Abstract
Schizosaccharomyces pombe carboxypeptidase Y (CPY) is synthesized as a zymogen and transported into the vacuole where maturation and activation occurs. The 110-kDa S. pombe CPY precursor is processed twice and finally converted to a mature form consisting of polypeptides of approximately 19 and 32 kDa linked by a single disulfide bond. In Saccharomyces cerevisiae, maturation of CPY occurs mostly through the activity of vacuolar aspartyl protease Pep4p, whereas a Pep4p homolog has not been found in the S. pombe genome database. Based on analysis of protease-deficient mutants, we found that S. pombe CPY was not able to be processed or activated in isp6Δpsp3Δ double disruptants. Both Isp6p and Psp3p are subtilase-type serine proteases with related sequences. Moreover, alkaline phosphatase of S. pombe was found to be localized at the vacuolar membrane and was also unprocessed in isp6Δpsp3Δ double disruptants. Vacuolar localization of GFP-fused Isp6p and Psp3p was determined by fluorescence microscopy. These results suggest that the two serine proteases Isp6p and Psp3p are functional in the vacuole and are involved in proteolytic processing of vacuolar proteins.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The vacuole of the yeast Saccharomyces cerevisiae has been shown to contain a multitude of hydrolytic enzymes and consequently has been proposed to be the lysosome of yeast cells (Matile and Wiemken 1967; Martinoia et al. 1979). The biosynthesis and function of a variety of vacuolar proteinases has been studied (Mechler et al. 1982; Jones and Cavanagh 1984; Achstetter and Wolf 1985). S. cerevisiae carboxypeptidase Y (CPY) is one of the best characterized vacuolar proteins, and its biosynthesis and transport into the vacuole has been studied in detail (Klionsky and Emr 1990). S. cerevisiae CPY is synthesized as a high molecular weight precursor which is translocated into the ER, where it is core-glycosylated to generate the so-called p1 precursor form. It next traverses the Golgi complex, where its oligosaccharides are elongated to generate precursor p2CPY. In the Golgi apparatus, a S. cerevisiae CPY-specific sorting signal is recognized which leads to formation of a receptor–ligand complex (Klionsky and Emr 1990). The receptor has been identified as being encoded by the VPS10 gene. VPS10 encodes a transmembrane sorting receptor that is responsible for the recognition and targeting of CPY to the vacuole (Marcusson et al. 1994). Precursor CPY can bind to Vps10p in the late-Golgi compartment (Marcusson et al. 1994; Cooper and Stevens 1996). Receptor–ligand complexes are delivered to an intermediate endosomal compartment, where CPY dissociates from Vps10p. Vps10p cycles back to the Golgi for additional rounds of sorting, while CPY continues on toward the vacuole (Cooper and Stevens 1996; Seaman et al. 1997, 1998). The propeptide of the inactive pro-CPY molecule is proteolytically cleaved in the vacuole, mainly by the vacuolar aspartyl protease Pep4p (Stevens et al. 1982).
The fission yeast, Schizosaccharomyces pombe, taxonomically and evolutionarily distant from the budding yeast (Russell and Nurse 1986), is genetically and physiologically well characterized. We found that S. pombe has many proteins homologous to the Vps proteins of S. cerevisiae (Takegawa et al. 2003b). To analyze vacuolar protein transport pathways in S. pombe, we isolated a CPY homolog (cpy1+) from S. pombe (Tabuchi et al. 1997). Using a S. pombe Cpy1-specific antibody, we analyzed the contribution of specific fission yeast VPS homologs to vacuolar protein transport: phosphatidylinositol 3-kinase (Takegawa et al. 1995; Onishi et al. 2003), sorting nexin homolog (Koga et al. 2004), homotypic vesicular protein sorting component (Iwaki et al. 2003; Koga et al. 2004), class E Vps proteins (Iwaki et al. 2003, 2007), dynamin-related Vps1p (Iwaki et al. 2007; Rothlisberger et al. 2009), soluble NSF attachment protein receptor proteins (Takegawa et al. 2003a; Rothlisberger et al. 2009), V-ATPase complex (Iwaki et al. 2004), and CPY receptor (Takegawa et al. 2003a; Iwaki et al. 2006). Through these analyses, we confirmed that the basic vacuolar protein transport machinery is conserved between the two yeast species.
We previously reported that S. pombe CPY is initially synthesized as a 110-kDa pro-precursor that is processed twice in the vacuole leading to loss of a pro-sequence and subsequent conversion of the 50-kDa single-polypeptide-chain intermediate form to the mature disulfide bound heterodimer consisting of polypeptides of about 19 and 32 kDa (Tabuchi et al. 1997). We examined the processing behavior of S. pombe CPY in S. cerevisiae (Takegawa et al. 2003c). The 32-kDa mature form was found in wild-type cells, while a pep4∆ mutant accumulated exclusively the 110-kDa precursor form of S. pombe CPY. This result suggested that maturation of S. pombe CPY in S. pombe requires an aspartyl protease homologous to ScPep4p. Unexpectedly, analysis of the S. pombe genome database did not indicate the presence of a ScPep4p homolog. In the present study, we identified the protease(s) responsible for maturation of S. pombe CPY in S. pombe.
Materials and methods
Strains, media, and growth conditions
The S. pombe strains used in this study were ARC039 (h − leu1-32 ura4-C190T) and A8 (h − ∆psp3, ∆isp6, ∆ppp53 (SPAP14E8.04; putative zinc metallopeptidase)), ∆ppp16 (SPBC1711.12 putative dipeptidyl peptidase), ∆ppp22 (SPBC14C8.03; putative methionine metallopeptidase), ∆sxa2, ∆ppp80 (SPAC19B12.08; putative peptidase), and ∆ppp20 (SPAC4F10.02; putative aspartyl aminopeptidase; leu1-32 ura4-C190T) lacking eight proteases (Idiris et al. 2009). Strains were grown in the following media: yeast extract medium with supplements (YES; 0.5% Bacto-yeast extract (BD), 3% glucose and SP supplements (Qbiogene, Montreal, QC, Canada)) or minimal medium (MM) which contains (per liter) 3 g potassium hydrogen phthalate, 2.2 g Na2HPO4, 5 g NH4Cl, 20 g glucose, 1.05 g MgCl2 6 H20, 11 mg CaCl2, 1 g KCl, 40 mg Na2SO4, 1 mg pantothenic acid, 10 mg nicotinic acid, 10 mg inositol, 10 μg biotin, 0.5 mg boric acid, 0.4 mg MnSO4, 0.4 mg ZnSO4 7H2O, 0.2 mg FeCl2 6H2O, 0.04 mg H2MoO4 H2O, 0.1 mg KI, 0.04 mg CuSO4 5H2O, 1 mg citric acid, and 75 mg each of adenine, histidine, and lysine. A total of 37.5 mg of uracil or 75 mg of leucine was added per liter to cover auxotrophies as needed. MM medium was also used as a thiamine-deficient medium as appropriate. S. pombe cells were transformed by the lithium acetate method or by electroporation as described (Okazaki et al. 1990; Suga and Hatakeyama 2001; Morita and Takegawa 2004). Standard genetic methods have been described (Alfa et al. 1993). Transformants were plated onto agar-based minimum media (MMA), MMA Ura− (MMA without uracil), or MMA Leu− (MMA without leucine) and grown at 30°C. Escherichia coli strain XL-1 Blue (Stratagene) was used as a host for plasmid preparations.
Preparation of S. pombe Isp6p-specific antiserum
A fusion between the glutathione-S-transferase (GST) and isp6+ genes was constructed by subcloning an 876-bp PCR fragment encoding amino acids 529 to 1404 of S. pombe isp6+ into BamHI digested pGEX5X-1, generating an in-frame fusion gene. Induction of the GST-isp6 fusion protein was accomplished as previously described (Kamada et al. 2000). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) revealed a prominent 60-kDa band representing the fusion protein which was excised and electroeluted from gel slices with an ATTO AE-3590 electrochamber. Approximately 1 mg of the fusion protein was emulsified with Freund’s complete adjuvant and injected intramuscularly and subcutaneously into a young male New Zealand White rabbit. Antiserum was collected and screened by immunoprecipitation.
Pulse-chase and immunoblot analysis of S. pombe CPY and Isp6p
For analyses of CPY and Isp6p processing, cells were pulse-labeled with Expres35S Protein Labelling Mix (NEN) for 15 min at 30°C and chased at the same temperature for given periods, as described (Tabuchi et al. 1997). Immunoprecipitation of CPY was performed using rabbit polyclonal antibody against S. pombe CPY as described previously (Tabuchi et al. 1997). Immunodetection analysis was performed on the blotted membrane with SNAP i.d. (Millipore). GFP-fused proteins were detected with mouse polyclonal anti-GFP antibody (Molecular Probes Inc., Eugene, OR, USA) and horseradish peroxidase-conjugated anti-rabbit IgG serum (Amersham Biosciences, Little Chalfont, UK). Western blot analysis of CPY was performed with rabbit antiserum against the trpE-cpy1 fusion protein (Tabuchi et al. 1997). Western blot analysis of Isp6p was performed with rabbit antiserum against the GST-isp6 fusion protein. Proteins were then detected with horseradish peroxidase-conjugated antibody against rabbit immunoglobulin G (Millipore). Signals were visualized by enhanced chemiluminescence (ECL-plus, GE Healthcare, USA) and were detected with an LAS4000 imaging system (Fuji Film Co. Ltd. Japan).
Vacuole staining, fluorescence microscopy, and enzyme assays
To visualize the fission yeast vacuole, cells were labeled with the lipophilic dye FM4-64 as described (Vida and Emr 1995). Briefly, 1 ml of exponentially growing cells in YES medium was harvested by centrifugation and suspended in 0.5 ml of YES medium containing 16 nM FM4-64 and was incubated at 30°C for 30 min with shaking for pulse labeling. The labeled cells were then washed once with fresh medium and resuspended in 1 ml of YES medium without dye and were incubated at 30°C for 90 min and then examined by fluorescence microscopy. Carboxypeptidase activity was measured in cell extracts using carboxybenzyloxy-Phe-Leu as substrate as described (Stevens et al. 1986).
Construction of expression vectors and gene disruptions
S. pombe isp6+, psp3+, and SPBC14F5.13c with 1,028, 1,011, and 500 bp, respectively, of upstream region were amplified by PCR using Primestar PCR polymerase (TaKaRa Co Ltd., Japan) and the following primers: 5′-GTTTTGTCGACCCTAAGCAGCGCAATGCGC-3′ and 5′-GTTTTGGATCCTCTTGAGCACCATTGAAAG-3′ (isp6+), or 5′-GTTTTGTCGACAGCTCCTGCAGGTAGCAAG-3′ and 5′-GTTTTGGATCCTCATAATTGTTAAGGC-3′ (psp3+), or 5′-GTTTTGTCGACTGTACTTTATTTTCAAGTAACAGTTC-3′ and 5′-GTTTTGGATCCAAACAAATCAGCACCGAAACCTACAA-3′ (SPBC14F5.13c). Each PCR product was then digested with SalI and BamHI, and the PCR-amplified EGFP sequence containing BamHI and NotI sites was subcloned in parallel into SalI and NotI sites of the yeast shuttle vector pAL-KS to construct 3′-terminal GFP fusion genes. Isp6 was disrupted by insertion of ura4+ into the isp6+ ORF. psp3 and isp6 psp3 double disruptants were constructed as described (Idiris et al. 2006a, 2009). cpy1+ was disrupted as described (Tabuchi et al. 1997). Isp6+ with 2,070 bp of upstream region was amplified by PCR from ARC039 genomic DNA using primers 5′-GTTTTGCGGCCGCAATTGCTGGGTGTACGGCGCCA-3′ and 5′-GTTTTGCGGCCGCGCGGCGGATAGAGAAGTATCAA-3′. The PCR product was digested with NotI and subcloned into identical site in the pAL-KS vector to generate pAL-isp6. pAL-isp6-D221S/H253S/S409A was constructed as described (Kunkel et al. 1991), using pAL-isp6 as a template and primers 5′-TTATGTTGTGTCGACCGGTCTAAGC-3′, 5′-ATAACAATGGATCCGGTACGCATGT-3′, and 5′-TCTCTGGTACCGCTATGGCAACCCC-3′.
RNA preparation and Northern blotting
RNA was prepared using a Qiagen RNeasy kit according to the manufacturer’s instructions for isolating total RNA from yeast. RNA was run on a formaldehyde gel, followed by blotting to a Pall Biodyne Transfer Membrane (Pall Corp., USA). Probes were PCR-amplified sequences of ∼500 bp consisting of the 5′ terminus of isp6+ and psp3+. Pre-hybridization and hybridization were performed as described (Cooper et al. 1997) with an AlkPhos Direct kit module (GE Healthcare, USA). Transcripts were visualized with a CDP-Star detection reagent (GE Healthcare, USA) and detected using an LAS4000 Imaging System (Fuji Film Co. Ltd., Japan).
Results
Protease disruptant mutants exhibit an S. pombe CPY maturation defect
We previously described an S. pombe mutant, designated A8, missing eight different protease-encoding genes, which was constructed for improved production of protease-sensitive heterologous proteins (Idiris et al. 2006a, b). No carboxypeptidase activity was detected in A8 even thought an intact cpy1+ gene was still present (Fig. 2a). We then assayed carboxypeptidase activity in single and multiple protease-deficient mutants (Idiris et al. 2006b). Interestingly, carboxypeptidase activity was absent in a double mutant carrying disrupted psp3+ (the third S. pombe serine protease, SPAC1006.01) and disrupted isp6+ (induced during sporogenesis in S. pombe, SPAC4A8.04) but was present in strains in which either psp3+ alone or isp6+ alone had been disrupted (Fig. 2a). These data suggest that both psp3+ and isp6+ gene products are necessary for S. pombe CPY function.
Isp6p has been reported to be a nitrogen starvation-specific vacuolar protease with broad substrate specificity that is transported through the cell secretory pathway to either the vacuole or extracellular medium (Sato et al. 1994). The molecular structure of Psp3p is very similar to Isp6p (43% identity and 64% similarity). Psp3p was originally identified with three other proteases, Isp6p, Pgp1p, and Yps1p, in a study of Krp1p-related proteases in S. pombe. Krp1p is the only kexin identified in S. pombe and is able to process the P-factor precursor into individual subunits. Individual overexpression of Psp3p, Isp6p, Pgp1p, or Yps1p has been shown to complement loss of Krp1p, which is an essential dibasic endopeptidase (Davey et al. 1994; Ladds and Davey 2000). Neither Psp3p nor Isp6p appear to be kexins but rather seem to be related to a group of general serine proteases of S. cerevisiae, e.g., Prb1p, a vacuolar endoprotease and subtilase-type serine peptidase with broad substrate specificity (Moehle et al. 1987a, b, 1989; Fig. 1). Both Psp3p and Isp6p show high homology with Prb1p (Psp3p—46% identity and 73% similarity, Isp6p—47% identity and 76% similarity).
Alignment of serine proteases, S. cerevisiae Prb1p, S. pombe Isp6p, and Psp3p. Sequences were aligned using the ClustalW program (version 1.83, gap opening penalty, 10; gap extension penalty, 0.2). The catalytic residues (D H N S) are lettered above the sequences. A dotted line represents peptidase inhibitor I9 domain. The arrowhead above the sequences indicates the region of mature Prb1p (from E281 to the region just upstream of N594)
We previously reported that the S. pombe cpy1+ gene product S. pombe CPY was detected as a 110-kDa protein by pulse-chase analysis. This 110-kDa form was transported to the endosome or vacuole, and its propeptide was proteolytically cleaved to give an intermediate 50-kDa form. The intermediate form was processed to the mature disulfide-linked heterodimer consisting of 32 and 19 kDa polypeptides (Tabuchi et al. 1997). Next, we performed pulse-chase analysis of S. pombe CPY in isp6∆, psp3∆, and isp6∆psp3∆ double disruptants. After 30 min of pulse labeling with 100 μCi of Expres35S-label, almost all of the 110-kDa pre-S. pombe CPY were converted to mature form of S. pombe CPY (32 kDa) in wild-type, isp6∆, and psp3∆ strains. In contrast, in the isp6∆ psp3∆ double disruptant, the only S. pombe CPY species detected was the pre-mature 110-kDa form (Fig. 2b). Moreover, the isp6∆ strain accumulated a 50-kDa S. pombe CPY form which may represent a single-polypeptide intermediate missing the pro-segment. These data suggest that both Psp3p and Isp6p are necessary for maturation of S. pombe CPY and that the contribution of Isp6p may be greater than that of Psp3p.
Maturation and activation of S. pombe CPY. a Relative activity of S. pombe CPY in wild-type (WT), isp6∆, psp3∆, isp6∆ psp3∆, and A8 strains. Cells were grown to mid-log phase in MM medium, harvested, and used as a source of cell-free extracts prepared by the glass bead method. Carboxypeptidase activity was measured in cell extracts using carboxybenzyloxy-Phe-Leu as substrate as described (Stevens et al. 1986). Carboxypeptidase activity of WT cell (2.8 mU/OD) was defined as 100%. b Processing of S. pombe CPY in vivo. WT (ARC039), isp6∆, psp3∆, and isp6∆ psp3∆ cells were pulse-labeled with Express-35S-label for 10 min at 30°C and chased for 30 min. Crude extracts of the cells and immunoprecipitates were prepared as described in “Materials and methods”. The upper arrow indicates pre-S. pombe CPY (110 kDa), the arrow in middle indicates intermediate missing the pro-segment of S. pombe CPY (50 kDa), and the lower arrow indicates mature S. pombe CPY (32 kDa). We used an antibody α-CPY that only reacted with the 32-kDa portion of the S. pombe CPY heterodimer (Tabuchi et al. 1997). The immunoprecipitate was separated on an SDS–10% polyacrylamide gel. The autoradiogram of the fixed dried gels is shown. M.W. molecular weight
Northern blot analysis of psp3+ and isp6+ and localization of GFP-fused Isp6p and Psp3p
Sato et al. reported that Isp6p was specifically induced during sexual differentiation. In the present study, we showed that Isp6p is necessary for maturation of S. pombe CPY and that it was active during vegetative growth. We then observed expression of isp6+ and psp3+ both during vegetative growth on minimal medium and under conditions of nitrogen starvation. Sato et al. identified genes expressed during sporulation and named them isp (genes induced during sporogenesis in S. pombe; Sato et al. 1994). They reported that isp6+ was strongly induced during nitrogen starvation. However, our Northern blot analysis revealed that both isp6+ and psp3+ were expressed during vegetative growth and that an increase in expression was not apparent following nitrogen starvation, whereas the expression level of psp3+ was much lower than that of isp6+ (Fig. 3a). We then analyzed expression of isp6+ in MM, YES, and YPD media (Fig. 3b). Northern blot analysis revealed that transcription of isp6+ was induced in all the media and was strongly induced in MM medium. A survey of the S. pombe transcriptome revealed that isp6+ mRNA levels were relatively high in minimum medium. In contrast, in a relatively rich YES medium, the abundance of isp6+ mRNA was much lower than in minimal medium (Wilhelm et al. 2008).
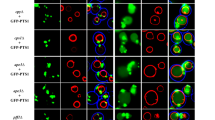
a Northern blot analysis of isp6+ and psp3+ gene expression. Cells were grown to mid-log phase in MM medium at 30°C and then shifted to MM-N medium. Vegetatively grown cells and nitrogen-starved cells were harvested, and total RNA was extracted as described in “Materials and methods”. +N indicates RNA from the vegetatively grown cells, −N indicates RNA from the nitrogen-starved cells. isp6+ and psp3+ mRNAs are shown in the upper panel, and ribosomal RNA (loading control) is shown in the lower panel. b Northern blot analysis of transcription levels of isp6+. Cells were grown to an OD = 2.0 in MM medium, YES or YPD at 30°C. isp6+ mRNA is shown in the upper panel, and ribosomal RNA (loading control) is shown in the lower panel. c Localization of Isp6p-GFP and Psp3p-GFP expressed in WT (ARC039) cells. Cells were grown to mid-log phase in MM medium. Staining of the vacuole with FM4-64 and observation by fluorescence microscopy was performed as described in the “Materials and methods”
In S. cerevisiae, Pep4p-dependent proteolytic processing and maturation of S. cerevisiae CPY occurs in the vacuole. To determine if processing of S. pombe CPY also occurred in the vacuole, we constructed C-terminal GFP-fused Isp6p and C-terminal GFP-fused Psp3p. Both GFP fusion proteins were found to localize in the vacuolar lumen (Fig. 3c), suggesting that both native Isp6p and Psp3p are also transported to the vacuole in S. pombe.
Propeptide of Isp6p precursor is processed autocatalytically
In S. cerevisiae, Prb1p is synthesized as a 69-kDa precursor protein containing large propeptides. This precursor is autocatalytically processed resulting in cleavage of the N-terminal propeptide in the ER to yield the smaller precursor pro-Prb1p, whose activity is subsequently inhibited by the non-covalently linked large propeptide. Upon transport of this complex to the vacuole, the large propeptide is degraded and the C-terminal pro-Prb1p propeptide is cleaved to yield the mature 31–32-kDa Prb1p (31–32 kDa; Nebes and Jones 1991; Hirsch et al. 1992). Moreover, a protein family motif search using the Pfam database revealed that Isp6p contains an N-terminal peptidase inhibitor I9 domain which is also found in the propeptide sequence of pro-Prb1p. These data suggest that Isp6p may also possess a propeptide sequence. We undertook an analysis to determine whether the maturation of Isp6p occurred by an analogous mechanism. Cells were pulse-labeled with the Expres35S-labeling mix for 15 min at 30°C and chased at the same temperature for given periods. Autoradiography of SDS-PAGE gels loaded with samples immunoprecipitated with anti-Isp6p serum detected two bands represented lager molecular weight and smaller molecular weight (Fig. 4b). Western blot analysis of a cell-free extract from wild-type cells revealed that the approximately 30-kDa protein was identical to mature Isp6p, but about 67 kDa and 30 kDa of band were detected in Western blot analysis of cell free extract of A8 cells (data not shown), and then smaller band detected in pulse-chase analysis may represent mature Isp6p. After a 30-min chase, the amount of the 67-kDa protein decreased while the amount of the 30 kDa increased correspondingly, consistent with the 67-kDa protein being a precursor of Isp6p. These results indicate that Isp6p was processed post-transcriptionally. Next, we determined whether Isp6p was autocatalytically processed like Prb1p. An amino acid alignment with Prb1p indicated that D221, H253, N344, and S409 in the isp6+ open reading frame were catalytic residues in the active site (Moehle et al. 1987b). Using site-directed mutagenesis, the codons for three of the catalytic residues D221, H253, and S409 were changed to S221, S253, and A409, respectively, to produce Isp6p-D221S/H253S/S409A. This mutant allele under the control of the native isp6+ promoter was subcloned into the S. pombe multi-copy vector, pAL-KS+, which was introduced into an isp6∆ mutant by transformation. Western blot analysis of a cell-free extract from the isp6∆ strain carrying the Isp6p-D221S/H253S/S409A protein only detected the 67-kDa Isp6p-reactive species (data not shown). After pulse labeling of cells harboring the Isp6p-D221S/H253S/S409A protein with 35S methionine for 30 min and subsequent immunoprecipitation by anti-Isp6p serum, only the 67-kDa band corresponding to pro-Isp6p was detected (Fig. 4c). We interpret this result as indicating that pro-Isp6p is processed autocatalytically. Moreover, because the isp6∆ mutant still possessed functional Psp3p, which is a serine protease highly homologous to Isp6p, we conclude that Psp3p is not involved in processing of Isp6p.
Processing of Isp6p in vivo. a Localization of S. pombe CPY-GFP expressed in WT (ARC039). b Pulse-chase analysis of Isp6p. WT (ARC039), isp6∆, and vps34∆ cells were pulse-labeled with Express-35S-label for 15 min at 30°C and chased for 30 min. Crude cell extracts and α-Isp6p immunoprecipitates were prepared as described in “Materials and methods”. The upper arrow indicates pro-Isp6p while the lower arrow indicates mature-Isp6p. The asterisk indicates the deduced Isp6p moiety, as the isp6∆ strain harbored a partial deletion of just the C-terminal peptidase domain essential for peptidase activity. c Pulse-chase analysis of Isp6p-D221S/H253S/S409A. A pulse-chase experiment was performed as described above. Lanes 1 and 2 indicate immunoprecipitate from an isp6∆ strain expressing Isp6p-D221S/H253S/S409A; lane 3 indicates immunoprecipitate from wild-type cells
We next undertook analysis of pro-Isp6p processing in a vps34∆ mutant. S. pombe Vps34p encodes a phosphatidylinositol (PtdIns) 3-kinase, and the vps34∆ strain exhibits a defect in vacuolar protein transport (Takegawa et al. 1995; Tabuchi et al. 1997). Therefore, the Vps34p/PtdIns 3-kinase facilitates anterograde protein transport from the Golgi to the vacuole through the regulated synthesis of PtdIns(3)P. Pulse-labeled pro-Isp6p and the processed form of Isp6p were detected in this mutant (Fig. 4b). These results suggest that pro-Isp6p was rapidly autocatalytically processed before transport to the vacuole.
Both Isp6p and Psp3p are also responsible for maturation of vacuolar alkaline phosphatase
The S. cerevisiae PHO8 gene product, repressible alkaline phosphatase (ALP), is a vacuolar glycoprotein. ALP is synthesized as an inactive precursor containing a C-terminal propeptide, and this C-terminal propeptide is also cleaved from the protein in a Pep4p-dependent manner (Klionsky and Emr 1989). ALP has the topology of a type-II integral membrane protein, and the precursor and mature protein are anchored in the vacuole membrane by an N-terminal hydrophobic domain. Analysis of the S. pombe genome database revealed that SPBC14F5.13c is a homolog of S. cerevisiae PHO8. SPBC14F5.13c (S. pombe pho8 +) encodes a 59-kDa protein with an alkaline phosphatase active site. S. pombe ALP shares 45.9% identity and 71.7% similarity with the S. cerevisiae PHO8 gene product and possesses a putative C-terminal propeptide sequence. We sought to determine whether Isp6p and Psp3p were also responsible for maturation of S. pombe ALP. We constructed a C-terminal GFP-fused S. pombe Pho8 expressed under the control of an nmt41 promoter which is induced by thiamine depletion. We then performed Western blot analysis of S. pombe ALP-GFP using an anti-GFP antibody in isp6∆, psp3∆, and isp6∆ psp3∆ double disruptants. In wild-type cells, isp6∆ and psp3∆, the GFP moiety of S. pombe Pho8-GFP was mostly observed, while in the isp6∆ psp3∆ double disruptant, about 90-kDa full-length S. pombe ALP-GFP band was the major species detected (Fig. 5a). Moreover, it is interesting that disruption of isp6+ did not influence maturation of S. pombe ALP, whereas disruption of psp3+ did. That is, disruption of psp3+ decreased the amount of mature S. pombe ALP and led to retention of a small amount of pro-S. pombe ALP (Fig. 5a).These data suggest that both Isp6p and Psp3p are essential for cleaving the C-terminal S. pombe ALP propeptide. We next determined the localization of S. pombe ALP-GFP. In both isp6∆ and psp3∆ cells, GFP fluorescence was observed in the lumen of the vacuole which corresponds to the localization observed in wild-type cells (Fig. 5b). Western blot analysis indicated that the fluorescence was derived from the cleaved GFP moiety of S. pombe ALP-GFP presumably due to processing of S. pombe ALP in the vacuolar lumen. In contrast, GFP fluorescence was observed in the vacuolar membrane in the isp6∆ psp3∆ double disruptant. Western blot analysis indicated the presence of unprocessed S. pombe Pho8-GFP in the same strain, suggesting that S. pombe ALP was also integrated in the vacuolar membrane as for S. cerevisiae ALP. In conclusion, both Isp6p and Psp3p are also necessary for processing of S. pombe ALP in the vacuole.
Cleavage and localization of S. pombe ALP-GFP. a Western blot analysis of S. pombe ALP-GFP. Wild-type (WT; lane 1), isp6∆ (lane 2), psp3∆ (lane 3), and isp6∆psp3∆ (lane 4) strains were grown to mid-log phase in MM medium. Full-length S. pombe ALP-GFP and the GFP moiety were detected by immunoblotting using anti-GFP antibody. b Localization of S. pombe ALP-GFP expressed in WT (ARC039), isp6∆, psp3∆, and isp6∆ psp3∆ cells. Cells were grown to mid-log phase in MM medium, and then staining of the vacuole with FM4-64 and observation by fluorescence microscopy was performed as described in the “Materials and methods”
Discussion
Several vacuolar enzymes in yeast are synthesized as inactive precursors. Most of these proteins are delivered to the vacuole via the early compartments of the secretory pathway and the endosome. After transit to the vacuole, these zymogens are activated by removal of propeptides. In S. pombe, we previously reported that carboxypeptidase Y is synthesized as a 110-kDa precursor zymogen, which is then transported to the vacuole via the secretory pathway involving several Vps proteins (Takegawa et al. 2003a, c; Iwaki et al. 2006). After cleavage of the propeptide sequence and signal processing in the mature region, precursor S. pombe CPY is converted to the mature form and activated in the vacuole as a disulfide bound 19-kDa and 32-kDa heterodimer (Tabuchi et al. 1997).
In the present study, we show that two subtilase-type serine peptidases, Isp6p and Psp3p, are vacuolar proteins and are critical for maturation of not only S. pombe CPY but also S. pombe ALP. In S. cerevisiae, both S. cerevisiae CPY and S. cerevisiae ALP are processed proteolytically largely by the vacuolar aspartyl protease Pep4p (Jones et al. 1982; Klionsky and Emr 1989). While the S. pombe genome lacks Pep4p homologs, it does contain two aspartyl proteases yps1+ and sxa1+, which are putative extracellular proteins harboring GPI attachment sites. These proteins share little homology with S. cerevisiae Pep4p. We found that CPY was processed normally and converted to the mature form in both yps1∆ and sxa1∆ mutants (data not shown).
Isp6p and Psp3p are related to each other (43% identity and 64% similarity) and also share similarity with S. cerevisiae Prb1p that mediates processing of various vacuolar proteins (Van Den Hazel et al. 1996). Isp6, which is not essential for cell viability, was identified in a screen for cDNAs preferentially expressed during sexual differentiation in S. pombe (Sato et al. 1994). Nakashima et al. (2002b, 2006) reported that Isp6p is essential for nitrogen starvation-induced autophagy in S. pombe. However, our Northern blot analysis showed that isp6+ was constitutively transcribed during vegetative growth in minimal medium and that a shift to conditions of nitrogen starvation did not induce an increase in transcription. A comprehensive survey of the S. pombe transcriptome revealed that isp6+ expression is dependent on culture medium as isp6+ mRNA levels were much higher in minimum medium than in YES, a relatively rich medium (Wilhelm et al. 2008). We also found that transcription levels differed between cells growing on nutrient medium MM, YES, and YPD, while Sato et al. (1994) reported low levels of isp6+ expression in cells growing on YPD. Other factors may also influence expression as the isp6+ gene has been reported to be responsive to environmental stress (Nakashima et al. 2002a). While Psp3p shares a high degree of sequence similarity with Isp6p, it is not essential for autophagy, and to our knowledge, its intracellular function has not yet been determined. Interestingly, processing of S. pombe CPY and S. pombe ALP were slightly delayed in isp6 disruptant (Figs. 2b and 5a), while expression level of psp3+ was much lower than that of isp6+ (Fig. 3a). We found that both Psp3p-GFP and Isp6p-GFP were sorted to the vacuole (Fig. 3c), which suggested that Psp3p and Isp6p may perform redundant functions in vacuolar protein degradation. Nakashima et al. (2006) reported that Isp6p is involved in both autophagy and sexual development in S. pombe. Therefore, Isp6p may be excessively expressed because Isp6p is required for not only processing of vacuolar enzymes but also bulk degradation of vacuolar proteins like autophagy.
We recently showed that disruption of psp3+ or isp6+ increased secretion of protease-sensitive heterologous proteins, human growth hormone, and human transferrin (Idiris et al. 2006b; Mukaiyama et al. 2009, 2010). Isp6p is probably transported through the cell secretory pathway to either the vacuole or the extracellular medium (Sato et al. 1994). Like Isp6p, Psp3p may also be transported to the extracellular medium and have an impact on the productivity of heterologous proteins. Moreover, Ladds and Davey reported that Psp3p overexpression complemented loss of Krp1p, which likely acts in the Golgi complex and cleaves proproteins on the C-terminal side of Lys–Arg and Arg–Arg peptide bonds (Ladds and Davey 2000). These observations suggest that Psp3p is responsible for some observed proteolytic activity.
Our Western blot analysis of S. pombe ALP revealed that disruption of isp6+ did not influence maturation of S. pombe ALP, whereas disruption of psp3+ did. That is, disruption of psp3+ decreased the amount of mature S. pombe ALP and led to retention of a small amount of pro-S. pombe ALP (Fig. 5a). With respect to maturation of S. pombe CPY, the pulse-chase analysis indicated that while the isp6Δ strain accumulated the 50-kDa S. pombe CPY intermediate missing the pro-segment, maturation of S. pombe CPY proceeded normally in psp3∆ cells (Fig. 2b). Ladds and Davey reported that Isp6p is specific for dibasic motifs such as Lys–Lys or Arg–Arg, although it appeared to prefer cleavage after lysine rather than arginine. In contrast, Psp3p has a broader substrate specificity than Isp6p (Ladds and Davey 2000). These data suggest that Isp6p and Psp3p may be involved in processing and activation of various vacuolar proteins, by analogy to Prb1p function in S. cerevisiae, while the substrate preferences of these proteases may differ. We are currently attempting to identify the precise processing sites within pro-S. pombe CPY and pro-S. pombe ALP that are cleaved by Isp6p and Psp3p.
References
Achstetter T, Wolf DH (1985) Proteinases, proteolysis and biological control in the yeast Saccharomyces cerevisiae. Yeast 1:139–157
Alfa C, Fantes P, Hyams JS, Mcleod M, Warbrick E (1993) Experiments with fission yeast: a laboratory course manual. Cold Spring Harbor Laboratory, Cold Spring Harbor
Cooper AA, Stevens TH (1996) Vps10p cycles between the late-Golgi and prevacuolar compartments in its function as the sorting receptor for multiple yeast vacuolar hydrolases. J Cell Biol 133:529–541
Cooper JP, Nimmo ER, Allshire RC, Cech TR (1997) Regulation of telomere length and function by a myb-domain protein in fission yeast. Nature 385:744–747
Davey J, Davis K, Imai Y, Yamamoto M, Matthews G (1994) Isolation and characterization of krp, a dibasic endopeptidase required for cell viability in the fission yeast Schizosaccharomyces pombe. EMBO J 13:5910–5921
Hirsch HH, Schiffer HH, Muller H, Wolf DH (1992) Biogenesis of the yeast vacuole (lysosome). Mutation in the active site of the vacuolar serine proteinase yscB abolishes proteolytic maturation of its 73-kDa precursor to the 41.5-kDa pro-enzyme and a newly detected 41-kDa peptide. Eur J Biochem 203:641–653
Idiris A, Bi K, Tohda H, Kumagai H, Giga-Hama Y (2006a) Construction of a protease-deficient strain set for the fission yeast Schizosaccharomyces pombe, useful for effective production of protease-sensitive heterologous proteins. Yeast 23:83–99
Idiris A, Tohda H, Bi KW, Isoai A, Kumagai H, Giga-Hama Y (2006b) Enhanced productivity of protease-sensitive heterologous proteins by disruption of multiple protease genes in the fission yeast Schizosaccharomyces pombe. Appl Microbiol Biotechnol 73:404–420
Idiris A, Tohda H, Sasaki M, Okada K, Kumagai H, Giga-Hama Y, Takegawa K (2009) Enhanced protein secretion from multiprotease-deficient fission yeast by modification of its vacuolar protein sorting pathway. Appl Microbiol Biotechnol 85:667–677
Iwaki T, Osawa F, Onishi M, Koga T, Fujita Y, Hosomi A, Tanaka N, Fukui Y, Takegawa K (2003) Characterization of vps33+, a gene required for vacuolar biogenesis and protein sorting in Schizosaccharomyces pombe. Yeast 20:845–855
Iwaki T, Goa T, Tanaka N, Takegawa K (2004) Characterization of Schizosaccharomyces pombe mutants defective in vacuolar acidification and protein sorting. Mol Genet Genomics 271:197–207
Iwaki T, Hosomi A, Tokudomi S, Kusunoki Y, Fujita Y, Giga-Hama Y, Tanaka N, Takegawa K (2006) Vacuolar protein sorting receptor in Schizosaccharomyces pombe. Microbiology 152:1523–1532
Iwaki T, Onishi M, Ikeuchi M, Kita A, Sugiura R, Giga-Hama Y, Fukui Y, Takegawa K (2007) Essential roles of class E Vps proteins for sorting into multivesicular bodies in Schizosaccharomyces pombe. Microbiology 153:2753–2764
Jones HB, Cavanagh JB (1984) Cytoplasmic vacuoles. Ultrastruct Pathol 6:359–362
Jones EW, Zubenko GS, Parker RR (1982) PEP4 gene function is required for expression of several vacuolar hydrolases in Saccharomyces cerevisiae. Genetics 102:665–677
Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y (2000) Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol 150:1507–1513
Klionsky DJ, Emr SD (1989) Membrane protein sorting: biosynthesis, transport and processing of yeast vacuolar alkaline phosphatase. EMBO J 8:2241–2250
Klionsky DJ, Emr SD (1990) A new class of lysosomal/vacuolar protein sorting signals. J Biol Chem 265:5349–5352
Koga T, Onishi M, Nakamura Y, Hirata A, Nakamura T, Shimoda C, Iwaki T, Takegawa K, Fukui Y (2004) Sorting nexin homologues are targets of phosphatidylinositol 3-phosphate in sporulation of Schizosaccharomyces pombe. Genes Cells 9:561–574
Kunkel TA, Bebenek K, McClary J (1991) Efficient site-directed mutagenesis using uracil-containing DNA. Methods Enzymol 204:125–139
Ladds G, Davey J (2000) Identification of proteases with shared functions to the proprotein processing protease Krp1 in the fission yeast Schizosaccharomyces pombe. Mol Microbiol 38:839–853
Marcusson EG, Horazdovsky BF, Cereghino JL, Gharakhanian E, Emr SD (1994) The sorting receptor for yeast vacuolar carboxypeptidase Y is encoded by the VPS10 gene. Cell 77:579–586
Martinoia E, Heck U, Boller T, Wiemken A, Matile P (1979) Some properties of vacuoles isolated from Neurospora crassa slime variant. Arch Microbiol 120:31–34
Matile P, Wiemken A (1967) The vacuole as the lysosome of the yeast cell. Arch Mikrobiol 56:148–155
Mechler B, Muller M, Muller H, Meussdoerffer F, Wolf DH (1982) In vivo biosynthesis of the vacuolar proteinases A and B in the yeast Saccharomyces cerevisiae. J Biol Chem 257:11203–11206
Moehle CM, Aynardi MW, Kolodny MR, Park FJ, Jones EW (1987a) Protease B of Saccharomyces cerevisiae: isolation and regulation of the PRB1 structural gene. Genetics 115:255–263
Moehle CM, Tizard R, Lemmon SK, Smart J, Jones EW (1987b) Protease B of the lysosomelike vacuole of the yeast Saccharomyces cerevisiae is homologous to the subtilisin family of serine proteases. Mol Cell Biol 7:4390–4399
Moehle CM, Dixon CK, Jones EW (1989) Processing pathway for protease B of Saccharomyces cerevisiae. J Cell Biol 108:309–325
Morita T, Takegawa K (2004) A simple and efficient procedure for transformation of Schizosaccharomyces pombe. Yeast 21:613–617
Mukaiyama H, Giga-Hama Y, Tohda H, Takegawa K (2009) Dextran sodium sulfate enhances secretion of recombinant human transferrin in Schizosaccharomyces pombe. Appl Microbiol Biotechnol 85:155–164
Mukaiyama H, Tohda H, Takegawa K (2010) Overexpression of protein disulfide isomerases enhances secretion of recombinant human transferrin in Schizosaccharomyces pombe. Appl Microbiol Biotechnol 86:1135–1143
Nakashima A, Ueno M, Ushimaru T, Uritani M (2002a) Involvement of a CCAAT-binding complex in the expression of a nitrogen-starvation-specific gene, isp6+, in Schizosaccharomyces pombe. Biosci Biotechnol Biochem 66:2224–2227
Nakashima A, Yoshida M, Nakayama K, Kato-Furuno A, Ueno M, Ushimaru T, Uritani M (2002b) Genes for a nuclease and a protease are involved in the drastic decrease in cellular RNA amount in fission yeast cells during nitrogen starvation. J Biochem 131:391–398
Nakashima A, Hasegawa T, Mori S, Ueno M, Tanaka S, Ushimaru T, Sato S, Uritani M (2006) A starvation-specific serine protease gene, isp6+, is involved in both autophagy and sexual development in Schizosaccharomyces pombe. Curr Genet 49:403–413
Nebes VL, Jones EW (1991) Activation of the proteinase B precursor of the yeast Saccharomyces cerevisiae by autocatalysis and by an internal sequence. J Biol Chem 266:22851–22857
Okazaki K, Okazaki N, Kume K, Jinno S, Tanaka K, Okayama H (1990) High-frequency transformation method and library transducing vectors for cloning mammalian cDNAs by trans-complementation of Schizosaccharomyces pombe. Nucleic Acids Res 18:6485–6489
Onishi M, Koga T, Morita R, Nakamura Y, Nakamura T, Shimoda C, Takegawa K, Hirata A, Fukui Y (2003) Role of phosphatidylinositol 3-phosphate in formation of forespore membrane in Schizosaccharomyces pombe. Yeast 20:193–206
Rothlisberger S, Jourdain I, Johnson C, Takegawa K, Hyams JS (2009) The dynamin-related protein Vps1 regulates vacuole fission, fusion and tubulation in the fission yeast, Schizosaccharomyces pombe. Fungal Genet Biol 46:927–935
Russell P, Nurse P (1986) Schizosaccharomyces pombe and Saccharomyces cerevisiae: a look at yeasts divided. Cell 45:781–782
Sato S, Suzuki H, Widyastuti U, Hotta Y, Tabata S (1994) Identification and characterization of genes induced during sexual differentiation in Schizosaccharomyces pombe. Curr Genet 26:31–37
Seaman MN, Marcusson EG, Cereghino JL, Emr SD (1997) Endosome to Golgi retrieval of the vacuolar protein sorting receptor, Vps10p, requires the function of the VPS29, VPS30, and VPS35 gene products. J Cell Biol 137:79–92
Seaman MN, McCaffery JM, Emr SD (1998) A membrane coat complex essential for endosome-to-Golgi retrograde transport in yeast. J Cell Biol 142:665–681
Stevens T, Esmon B, Schekman R (1982) Early stages in the yeast secretory pathway are required for transport of carboxypeptidase Y to the vacuole. Cell 30:439–448
Stevens TH, Rothman JH, Payne GS, Schekman R (1986) Gene dosage-dependent secretion of yeast vacuolar carboxypeptidase Y. J Cell Biol 102:1551–1557
Suga M, Hatakeyama T (2001) High efficiency transformation of Schizosaccharomyces pombe pretreated with thiol compounds by electroporation. Yeast 18:1015–1021
Tabuchi M, Iwaihara O, Ohtani Y, Ohuchi N, Sakurai J, Morita T, Iwahara S, Takegawa K (1997) Vacuolar protein sorting in fission yeast: cloning, biosynthesis, transport, and processing of carboxypeptidase Y from Schizosaccharomyces pombe. J Bacteriol 179:4179–4189
Takegawa K, DeWald DB, Emr SD (1995) Schizosaccharomyces pombe Vps34p, a phosphatidylinositol-specific PI 3-kinase essential for normal cell growth and vacuole morphology. J Cell Sci 108:3745–3756
Takegawa K, Hosomi A, Iwaki T, Fujita Y, Morita T, Tanaka N (2003a) Identification of a SNARE protein required for vacuolar protein transport in Schizosaccharomyces pombe. Biochem Biophys Res Commun 311:77–82
Takegawa K, Iwaki T, Fujita Y, Morita T, Hosomi A, Tanaka N (2003b) Vesicle-mediated protein transport pathways to the vacuole in Schizosaccharomyces pombe. Cell Struct Funct 28:399–417
Takegawa K, Tokudomi S, Bhuiyan MS, Tabuchi M, Fujita Y, Iwaki T, Utsumi S, Tanaka N (2003c) Heterologous expression and characterization of Schizosaccharomyces pombe vacuolar carboxypeptidase Y in Saccharomyces cerevisiae. Curr Genet 42:252–259
Van Den Hazel HB, Kielland-Brandt MC, Winther JR (1996) Review: biosynthesis and function of yeast vacuolar proteases. Yeast 12:1–16
Vida TA, Emr SD (1995) A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J Cell Biol 128:779–792
Wilhelm BT, Marguerat S, Watt S, Schubert F, Wood V, Goodhead I, Penkett CJ, Rogers J, Bahler J (2008) Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature 453:1239–1243
Acknowledgments
We thank Yoko Kusunoki, Yukio Ozaka, and Naotaka Tanaka for excellent technical assistance. This work was partly supported by the Project for Development of a Technological Infrastructure for Industrial Bioprocesses on R&D of New Industrial Science and Technology Frontiers by the Ministry of Economy, Trade & Industry, as supported by the New Energy and Industrial Technology Development Organization.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Mukaiyama, H., Iwaki, T., Idiris, A. et al. Processing and maturation of carboxypeptidase Y and alkaline phosphatase in Schizosaccharomyces pombe . Appl Microbiol Biotechnol 90, 203–213 (2011). https://doi.org/10.1007/s00253-010-3031-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-010-3031-3