Abstract
Extremely acidic soils of natural forests in Nanling National Nature Reserve have been previously investigated and revisited in two successive years to reveal the active ammonia oxidizers. Ammonia-oxidizing archaea (AOA) rather than ammonia-oxidizing bacteria (AOB) were found more functionally important in the extremely acidic soils of the natural forests in Nanling National Nature Reserve. The relative abundances of Nitrosotalea, Nitrososphaera sister group, and Nitrososphaera lineages recovered by ammonia monooxygenase subunit A (amoA) transcripts were reassessed and compared to AOA communities formerly detected by genomic DNA. Nitrosotalea, previously found the most abundant AOA, were the second-most-active lineage after Nitrososphaera sister group. Our field study results, therefore, propose the acidophilic AOA, Nitrosotalea, can better reside in extremely acidic soils while they may not contribute to nitrification proportionately according to their abundances or they are less functionally active. In contrast, the functional importance of Nitrososphaera sister group may be previously underestimated and the functional dominance further extends their ecological distribution as little has been reported. Nitrososphaera gargensis–like AOA, the third abundant lineage, were more active in summer. The analyses of AOA community composition and its correlation with environmental parameters support the previous observations of the potential impact of organic matter on AOA composition. Al3+, however, did not show a strong adverse correlation with the abundances of functional AOA unlike in the DNA-based study. The new data further emphasize the functional dominance of AOA in extremely acidic soils, and unveil the relative contributions of AOA lineages to nitrification and their community transitions under the environmental influences.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Ammonia-oxidizing archaea (AOA) have received increasing interests since the first discovery of archaeal ammonia monooxygenase (amo)–like sequences and the first AOA culture, Nitrosopumilus maritimus SMC1, through metagenomic and isolation studies (Konneke et al. 2005; Schleper et al. 2005; Venter et al. 2004). The high abundances of the marine AOA lineages were counted for 20–30% of the marine microorganisms, challenging the perspective of ammonia-oxidizing bacteria (AOB)–driven aerobic ammonia oxidation established over a century (Herndl et al. 2005; Karner et al. 2001; Wuchter et al. 2006). As the distinctive physiology of AOA, the group of archaea capable of oxidizing ammonia initiating the first step of nitrification was proposed as a novel phylum, Thaumarchaeota.
During the past 10 years, a myriad of environmental surveys of AOA reveals their wide ecological distributions in estuarine sediments (Beman and Francis 2006), wastewater bioreactors (Park et al. 2006), soils (Leininger et al. 2006), corals (Beman et al. 2007), terrestrial hot springs (Weidler et al. 2007), sponges (Steger et al. 2008), and wetland (Wang and Gu 2013). Fueled by the information of AOA niches differentiation, more enriched AOA cultures or isolates besides the marine representatives were obtained, offering more detailed physiological features. Candidatus Nitrososphaera gargensis (Hatzenpichler et al. 2008) and Candidatus Nitrosocaldus yellowstonii (de la Torre et al. 2008), belonging to Group 1.1b and ThAOA, were described thermophilic thriving at the temperature up to 46 °C and 74 °C, respectively, and the growth of Nitrososphaera viennensis EN76, a soil inhabiting AOA, can be stimulated by urea in addition to ammonia (Tourna et al. 2011). Acidophilic Candidatus Nitrosotalea devanaterra (Group 1.1a–associated AOA) (Lehtovirta-Morley et al. 2011) and neutrophilic Candidatus Nitrosocosmicus franklandus (Nitrososphaera sister group) (Lehtovirta-Morley et al. 2016) were discovered recently implying pH can also impact on AOA specialization, as it can interact with the availability of ammonia and toxicity of NO and N2O (De Boer et al. 1991; De De Boer and Kowalchuk 2001). Apart from their potential contributions to nitrification in the diverse ecosystems, AOA like AOB were also responsible for the emission of N2O with 310 times more potent greenhouse effect than CO2 (Loscher et al. 2012; Santoro et al. 2011; Stieglmeier et al. 2014). Studying the community of functional ammonia oxidizers would also serve the purpose of delineating the greenhouse gas emission patterns.
Nevertheless, few cultures can be obtained, and it is still questionable if they can represent the dominant and functional AOA species, and the physiological characteristics detected in the cultures can hardly apply to microbial communities under multifactorial influences of the natural environments. Therefore, more field studies with activity-based analysis are needed to further expend the knowledge of AOA niche specialization and decipher the potential physiology of the functional species.
Our previous study has first explored the seasonal and spatial dynamics of ammonia oxidizers in extremely acidic natural forest soils with pH ranging from 3.80 to 4.59 at Guangdong Nanling National Nature Reserve (unpublished data). Detection of amoA abundance based on genomic DNA demonstrated the potential functional dominance of AOA over AOB in both winter and summer. In this study, amoA transcripts were directly detected to probe the relative dominance of functional ammonia oxidizers, in order to re-evaluate the relative contributions of functional phylotypes to nitrification and re-assess the previously proposed potential toxicity of organic acids and aluminum on ammonia oxidizer communities. This study would provide more evidence of niche differentiation and specialization of ammonia oxidizers and hint the possible survival and function strategies of AOA in acidic soils.
Materials and methods
Site description and sample collection
The three natural forests in Guangdong Nanling National Nature Reserve (24°37′–24°57′N, 112°30′–113°04′E) were revisited on August 5, 2015 and January 8, 2016. The climate is subtropical monsoon with an annual precipitation of 2108.4 mm (NNNR 2015, 2016), and the soils are characterized as extremely acidic with pH lower than 4.6 according to Natural Resources Conservation Service (NRCS).
Same sampling plan was followed as collecting soil samples from mountainous dwarf forest (1858 m, MD) with main tree species of Rhododendron simiarum, Cyclobalanopsis stewardiana, Schima superba mixed forest of needle and broadleaf trees (1371 m, MF) such as Pinus kwangtungensis, Tsuga longibracteata, Michelia maudiae, and Fokienia hodginsii, and evergreen broadleaf forest (1009 m, EB) with Schima superba, Machilus thunbergii, and Acer davidii. For each forest type, both surface (A0 layer, 0–2 cm, with removal of the litter coverage) and lower layer (B layer, 18–20 cm) soil samples were collected in triplicates. Approximately 2 g homogenized soil samples for total RNA extraction were preserved by 5 mL LifeGuard Soil Preservation Solution (MO BIO, Carlsbad, CA, USA) on site. The rest of the soil samples from each replicate (ca. 1 kg) were kept in a cooler with ice-bags during sampling and transported back to laboratory immediately, subjected to physicochemical analysis.
Physicochemical analysis
Physicochemical analysis of soil samples was performed according to the guidelines of Methods of Agriculture Chemical Analysis (Lu 2000) as mentioned in the previous study (Wu et al. 2017). Briefly, a slurry (soil-to-water ratio 1:1) was prepared for pH measurement (Starter 3C, OHAUS, Pine Brook, NJ, USA). Organic carbon was quantified by the sulfuric acid dichromate digestion method, and organic matter was calculated with ratio of organic matter–to–organic carbon of 1.724 (Nelson and Sommers 1996). Kjeldahl method was applied to measure total nitrogen (N). NH4+-N and NO3−-N were extracted with 2 M KCl and analyzed by Nessler’s reagent colorimetry and ultraviolet spectrophotometry (UV–Vis spectrophotometer, 752 N type, Shanghai Jingke Co., China), respectively. Exchangeable Al3+ was extracted with 2 M KCl, and the concentration was determined by ICP-OES (Perkin Elmer Optima 8300, Waltham, MA, USA).
RNA extraction and reverse transcription
Total RNA was extracted from the preserved soil following the manual of RNA PowerSoil Total RNA Isolation Kit (MO BIO, Carlsbad, CA, USA) with modifications including optimizing bead beating duration for 20 mins, adjusting elution buffer amount to 50 μL, and applying DNase I (ThermoFisher Scientific, Basingstoke, UK) incubation to maximize the final yield and quality. RNA quality and concentration were accessed by Nanodrop Spectrophotometer ND-1000 and RNA gel electrophoresis. The purified RNA was reversely transcripted into complementary DNA (cDNA) by random hexamers using PrimeScript First Strand cDNA Synthesis Kit (TaKaRa Bio, Shiga, Japan).
amoA gene amplification
Amplification of archaeal amoA and bacterial amoA genes was performed using primer sets as shown in Table 1. The reaction mixture and the PCR condition were specified in the previous study (Wu et al. 2017). Nested PCR was carried out for bacterial amoA which cannot be amplified by optimized general PCR routine, using primer set A189 and amoA-2R for the first round amplification and amoA-1F/2R for the second (Horz et al. 2004) (Table 1).
Clone library construction and sequencing
PCR products were purified from size-verified gel bands by Illustra GFX PCR DNA and Gel Band Purification Kits (GE Healthcare Sciences, Little Chalfont, UK). The archaeal and bacterial amoA fragments were ligated into vector PMD18 T (pMD™18-T VectorCloning Kit, Takara Bio Inc., Shiga, Japan) and transformed into Escherichia coli DH5α competent cells (pMD™18-T Vector Cloning Kit, Takara International, Hong Kong, China) ready for clone library. Ninety colonies for each samples were randomly selected after size verification of the PCR amplicons using M13 F/R primer set and sequenced by ABI 3730xl DNA analyzer (Applied Biosystems, Foster City, CA, USA).
Phylogenetic analysis
Sequences from short or failed reads or of apparent chimeric origin were excluded after the quality control. The remaining trimmed sequences were fed to RDP FunGene analyzing platform (Fish et al. 2013). The nucleotide sequences were translated into amino acid sequences using Framebot to detect and correct frameshifts, following with alignment by HMMER3 aligner. The deduced amino acid sequences were clustered into operational taxonomic units (OTUs) by mcCLUST (RDP) coded by complete-linkage clustering method, and the distance cutoff was set as 3% (a conservative cutoff was determined based on Supplemental Table S1). All the representative sequences of the collected OTUs from each samples were further pooled together and clustered by CD-hit (Huang et al. 2010), generating a set of non-redundant representative sequences. The representative sequences were aligned with well-defined phylogenetic relatives including both cultured and uncultured reference sequences by ClusterW in MEGA 6 software (Tamura et al. 2013; Thompson et al. 1994). The aligned sequences were used to construct the phylogenetic trees by neighbor-joining method in MEGA 6 with 1000 bootstraps.
Statistical analysis
Chao1 and Shannon indexes of each sample were calculated with a cutoff of 3% by RDP FunGene analyzing platform (Fish et al. 2013). The significance of the differences between two groups was compared by Student’s t test, and multiple group comparisons were made by one-way ANOVA with Tukey’s post hoc test (Version 5.0, GraphPad Software). Data were expressed as mean ± standard deviation (SD). Redundancy analysis (RDA) using CANOCO 4.5 package (Ter Braak and Smilauer 2002) was performed to display the relationship between community compositions and the environmental variables, and the strength of correlation in between was further tested by Pearson product-moment correlation method and was considered significant at p < 0.05 (SPSS, 16.0).
DNA sequence accession numbers
The archaeal amoA-AOA cDNA sequences reported in this study have been deposited in GenBank under accession numbers KX650440–KX650476.
Results
Physicochemical characteristics of the forest soils
The seasonal and spatial changes of pH, organic matter, total N, NO3−-N, and NH4+-N, and exchangeable Al3+ are shown in Table 2, and the C:N ratio of each site was calculated to indicate the rate of nutritional conversion of the detected ecosystems.
Soil pH ranged from 3.66 to 4.52 and was classified as extremely acidic. Similar to the previous results (Wu et al. unpublished), organic matter in summer tended to be more abundant than in winter at each sampling site while not shown statistically significant which may be due to the soil heterogeneity among the triplicates. Clear spatial variations by soil depth were observed in all detected environmental factors. Surface soil was normally found lower in pH while higher in concentrations of organic matter, total N, NO3−-N, and NH4+-N, and exchangeable Al3+ compared to the corresponding lower layer (p < 0.05). Organic matter deposition in the surface layer of mixed forest (MF) of needle and broadleaf trees was significantly higher than the layers of mountain dwarf (MD) forest and evergreen broadleaf (EB) forest (p < 0.05) in both winter and summer. In contrast to forest MD and MF, forest EB was characterized by higher exchangeable Al3+ quantity (p < 0.05). The calculated C:N ratios of MD and EB were lower than 20 but not for MF, implying the higher nutritional turnover rate in MD and EB.
amoA amplification and phylogenetic reconstruction of AOA communities
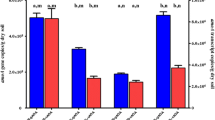
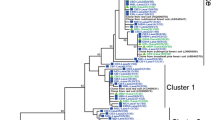
Bacterial amoA transcripts were not detected at all sampling sites in winter and summer, even by nested PCR. Archaeal amoA transcript, on the contrary, can be retrieved and used to recover the phylogeny of the functionally active ammonia oxidizers. All archaeal amoA representative sequences in summer and winter communities were merged in 37 OTUs, and the representative sequences from each OTU were grouped into 8 clusters based on the relative similarities to phylogenetic reference sequences (Fig. 1). Three out of the five AOA lineages have been identified, and the similarity indexes within each lineage were calculated from pairwise P distance by MEGA 6 and are shown in Supplemental Table S1.
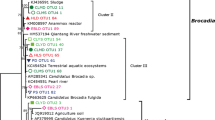
Sequences which showed the same sequence identity with all enriched or isolated references in Nitrososphaera/Group 1.1b/soil AOA were collectively identified as Nitrososphaera sister group, which further branched into Cluster 1 and Cluster 2 and could be retrieved from all sampling sites. Cluster 3 sequences were affiliated to Nitrososphaera/Group 1.1b/soil AOA with identity index of 93–94%, lower than the lower bound of the similarity range of the reference sequences within the same lineage (Supplemental Table S1) and therefore suggested a new cluster detected only in summer. Sequences with high identity indexes to Nitrososphaera gargensis and Nitrososphaera sp. JG1 (95–98%) were grouped into Cluster 4 and recovered mainly from summer. Cluster 5 and Cluster 6 contained the sequences with low identity indexes to N. gargensis (93–95% and 93%, respectively, lower than the lower bound of the similarity range within Nitrososphaera, Supplemental Table S1) and most importantly displayed distinct branching orders from lineages Nitrososphaera and Nitrososphaera sister group, indicating differentiated phylogenetic placements and are classified as new clusters. The majority of sequences in Cluster 5 and Cluster 6 were detected in forests MF and EB. The rest of the sequences were grouped with Nitrosotalea devanaterra and Nitrosotalea Nd2 and divided into two clusters, Cluster 7 and Cluster 8, with identity indexes to N. devanaterra or Nitrosotalea Nd2, 91–92% and 96–98%, respectively. Nitrosotalea-like AOA were widely distributed among all the sampling sites in both winter and summer.
Seasonal and spatial variations of AOA diversity and community structures
The sufficient sequencing depth was verified by rarefaction curves shown in Supplemental Fig. S1 for both winter and summer AOA communities. Same as previous results, diversity (Shannon) and richness (Chao1) indexes of AOA communities in summer were much higher than those in winter (Table 3). The lower layers in winter were more diverse than the surface layers, while a generally reverse trend was observed in summer (Table 3).
All verified AOA sequences in summer and winter were pooled to compare the community structures through principal coordinates analysis (PCoA) using Mothur program (Schloss et al. 2009). Community structure of EBS-W remarkably differed from the other AOA communities (Fig. 2a). To better resolve the remaining AOA communities, EBS-W was excluded and seasonal variations of the community structures were observed as the summer and winter samples were separated at the different sides of P1, the axis with the highest percentage of variation explained, i.e., 42.17%. Therefore, seasonality of the environments indeed impact on shaping AOA structures. Moreover, summer AOA tended to group closer in contrast to winter AOA, demonstrating a similar microbial structure shared among summer AOA communities (Fig. 2b). AOA communities from the same natural forest were generally grouped together, and AOA from MD (highest latitude) were separated from the other two locations on P2 axis (Fig. 2c), suggesting geographical location and/or vegetation types may influence AOA community structures as well. Studies with more sampling sites should be designed to better address this question. Soil stratification also differentiated AOA communities in MF and EB but not in MD (Fig. 2d).
Seasonal and spatial variation of AOA community compositions
Calculating the frequencies of 37 OTUs in each AOA communities (Fig. 3a) and projecting the OTUs back to the phylogenetic tree, the relative abundances of identified AOA lineages and new clusters are shown in Fig. 3b.
Consistent with PCoA results, EBS-W with the largest proportion of the Nitrososphaera sister group demonstrated a distinctive community composition from the others. In addition, summer AOA communities were relatively more alike with each other as they shared more OTUs. In winter, the differences of community compositions between surface and lower layers were more drastic in the MF and EB.
All AOA communities detected in winter and summer, except for EBL-W, were dominated by Nitrososphaera sister group–like AOA, among which OTU3 was the most abundant. The other common OTU was OTU33 (Nitrosotalea-like AOA). Nitrosotalea was the second dominant AOA lineage found in the majority of the communities. OTU2, OTU5–8, OTU11, and OTU13–14 (Nitrososphaera sister group–like AOA); OTU15–18, OTU20, OTU22, and OTU23–25 (Nitrososphaera-like AOA); OTU27 and OTU29–31 (new cluster); and OTU32 and OTU37 (Nitrosotalea-like AOA) were specific to summer AOA communities. Only OTU9 (Nitrososphaera sister group–like AOA), OTU19 (Nitrososphaera-like AOA), and OTU34 (Nitrosotalea-like AOA) were detected in winter AOA. During the transition from summer to winter, the relative abundance of Nitrosotalea was generally decreased while the relative abundance of Nitrososphaera sister group increased in summer samples, except for MFL and EBS. Therefore, the summer-specific OTUs and the seasonal changes of dominant AOA relative abundances imply the functional AOA respond to the seasonality of the environments.
Discussion
The similar seasonal and spatial shifts of environmental parameters in two successive years grant a possibility of cross-comparison between the genomic DNA- and RNA-revealed ammonia oxidizer communities. The availability of substrate for AOA and AOB in forest soils was restricted by both soil acidity and mineralization of organic matter. The decomposition of organic matter was proposed as the main source of microbial available nitrogen supported by the strong positive correlations of organic matter with total N and NH4+ (p < 0.01, Table 4; redundancy analysis, Fig. 4) and also evidenced in other soil studies as high mineralization rate was frequently coupled with high net nitrification rate (Booth et al. 2005; Stopnisek et al. 2010). The extremely acidic soil pH (3.66–4.52) of the three natural forests further limits NH3 concentration owing to the high pKa (NH3 + H+ ↔ NH4+; pKa = 9.25). Therefore, the slowly released nitrogen from organic matter mineralization and the low level of available ammonia in extremely acidic soils would pose great challenges for the growth and the function of ammonia oxidizers.
AOA, however, featured with relatively lower substrate requirement, higher enzyme affinity to ammonia, and flexible metabolic regime, were regarded as a group of abundant ammonia oxidizers in acidic soils (Jung et al. 2011; Martens-Habbena et al. 2009). Our results indeed provide more field study evidences to support that AOA are not only numerically but also functionally dominant over AOB in extremely acidic soils (pH < 4.5). Bacterial amoA transcripts were not able to retrieve from the three natural forests in contrast to the diversified AOA communities detected in both summer and winter.
Moreover, AOA affiliated to N. devanaterra, the first cultured obligate acidophilic ammonia oxidizer, were previously found to be the dominant ammonia oxidizers recovered from the extremely acidic soils of the three natural forests (Lehtovirta-Morley et al. 2011). Different from the genomic DNA–revealed relative abundance, archaeal amoA transcripts belonging to Nitrosotalea were found the second abundant after Nitrososphaera sister group, implying Nitrosotalea-like AOA may not contribute to the nitrification proportionately according to their abundances in extremely acidic forest soils (Wu et al. 2017). The advantages of N. devanaterra, capable of transporting NH4+ through ammonium transporter (Amt) with high affinities and maintaining pH homeostasis by cation regulation and proton scavenging (Lehtovirta-Morley et al. 2016), would support their growth in the extremely acidic niches. The regulations of ion gradients, nevertheless, are energy demanding, and culture studies also indicated that their cellular activities were generally 10 times slower than the other ammonia oxidizers (Jung et al. 2011; Konneke et al. 2005; Lehtovirta-Morley et al. 2014). The re-evaluated relative abundance of Nitrosotalea by archaeal amoA transcripts further endorses the inefficient nitrification of Nitrosotalea at community levels and proposes the possibility of overestimation of Nitrosotalea-mediated nitrification in extremely acidic soils.
On the contrary to Nitrosotalea, the relative abundances of the Nitrososphaera sister group at all sampling sites in both summer and winter were higher than the ones revealed by genomic DNA, implying greater functional importance of the Nitrososphaera sister group in extremely acidic soil nitrification. The Nitrososphaera sister group branching out from Nitrososphaera was first described in 2012 (Pester et al. 2012) and later reported to show active nitrification in acidic red soils (Gubry-Rangin et al. 2015; Wu and Conrad 2014). But still, there are only few reports about the presence of the Nitrososphaera sister group in ecosystems and one recently isolated Nitrososphaera sister group representative was from neutral arable soil (pH 7.5) (Lehtovirta-Morley et al. 2016). Our results may contribute to reveal a wider distribution and functional importance of the sister group especially in extremely acidic forest soils. Among the sampling sites, soils from the surface layers of forest EB in summer were detected to have the most abundant Nitrososphaera sister group–like AOA (46%), making soils from EBS the most promising inoculum for the potential acidophilic Nitrososphaera sister group species enrichment or isolation.
For Nitrososphaera/Group 1.1b/soil AOA, N. gargensis and Nitrososphaera sp. JG1–like AOA were retrieved. It is interesting to notice that Nitrososphaera-like AOA were relatively more abundant (DNA inferred) in winter while they were more functionally active (RNA inferred) in summer, suggesting that Nitrososphaera-like AOA are sensitive to seasonal changes of environmental factors and that summer is preferred for the ones with higher activities in ammonia oxidation even though they are not numerically abundant. Higher organic matter loading and/or temperature in summer might impact on the cellular activities of Nitrososphaera-like AOA, as the genomes of N. gargensis and Nitrososphaera sp. JG1 evidenced their mesophilic lifestyles and moderate thermophiles (Kim et al. 2012; Spang et al. 2012). Therefore, summer environments may select and support the growth of newly evolved functional Nitrososphaera phylotypes in extremely acidic soils. Moreover, genomic DNA detection merely would largely underestimate the functional importance of Nitrososphaera in extremely acidic soils.
Compared to Nitrosotalea, Nitrososphaera lineage and Nitrososphaera sister group were relatively more diverse with more OTUs. Nitrososphaera sister group was carefully annotated as the well-aligned sequence in Cluster 1 and Cluster 2 which showed the same identity index with all the reference sequences (Nitrososphaera viennensis, Nitrososphaera JG1, Nitrososphaera evergladensis, and N. gargensis) in Nitrososphaera/Group 1.1b/soil AOA and therefore should form a new mirrored structure separated from Nitrososphaera. The coupling of Thaumarchaeota diversification with their pH adaptation has been reported (Gubry-Rangin et al. 2015), and Nitrososphaera sister group was found to undergo a lower rate of diversification from its sister group, implying a gradual adaptation process of selecting more specialized phylotypes in specific pH. Furthermore, two new clusters, Cluster 5 and Cluster 6 were found further branching from the clusters of Nitrososphaera and Nitrososphaera sister group. The diversification within and later separating from Nitrososphaera lineages to establish more phylotypes adapting in the extremely acidic forest soils may further guarantee AOA a greater ecological distribution and functional importance in various ecosystems.
Consistent to the seasonal AOA composition shifts detected by genomic DNA, archaeal amoA transcripts inferred that summer AOA differed from the corresponding winter AOA and shared more summer-specific OTUs, such as OTU2, OTU5–8, OTU11, and OTU13–14 (Nitrososphaera sister group–like AOA); OTU15–18, OTU20, OTU22, and OTU23–25 (Nitrososphaera-like AOA); OTU27 and OTU29–31 (new cluster); and OTU32 and OTU37 (Nitrosotalea-like AOA), in contrast to OTU9 (Nitrososphaera sister group–like AOA), OTU19 (Nitrososphaera-like AOA), and OTU34 (Nitrosotalea-like AOA) which were winter-AOA-specific. The drastic transitions of community compositions from summer to winter demonstrate that AOA actively responded to the seasonality of the forest environments by developing new and more adaptive phylotypes within the three main AOA lineages detected. OTU3 (Nitrososphaera sister group–like AOA) and OTU33 (Nitrosotalea-like AOA) were two abundant OTUs prevalently found among all sampling sites in both winter and summer. These may represent the core functional AOA species, further supporting that Nitrososphaera sister group and Nitrosotalea are the two main groups of AOA functionally active in extremely acidic forest soils.
Besides the extremely acidic pH of the natural forest soils, the fluctuating physicochemical parameters would also exert environmental stresses of selection and preference to the functional AOA. In the previous DNA-based study (Wu et al. unpublished), organic matter and exchangeable Al3+ were proposed as the two main environmental factors shaping ammonia oxidizer. Similar to the former observation, the loading of organic matter as the main nitrogen source was generally higher in summer than in winter. Summer AOA communities with higher diversity were more alike as they shared more common OTUs, again suggesting organic matter may impact on shaping AOA community compositions.
Exchangeable Al3+, more easily dissolved from acidic soils (p < 0.05, Table 4), was formerly found to negatively affect the abundance of both AOA and AOB (Wu et al. 2017). The relative abundance of functional Nitrososphaera, however, showed a positive correlation with the quantity of exchangeable Al3+ (p < 0.05, Table 4; redundancy analysis, Fig. 4), suggesting that the functional Nitrososphaera phylotypes detected may be less susceptible to Al3+ toxicity or even evolve ones similar to N. gargensis with heavy metal resistance (Spang et al. 2012). But the quantity of exchangeable Al3+ did not show significant correlations with the relative abundances of the other two dominant AOA lineages. Contradictory to the former observations, the relative abundances of functional AOA were not affected by the quantity of exchangeable Al3+. This may result from two possibilities, the non-selective toxicity of Al3+ to microbial cells and phylotype redundancy (Wu et al. 2017; Yin et al. 2000). In other words, all AOA are vulnerable to the profound Al3+ toxicity in extremely acidic forest soils making the relative abundance of the two dominant to remain the same. Alternatively, the functionally redundant phylotypes within the three AOA lineages were sacrificed under the stress of Al3+ toxicity leaving the main functional phylotype unchanged. More in-depth studies are needed to further explore the physiological effect of Al3+ on AOA abundances and functions.
References
Beman JM, Francis CA (2006) Diversity of ammonia-oxidizing archaea and bacteria in the sediments of a hypernutrified subtropical estuary: Bahia del Tobari, Mexico. Appl Environ Microbiol 72:7767–7777. https://doi.org/10.1128/AEM.00946-06
Beman JM, Roberts KJ, Wegley L, Rohwer F, Francis CA (2007) Distribution and diversity of archaeal ammonia monooxygenase genes associated with corals. Appl Environ Microbiol 73:5642–5647. https://doi.org/10.1128/AEM.00461-07
Booth MS, Stark JM, Rastetter E (2005) Controls on nitrogen cycling in terrestrial ecosystems: a synthetic analysis of literature data. Ecol Monogr 75:139–157. https://doi.org/10.1890/04-0988
De Boer W, Kowalchuk GA (2001) Nitrification in acid soils: micro-organisms and mechanisms. Soil Biol Biochem 33:853–866. https://doi.org/10.1016/S0038-0717(00)00247-9
De Boer W, Gunnewiek PK, Veenhuis M, Bock E, Laanbroek H (1991) Nitrification at low pH by aggregated chemolithotrophic bacteria. Appl Environ Microbiol 57:3600–3604
de la Torre JR, Walker CB, Ingalls AE, Konneke M, Stahl DA (2008) Cultivation of a thermophilic ammonia oxidizing archaeon synthesizing crenarchaeol. Environ Microbiol 10:810–818. https://doi.org/10.1111/j.1462-2920.2007.01506.x
Fish JA, Chai B, Wang Q, Sun Y, Brown CT, Tiedje JM, Cole JR (2013) FunGene: the functional gene pipeline and repository. Front Microbiol 4:291
Francis C, Roberts K, Beman J, Santoro A, Oakley B, (2005) Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc Natl Acad Sci 102 (41):14683-14688
Gubry-Rangin C, Kratsch C, Williams TA, McHardy AC, Embley TM, Prosser JI, Macqueen DJ (2015) Coupling of diversification and pH adaptation during the evolution of terrestrial Thaumarchaeota. Proc Natl Acad Sci U S A 112:9370–9375. https://doi.org/10.1073/pnas.1419329112
Hatzenpichler R, Lebedeva EV, Spieck E, Stoecker K, Richter A, Daims H, Wagner M (2008) A moderately thermophilic ammonia-oxidizing crenarchaeote from a hot spring. Proc Natl Acad Sci U S A 105:2134–2139. https://doi.org/10.1073/pnas.0708857105
Herndl GJ, Reinthaler T, Teira E, van Aken H, Veth C, Pernthaler A, Pernthaler J (2005) Contribution of archaea to total prokaryotic production in the deep Atlantic Ocean. Appl Environ Microbiol 71:2303–2309. https://doi.org/10.1128/AEM.71.5.2303-2309.2005
Horz HP, Barbrook A, Field CB, Bohannan BJM (2004) Ammonia-oxidizing bacteria respond to multifactorial global change. Proc Natl Acad Sci U S A 101:15136–15141. https://doi.org/10.1073/pnas.0406616101
Huang Y, Niu BF, Gao Y, Fu LM, Li WZ (2010) CD-HIT suite: a web server for clustering and comparing biological sequences. Bioinformatics 26:680–682. https://doi.org/10.1093/bioinformatics/btq003
Jung MY, Park SJ, Min D, Kim JS, Rijpstra WI, Damsté JS, Kim GJ, Madsen EL, Rhee SK (2011) Enrichment and characterization of an autotrophic ammonia-oxidizing archaeon of mesophilic crenarchaeal group i.1a from an agricultural soil. Appl Environ Microbiol 77:8635–8647. https://doi.org/10.1128/Aem.05787-11
Karner MB, DeLong EF, Karl DM (2001) Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature 409:507–510. https://doi.org/10.1038/35054051
Kim JG, Jung MY, Park SJ, Rijpstra WI, Sinninghe Damsté JS, Madsen EL, Min D, Kim JS, Kim GJ, Rhee SK (2012) Cultivation of a highly enriched ammonia-oxidizing archaeon of thaumarchaeotal group I. 1b from an agricultural soil. Environ Microbiol 14:1528–1543
Konneke M, Bernhard AE, de la Torre JR, Walker CB, Waterbury JB, Stahl DA (2005) Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437:543–546. https://doi.org/10.1038/nature03911
Lehtovirta-Morley LE, Stoecker K, Vilcinskas A, Prosser JI, Nicol GW (2011) Cultivation of an obligate acidophilic ammonia oxidizer from a nitrifying acid soil. Proc Natl Acad Sci U S A 108:15892–15897. https://doi.org/10.1073/pnas.1107196108
Lehtovirta-Morley LE, Ge CR, Ross J, Yao HY, Nicol GW, Prosser JI (2014) Characterisation of terrestrial acidophilic archaeal ammonia oxidisers and their inhibition and stimulation by organic compounds. FEMS Microbiol Ecol 89:542–552. https://doi.org/10.1111/1574-6941.12353
Lehtovirta-Morley LE, Ross J, Hink L, Weber EB, Gubry-Rangin C, Thion C, Prosser JI, Nicol GW (2016) Isolation of ‘Candidatus Nitrosocosmicus franklandus’, a novel ureolytic soil archaeal ammonia oxidiser with tolerance to high ammonia concentration. FEMS Microbiol Ecol 92:fiw057. https://doi.org/10.1093/femsec/fiw057
Leininger S, Urich T, Schloter M, Schwark L, Qi J, Nicol GW, Prosser JI, Schuster SC, Schleper C (2006) Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442:806
Loscher CR, Kock A, Konneke M, LaRoche J, Bange HW, Schmitz RA (2012) Production of oceanic nitrous oxide by ammonia-oxidizing archaea. Biogeosciences 9:2419–2429. https://doi.org/10.5194/bg-9-2419-2012
Lu R (2000) Agricultural chemical analysis methods of soil. China Agriculture Science and Technology Press, Beijing, pp 107–108
Martens-Habbena W, Berube PM, Urakawa H, de la Torre JR, Stahl DA (2009) Ammonia oxidation kinetics determine niche separation of nitrifying archaea and bacteria. Nature 461:976–979. https://doi.org/10.1038/nature08465
Nelson DW, Sommers LE (1996) Total carbon, organic carbon, and organic matter. Methods of soil analysis: part 3 - chemical methods: 961–1010
NNNR (2015, 2016) Nanling National Nation Reserve. http://www.gdnl.org. Accessed 12th Aug 2015 and 15th Jan 2016
Park H-D, Wells GF, Bae H, Criddle CS, Francis CA (2006) Occurrence of ammonia-oxidizing archaea in wastewater treatment plant bioreactors. Appl Environ Microbiol 72:5643–5647
Pester MRT, Flechl S, Gröngröft A, Richter A, Overmann J, Reinhold-Hurek B, Loy A, Wagner M (2012) amoA-based consensus phylogeny of ammonia-oxidizing archaea and deep sequencing of amoA genes from soils of four different geographic regions. Environ Microbiol 14:525–539. https://doi.org/10.1111/j.1462-2920.2011.02666.x
Rotthauwe J-H, Witzel K-P, Liesack W (1997) The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl Environ Microb 63(12):4704–4712
Santoro AE, Buchwald C, McIlvin MR, Casciotti KL (2011) Isotopic signature of N2O produced by marine ammonia-oxidizing archaea. Science 333:1282–1285. https://doi.org/10.1126/science.1208239
Schleper C, Jurgens G, Jonuscheit M (2005) Genomic studies of uncultivated archaea. Nat Rev Microbiol 3:479–488. https://doi.org/10.1038/nrmicro1159
Schloss PDWS, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. https://doi.org/10.1128/Aem.01541-09
Spang A, Poehlein A, Offre P, Zumbrägel S, Haider S, Rychlik N, Nowka B, Schmeisser C, Lebedeva EV, Rattei T, Böhm C (2012) The genome of the ammonia-oxidizing Candidatus Nitrososphaera gargensis: insights into metabolic versatility and environmental adaptations. Environ Microbiol 14:3122–3145
Steger D, Ettinger-Epstein P, Whalan S, Hentschel U, de Nys R, Wagner M, Taylor MW (2008) Diversity and mode of transmission of ammonia-oxidizing archaea in marine sponges. Environ Microbiol 10:1087–1094. https://doi.org/10.1111/j.1462-2920.2007.01515.x
Stieglmeier M, Mooshammer M, Kitzler B, Wanek W, Zechmeister-Boltenstern S, Richter A, Schleper C (2014) Aerobic nitrous oxide production through N-nitrosating hybrid formation in ammonia-oxidizing archaea. ISME J 8:1135–1146. https://doi.org/10.1038/ismej.2013.220
Stopnisek N, Gubry-Rangin C, Hofferle S, Nicol GW, Mandic-Mulec I, Prosser JI (2010) Thaumarchaeal ammonia oxidation in an acidic forest peat soil is not influenced by ammonium amendment. Appl Environ Microbiol 76:7626–7634. https://doi.org/10.1128/AEM.00595-10
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. https://doi.org/10.1093/molbev/mst197
Ter Braak CJ, Smilauer P (2002) CANOCO reference manual and CanoDraw for windows user’s guide: software for canonical community ordination (version 4.5). http://www.canoco.com. Accessed 1st Feb 2016
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Tourna M, Stieglmeier M, Spang A, Könneke M, Schintlmeister A, Urich T, Engel M, Schloter M, Wagner M, Richter A, Schleper C (2011) Nitrososphaera viennensis, an ammonia oxidizing archaeon from soil. Proc Natl Acad Sci U S A 108:8420–8425. https://doi.org/10.1073/pnas.1013488108
Van Hoek A, Van Alen T, Sprakel V, Hackstein J, Vogels G, (1998) Evolution of anaerobic ciliates from the gastrointestinal tract: phylogenetic analysis of the ribosomal repeat from Nyctotherus ovalis and its relatives. Mol Biol Evol 15(9):1195-1206
Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu D, Paulsen I, Nelson KE, Nelson W, Fouts DE (2004) Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66–74. https://doi.org/10.1126/science.1093857
Wang YF, Gu J-D (2013) Higher diversity of ammonia/ammonium-oxidizing prokaryotes in constructed freshwater wetland than natural coastal marine wetland. Appl Microbiol Biotechnol 97:7015–7033. https://doi.org/10.1007/s00253-012-4430-4
Weidler GW, Dornmayr-Pfaffenhuemer M, Gerbl FW, Heinen W, Stan-Lotter H (2007) Communities of archaea and bacteria in a subsurface radioactive thermal spring in the Austrian Central Alps, and evidence of ammonia-oxidizing Crenarchaeota. Appl Environ Microbiol 73:259–270. https://doi.org/10.1128/AEM.01570-06
Wu Y, Conrad R (2014) Ammonia oxidation-dependent growth of group I.1b Thaumarchaeota in acidic red soil microcosms. FEMS Microbiol Ecol 89:127–134. https://doi.org/10.1111/1574-6941.12340
Wu R-N, Meng H, Wang Y-F, Lan W, Gu J-D (2017) A more comprehensive community of ammonia-oxidizing archaea (AOA) revealed by genomic DNA and RNA analyses of amoA gene in subtropical acidic forest soils. Microb Ecol 74(4):910–922. https://doi.org/10.1007/s00248-017-1045-4
Wuchter C, Abbas B, Coolen MJ, Herfort L, van Bleijswijk J, Timmers P, Strous M, Teira E, Herndl GJ, Middelburg JJ, Schouten S (2006) Archaeal nitrification in the ocean. Proc Natl Acad Sci U S A 103:12317–12322. https://doi.org/10.1073/pnas.0600756103
Yin B, Crowley D, Sparovek G, De Melo WJ, Borneman J (2000) Bacterial functional redundancy along a soil reclamation gradient. Appl Environ Microbiol 66:4361–4365. https://doi.org/10.1128/Aem.66.10.4361-4365.2000
Acknowledgments
We would like to thank the comments and revision of Miss Jennifer Li, and the reviewers and editor for their assistance in the publication process.
Funding
This research was supported by a PhD graduate studentship (RW) from The University of Hong Kong Graduate School and a research grant (Yong-Feng Wang) from National Natural Science Foundation of China (31470562).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(PDF 437 kb)
Rights and permissions
About this article
Cite this article
Wu, RN., Meng, H., Wang, YF. et al. Functional dominance and community compositions of ammonia-oxidizing archaea in extremely acidic soils of natural forests. Appl Microbiol Biotechnol 103, 4229–4240 (2019). https://doi.org/10.1007/s00253-019-09721-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-019-09721-2