenot S, Rattei T, Lehner A, Taylor MW, Horn M, Daims H, Bartol-Mavel D, Wincker P, Barbe V, Fonknechten N, Vallenet D, Segurens B, Schenowitz-Truong C, Medigue C, Collingro A, Snel B, Dutilh BE, Op den Camp HJ, van der Drift C, Cirpus I, van de Pas-Schoonen KT, Harhangi HR, van Niftrik L, Schmid M, Keltjens J, van de Vossenberg J, Kartal B, Meier H, Frishman D, Huynen MA, Mewes HW, Weissenbach J, Jetten MS, Wagner M, Le Paslier D (2006) Deciphering the evolution and metabolism of an anammox bacterium from a community genome. Nature 440(7085):790–794
Tal Y, Watts JE, Schreier HJ (2005) Anaerobic ammonia-oxidizing bacteria and related activity in Baltimore inner harbor sediment. Appl Environ Microbiol 71(4):1816–1821
Tal Y, Watts JE, Schreier HJ (2006) Anaerobic ammonium-oxidizing (anammox) bacteria and associated activity in fixed-film biofilters of a marine recirculating aquaculture system. Appl Environ Microbiol 72(4):2896–2904
van de Graaf AA, Mulder A, de Bruijn P, Jetten MS, Robertson LA, Kuenen JG (1995) Anaerobic oxidation of ammonium is a biologically mediated process. Appl Environ Microbiol 61(4):1246–1251
van de Vossenberg J, Rattray JE, Geerts W, Kartal B, van Niftrik L, van Donselaar EG, Sinninghe Damste JS, Strous M, Jetten MS (2008) Enrichment and characterization of marine anammox bacteria associated with global nitrogen gas production. Environ Microbiol 10(11):3120–3129
Wang CC, Lee PH, Kumar M, Huang YT, Sung S, Lin JG (2010) Simultaneous partial nitrification, anaerobic ammonium oxidation and denitrification (SNAD) in a full-scale landfill-leachate treatment plant. J Hazard Mater 175(1–3):622–628
Wang J, Gu J-D (2012a) Dominance of Candidatus Scalindua species in anammox community revealed in soils with different duration of rice paddy cultivation in Northeast China. Appl Microbiol Biotechnol. doi:10.1007/s00253-012-4036-x
Wang S, Zhu G, Peng Y, Jetten MS, Yin C (2012a) Anammox bacterial abundance, activity, and contribution in riparian sediments of the pearl river estuary. Environ Sci Technol 46(16):8834–8842
Wang YF, Feng YY, Ma X, Gu J-D (2012b) Seasonal dynamics of ammonia/ammonium-oxidizing prokaryotes in oxic and anoxic wetland sediments of subtropical coastal mangrove. Appl Microbiol Biotechnol. doi:10.1007/s00253-012-4510-5
Wang YF, Gu J-D (2012b) Higher diversity of ammonia/ammonium-oxidizing prokaryotes in constructed freshwater wetland than natural coastal marine wetland. Appl Microbiol Biotechnol. doi:10.1007/s00253-012-4430-4
Weidner S, Arnold W, Puhler A (1996) Diversity of uncultured microorganisms associated with the seagrass Halophila stipulacea estimated by restriction fragment length polymorphism analysis of PCR-amplified 16S rRNA genes. Appl Environ Microbiol 62(3):766–771
Woebken D, Lam P, Kuypers MM, Naqvi SW, Kartal B, Strous M, Jetten MS, Fuchs BM, Amann R (2008) A microdiversity study of anammox bacteria reveals a novel Candidatus Scalindua phylotype in marine oxygen minimum zones. Environ Microbiol 10(11):3106–3119
Yoshinaga I, Amano T, Yamagishi T, Okada K, Ueda S, Sako Y, Suwa Y (2011) Distribution and diversity of anaerobic ammonium oxidation (anammox) bacteria in the sediment of a eutrophic freshwater lake, Lake Kitaura, Japan. Microbes Environ 26(3):189–197
Zhang Y, Ruan XH, Op den Camp HJ, Smits TJ, Jetten MS, Schmid MC (2007) Diversity and abundance of aerobic and anaerobic ammonium-oxidizing bacteria in freshwater sediments of the Xinyi River (China). Environ Microbiol 9(9):2375–2382
Zopfi A, Humbert S, Bagnoud A, Tarnawski S, Conen F, Seth B (2009) Anammox in terrestrial ecosystems: distribution, diversity and activity. Geochim Cosmochim Acta 73(13):A1538–A1538
Acknowledgments
This research project was supported in part by a Ph.D. studentship from The University of Hong Kong (PH), Environmental and Conservation Fund (Grant No.15/2011) (J-DG). Additional financial support was given by the Environmental Toxicology Education and Research Fund of this laboratory. Ms. Jessie Lai and Ms. Kelly Lau were thanked for their laboratory assistance.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 55 kb)
Rights and permissions
About this article
Cite this article
Han, P., Gu, JD. More refined diversity of anammox bacteria recovered and distribution in different ecosystems. Appl Microbiol Biotechnol 97, 3653–3663 (2013). https://doi.org/10.1007/s00253-013-4756-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-013-4756-6