Abstract
The major histocompatibility complex (MHC) genes are the most polymorphic genes found in the vertebrate genome, and they encode proteins that play an essential role in the adaptive immune response. Many songbirds (passerines) have been shown to have a large number of transcribed MHC class I genes compared to most mammals. To elucidate the reason for this large number of genes, we compared 14 MHC class I alleles (α1–α3 domains), from great reed warbler, house sparrow and tree sparrow, via phylogenetic analysis, homology modelling and in silico peptide-binding predictions to investigate their functional and genetic relationships. We found more pronounced clustering of the MHC class I allomorphs (allele specific proteins) in regards to their function (peptide-binding specificities) compared to their genetic relationships (amino acid sequences), indicating that the high number of alleles is of functional significance. The MHC class I allomorphs from house sparrow and tree sparrow, species that diverged 10 million years ago (MYA), had overlapping peptide-binding specificities, and these similarities across species were also confirmed in phylogenetic analyses based on amino acid sequences. Notably, there were also overlapping peptide-binding specificities in the allomorphs from house sparrow and great reed warbler, although these species diverged 30 MYA. This overlap was not found in a tree based on amino acid sequences. Our interpretation is that convergent evolution on the level of the protein function, possibly driven by selection from shared pathogens, has resulted in allomorphs with similar peptide-binding repertoires, although trans-species evolution in combination with gene conversion cannot be ruled out.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The major histocompatibility complex (MHC) genes are the most polymorphic genes in the vertebrate genome and the proteins that they encode play a pivotal role in the adaptive immune system. Two clusters of classical antigen presenting genes can be found within the MHC complex, the MHC class I and II genes. MHC genes are classified as classical based on high polymorphism and ability to bind antigenic peptides. The main function of the classical MHC molecules is to present pathogenic peptides on the cell surface to circulating T cells (Apanius et al. 1997). In humans, the classical MHC class I and II are each encoded by three genes (Neefjes et al. 2011).
Compared to the human MHC region, which consists of over 200 genes spanning four megabases, the classical chicken MHC region (MHC-B) is small and simple with just 19 genes over 92 kb encoding two MHC class I and two MHC class IIβ genes (Guillemot et al. 1988; Kaufman et al. 1999). Studies of other species of the order Galliformes, such as the turkey (Meleagris gallopavo), show a similar constricted MHC-B region with the exception that the turkey has three MHC class IIβ genes compared to only two in the chicken (Chaves et al. 2009). In the quail (Coturnix japonica), another species belonging to the order Galliformes, seven MHC class I and ten MHC class IIβ have been described. However, only two of each of the class I and class IIβ genes are transcribed and are considered to be classical MHC genes (Shiina et al. 2004).
Studies in chickens have shown that only one of the two MHC class I genes is highly transcribed and expressed as a protein on the cell surface (Kaufman 1999; Shaw et al. 2007; Wallny et al. 2006). The reason for having just one dominantly expressed classical MHC class I molecule is still unclear. However, it has been proposed that the co-evolution with polymorphic variants of the transporter associated with antigen processing (TAP) could be one explanation. This co-evolution may have favoured the expression of a single MHC class I molecule that shares peptide-binding specificity with TAP and disfavoured expression in the same individual of MHC class I molecules with other specificities (Walker et al. 2011). This kind of co-evolution can partly explain the prominent associations between certain MHC class I haplotypes and resistance or susceptibility to infectious diseases in chickens (Kaufman et al. 1995, 1999; Walker et al. 2005; Wallny et al. 2006).
Although the genomic organisation of avian MHC is best characterized in birds that belong to the order Galliformes (Chaves et al. 2009; Guillemot et al. 1988; Kaufman et al. 1999; Reed et al. 2011; Shiina et al. 2004), there has been recent progress towards understanding the MHC gene organisation of the zebra finch (Taeniopygia guttata) (Balakrishnan et al. 2010; Ekblom et al. 2011). The zebra finch is a small songbird belonging to the order Passeriformes. The orders Galliformes and Passeriformes diverged approximately 125 million years ago (MYA) (Brown et al. 2008) and, although the understanding of the genomic organisation of the zebra finch MHC genes is still fragmented, it has been shown that the MHC genes that are linked in the chicken are dispersed over two (Ekblom et al. 2011) or even four (Balakrishnan et al. 2010) different chromosomes in the zebra finch. It has also been shown that the MHC genes of zebra finches contain several putative pseudo genes (Warren et al. 2010).
The MHC genes of a variety of other passerines have also been studied. Most work in this respect has focused on the MHC class IIB genes (Aguilar et al. 2006; Bollmer et al. 2010; Miller and Lambert 2004; Sato et al. 2001; Westerdahl et al. 2000; Zagalska-Neubauer et al. 2010). For example, a study by Zagalska-Neubauer et al. (2010) used 454-amplicon sequencing to screen MHC diversity and found up to 18 exon 2 sequences (at least nine loci) per individual in collared flycatchers (Ficedula albicollis). Studies on MHC class I have shown that, in general, passerines have a larger number of MHC class I genes compared to both the chicken (Shaw et al. 2007; Wallny et al. 2006) and zebra finch (Warren et al. 2010). Blue tits (Cyanistes caeruleus) have been shown to have at least four MHC class I genes per individual (Schut et al. 2011), scarlet rose finches (Carpodacus erythrinus) have at least seven genes (Promerova et al. 2012), house sparrows (Passer domesticus) have at least six genes (Bonneaud et al. 2004) and great reed warblers (Acrocephalus arundinaceus) have at least eight different MHC class I genes per individual (Westerdahl 2007; Westerdahl et al. 1999). Previous studies using the Southern Blot technique suggested a considerably larger number of class I genes in passerines and a recent study on great tits (Parus major) using 454-amplicon sequencing confirmed this suggestion, reporting 32 exon 3 sequences (at least 16 loci) in a single individual (Sepil et al. 2012; Westerdahl 2007; Wittzell et al. 1998).
The high number of MHC genes found in passerines compared to the chicken is intriguing and to better understand this difference it is important to understand the function of the MHC molecules. All MHC molecules have a limited repertoire of peptides that they bind and present on the cell surface. This functional specificity is defined in the peptide-binding cleft of the MHC molecule. To understand the peptide-binding of a MHC protein it is essential not only to know the sequence of the protein but also the structure of the molecule. Several different approaches have been developed for predicting peptide-binding specificities and defining functional similarities between MHC molecules spanning from simple sequence-based phylogenetic analysis to clustering approaches based on advanced mathematical models predicting receptor-ligand binding specificities (reviewed by Lundegaard et al. 2010).
In the present study, we aim to investigate MHC class I genes from three passerines, one house sparrow, one tree sparrow (Passer montanus) and one great reed warbler to get a better understanding of the high number of MHC class I genes found in these birds. We base our analyses on cDNAs (exons 2–4) and use a panel of methods to investigate peptide-binding properties of passerine MHC class I molecules including homology modelling and in silico peptide-binding prediction methods in combination with classical selection and phylogenetic analyses. By using these approaches we are able to study MHC class I genes not only from a genetic perspective by doing sequence analyses but are also able to link our findings with the actual function of the protein in regards to its peptide-binding specificity.
Methods
Study species
The great reed warbler is a long-distance migrating, passerine bird that winters in Africa, south of the Sahara desert. It breeds in reed beds of marshes and lakes in the northern and central Palaearctic (Cramp 1992). The DNA sample used in this study has been collected from one great reed warbler at Lake Kvismaren just east of Örebro in Sweden (59°10′N,15°25′E). The six MHC class I cDNAs used in this study have been published previously (Westerdahl et al. 1999). The house sparrow and tree sparrow are both sedentary bird species. The house sparrow sample used in this study has been collected from one individual on Lundy Island (51°10′N, 4°40′W), which is a small island in the Bristol Channel in the UK, and the tree sparrow individual used in this study was caught in Båstad (56°24′N, 12°56′E) in northwest Skåne in Sweden. The four house sparrow MHC class I cDNAs used in this study is from Karlsson and Westerdahl, unpublished data, while the four tree sparrow MHC class I cDNAs are novel to this study.
Isolation of RNA
A blood sample of 30 μl was collected from a tree sparrow captured in a mist net. Blood for isolation of RNA was collected in 100 μl K2EDTA (0.2 M), after which 500 μl TRIzol-LS (Invitrogen, Paisley, UK) was added. Isolation of total RNA was performed using a TRIzol/chloroform extraction protocol, carefully collecting only the RNA containing aqueous phase to avoid DNA contamination and then precipitating the RNA using isopropanol (Invitrogen); for details, see Strandh et al. (2011). The dried and cleaned RNA pellet was then dissolved in 0.5 μl RNasin Plus RNase inhibitor (Promega, USA) and 40 μl of ddH2O and stored at −80 °C.
PCR and DNA sequencing
RT-PCR on mRNA (1 μg) was used to synthesize cDNA according to the manufactures protocol (RETROscript kit; Ambion, Austin, TX, USA). The cDNA from the RT-PCR was diluted five times before it was used as template in a standard PCR using the primers PadoM2 (forward): 5′-GTT CTC CAC TCC CTG CRT YAC CTG-3′ and Pado4M (reverse): 5′-CA AGC RAA GAT CCC GGG CTC CAG C-3′ (Applied Biosystems, New Jersey, USA). PCR products were cloned according to protocol (2.1 TOPO-TA cloning kit; Invitrogen, USA). Next, 10–20 clones were picked per PCR run and three independent PCR runs were performed in total. Colonies containing inserts of the correct length (approximately 850 bp) were DNA sequenced using standard procedures (BigDye terminator kit v.3.1) on an ABI PRISM 3130 genetic analyser (Applied Biosystems). We did not aim to get a complete coverage of the number of MHC class I alleles in one individual, this is better done using 454 amplicon sequencing. However, all DNA sequences used in the present study have been identified in at least two independent PCRs to confirm the existence of the alleles.
Homology modelling
Structural homology models of the α1 to α3 domains of each of the 14 MHC class I sequences were built using the CPHmodels-3.2 server (Nielsen et al. 2010). The structural templates with the highest sequence identity with the passerine sequences were used, with the chicken MHC class I structures in the Protein Data Bank (PDB); IDs 3BEV (Koch et al. 2007) for Acar_2, Acar_15, Acar_19, Pamo_01, Pamo_03, and Pado_242, 3BEW (Koch et al. 2007) for Acar_20 and Pamo_04, 2YF1 (Chapell et al. unpublished data) for Acar_5 and Pado_230, 2YF6 (Chapell et al. unpublished data) for Pado_240, Pado_241 and Pamo_02 and finally 3P73 (Hee et al. 2010) for Acar_3.
Pseudo sequence deduction and prediction of peptide-binding motifs
The pseudo sequence is a sequence of 34 amino acid residues in the MHC class I peptide-binding cleft that is estimated to be in contact with the bound peptide. The definition is based on human HLA-A and -B structures and a residue is said to have contact when it is within 4.0 Å of the bound peptide (Nielsen et al. 2007). To estimate the pseudo sequences in this study (i.e., the amino acids lining the peptide-binding cleft that are in contact with the bound peptide), the passerine sequences were aligned with 1,524 different HLA-A and -B allele sequences using the progressive method (FFT-NS-2) in MAFFT version 6 (Katoh et al. 2002). The residue positions of the passerine sequences were then deduced. The NetMHCcons method (Karosiene et al. 2012) was used to predict the peptide-binding motifs of the 14 passerine allomorphs (allele specific proteins). This method is a consensus of three previously described MHC class I peptide-binding prediction methods, the NetMHC (Lundegaard et al. 2008), the NetMHCpan (Hoof et al. 2009; Nielsen et al. 2007) and the PickPocket (Zhang et al. 2009). Briefly, the method assumes all MHC class I molecules to adopt an identical structure. Consequently, the differences in peptide-binding specificity between different MHC class I molecules is dependent only on the pseudo sequence. Since the structure of the MHC class I molecules is assumed to be invariant, the mapping of the pseudo sequence comes down to aligning the query sequence on to the sequence of the set of MHC class I molecules with characterised binding specificity. The predictions performed here were made using a set of 100,000 9mer peptides. The top 0.5 % binders were summarised and presented in peptide-binding logos using Seq2Logo 1.0 (Thomsen and Nielsen 2012).
In silico peptide-binding predictions and functional clustering of MHC class I allomorphs
By clustering the MHC class I allomorphs based on their peptide-binding specificity, it is possible to study the functional relationship between them. The MHCcluster method (Nielsen et al. 2012) was used to predict and functionally cluster the peptide-binding specificities of the 14 MHC class I allomorphs. This method is based on NetMHCconc predictions (Karosiene et al. 2012). The method predicts the binding of a number of predefined peptides to the MHC class I allomorphs of interest. A set of 100,000 natural random 9mer peptides and each of the 14 passerine MHC class I pseudo sequences was used to calculate peptide-binding specificities. The method then estimates the similarities between any two MHC class I allomorphs in the data set by correlating the union of the predicted top 10 % strongest binding peptides for each of the defined allomorphs. The similarity is defined as 1 if the two MHC class I allomorphs are predicted to have a perfect overlap in regards to their peptide-binding specificities and 0 if there is no overlap at all. A value of −1 will indicate perfect anti-correlation. The distance between two allomorphs is defined as 1–similarity. By using UPGMA clustering the distance matrix can be converted to a distance tree. The significance of the MHC class I distance tree is estimated by generating 1,000 distance trees using the bootstrap method. The bootstrapping is performed at the peptide level, i.e., for each distance tree a new set of 100,000 peptides and the correlating predictions is selected from the original pool of 100,000 peptides with replacement. The trees are then summarised, and a consensus tree is made with branch bootstrap values.
Genetic analysis
All sequences were aligned in BioEdit 7.0.9.0 Sequence Alignment Editor (Hall 1999). The amino acids in the peptide-binding region (PBR) (Bjorkman et al. 1987) and the pseudo sequence (Nielsen et al. 2007) were superimposed from human HLA. The tree based on tree sparrow (Pamo), house sparrow (Pado) and great reed warbler (Acar) MHC class I amino acid sequences was made in RAxML BlackBox, using Maximum Likelihood (ML) for phylogenetic reconstruction and bootstrap (bt) with 100 replicates (Jones–Taylor–Thornton [JTT] model, standard settings) (Stamatakis et al. 2008). Tests to find codon sites under positive selection were performed on six great reed warbler sequences using phylogeny-based ML analyses (CODEML) implemented in PAML v.4 (Yang 2007). Note that one additional great reed warbler sequence Acar_1 was added here to get better power in the selection analysis; this sequence lacks 13 amino acids in the 5′ region and was therefore excluded from all other analysis, since Acar_2 and Acar_19 have identical amino acid sequences only Acar_2 was included here. The tree topology used in the CODEML analysis was constructed in MEGA 5.05 (Tamura et al. 2011), using the neighbor-joining (NJ) method with Kimura’s two-parameter evolutionary distances, but the branch lengths were re-estimated under codon models using CODEML (Yang 1997). This tree topology is found in the supplementary material (Online Resource 1), and there were high bootstrap support for two out of three nodes (100, 97, 56). NJ tree topologies used in previous CODEML studies (e.g., Yang and Bielawski 2000) have suggested that minor differences in the tree topology make little impact on inference of positive selection under site models. Likelihood analysis was conducted under the random-sites models: M1a (nearly neutral), M2a (positive selection), M7 (β) and M8 (β + ω). When the selection models (M2a and M8) were significantly better than the neutral models (M1a and M7) we used Bayesian statistics integrated in CODEML (Empirical Bayes [BEB] method) to identify codon sites under positive selection (Yang et al. 2005).
Results
Pseudo sequences hold sites subject to positive selection
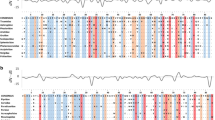
We amplified the major part of the MHC class I α1 to α3 domains in one tree sparrow (Pamo) and verified four MHC class I amino acid sequences (255–257 amino acids). The 14 MHC class I sequences used in the present study, four tree sparrow (Pamo), four house sparrow (Pado) and six great reed warbler (Acar), were aligned with a two amino acid insert at position 149 in the sequences Pamo_01, Pamo_02 and Pado_230 (Fig. 1). We used three independent approaches to find sites subject to positive selection/sites important for peptide-binding in great reed warbler MHC class I allomorphs; (1) a phylogeny-based ML approach to find sites subject to positive selection (CODEML; Yang 2007), (2) inferred the peptide-binding region from human HLA sequences (Bjorkman et al. 1987) and (3) deduced the pseudo sequence by estimating the contact between the residues in the peptide-binding cleft and the bound peptide (Nielsen et al. 2007). Positive selection occurred in the great reed warbler MHC class I alleles, since the models that allow selection at a subset of the sites (M2a and M8) fit the great reed warbler data better than the nearly neutral models (M2a: 2Δl = 62.2, df = 2, p < 0.001; M7 vs. M8: 2Δl = 62.4, df = 2, p < 0.001; Table 1). Model M2a showed that seven sites are subject to positive selection ('S') (cut-off p > 99 %; Table 1), six of which are found in both the inferred PBR ('P') and the pseudo sequence ('E') (Fig. 1, see 'S', 'P' and 'E'). Model M8 showed that nine sites are subject to positive selection ('S') (cut-off p > 99 %; Table 1), eight of which are found in both the inferred PBR ('P') and the pseudo sequence ('E') (Fig. 1, see 'S', 'P' and 'E').
Aligned MHC class I amino acid sequences, covering the α1, α2 and α3 regions, from great reed warbler (Acar, in Westerdahl et al. 1999), house sparrow (Pado, in Karlsson and Westerdahl under review), tree sparrow (Pamo, present study), zebra finch (Tagu XP_002186567), domestic chicken (MHC-B GagaBF2 HQ141386 and MHC-Y GagaY NM_001030675) and human (HLA:B*39:01 CCA63229). Identity with the Acar_3 is indicated with dots, codons corresponding to the peptide binding region (PBR) superimposed from the PBR of human HLA with a ‘P’, sites under positive selection in great reed warblers (Acar) according to PAML (p < 0.01) are marked with an ‘S’ (p < 0.05; with ‘s’) and sites included in the pseudo sequence are indicated with an ‘E’
The deduced pseudo sequences matched the inner lining of the peptide-binding cleft in the homology models
The chicken templates used to build the homology models had sequence identities with our 14 MHC class I sequences of between 45 % and 49 %. The pseudo sequences were superimposed on the peptide-binding cleft of these modelled molecules in an attempt to verify the accuracy of the pseudo sequences. The superimposed pseudo sequences from the 14 MHC class I allomorphs matched the inner lining of the peptide-binding cleft as illustrated by one representative great warbler allomorph, Acar_3 (Fig. 2). This indicates that the pseudo sequences are accurately deduced however to fully confirm their accuracy more detailed analysis of eluted peptide–MHC class I complexes are needed. We also superimposed the positively selected sites from the great reed warblers and found an overlap in the peptide-binding cleft of the modelled allomorph (Fig. 2).
Homology model illustrating the overlap (green) between the positively selected sites (yellow) and the pseudo sequence (blue) of one representative MHC class I allomorph Acar_3. The pseudo sequence was deduced by estimation of contact between the residues in the peptide-binding cleft and the bound peptide (Nielsen et al. 2007). The positively selected sites were obtained using PAML analysis
In silico peptide-binding predictions reveal separate functional clusters of MHC class I allomorphs based on their peptide-binding specificities
The peptide-binding motif of an MHC class I allomorph can be characterized from the peptides they bind. The peptide-binding motifs are generated by calculating the frequency of occurrence of any given amino acid at a specific position in the bound peptide. If a specific position within the bound peptide is crucial for peptide-binding only a limited set of amino acids will be present at that position. These crucial positions within the bound peptide are referred to as anchor positions and are indicated by high bars in the peptide-binding motifs. The tree based on functional clustering (MHC class I peptide-binding specificities) graphically illustrates the specificity of the 14 different MHC class I allomorphs and their overlap in peptide-binding specificity is further illustrated in a heat map (darker colour indicates more overlap in peptide-binding specificity; Figs. 3a and 4). The peptide-binding motifs revealed one distinct hydrophobic anchor position in the C-terminal part of the bound peptide at position nine for all 14 MHC class I allomorphs (Fig. 3a). Moreover, for eight of the allomorphs (Acar_2, Acar_19, Pamo_01, Pamo_02, Pamo_03, Pamo_04, Pado_230 and Pado_242) the peptide-binding motifs indicate that a positively charged amino acid, arginine or lysine, is preferred at position five (Fig. 3a). All the tree sparrow allomorphs and two out of four house sparrow allomorphs are found in two distinct clusters, one cluster with Pamo_01, Pamo_02 and Pado_230 and one with Pamo_03, Pamo_04 and Pado_242 (Fig. 3a). The peptide-binding motifs indicate that the allomorphs in the cluster with Pamo_01, Pamo_02 and Pado_230 have a preference for two anchor positions in the N-terminal part of the bound peptide with phenylalanine at position one and glutamine at position two. The allomorphs in the other cluster (Pamo_03, Pamo_04 and Pado_242) show preference for three anchor positions in the N-terminal, for Pamo_03, Pamo_04 (phenylalanine at position one, glutamine at position two and alanine at position three) and two anchor positions for Pado_242 (phenylalanine at position one and leucine at position two) (Fig. 3a). The overlap in the peptide-binding specificities within each of these two clusters is also seen in the heat map (Fig. 4). Interestingly, the allomorphs in these two clusters also share some peptide-binding specificity with the great reed warbler allomorphs, Acar_2 and Acar_19 (Fig. 4). The house sparrow allomorphs Pado_240 and Pado_241 are found among the great reed warbler allomorphs close to the great reed warbler allomorph Acar_3, again indicating an overlap in peptide-binding specificities across species (Figs. 3a and 4). The great reed warbler allomorphs Acar_5, Acar_15 and Acar_20 have more unique peptide-binding preferences as suggested by their peptide-binding motifs and that they do not cluster with any other allomorphs (Figs. 3a and 4).
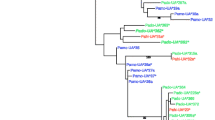
Phylogenetic relationships of great reed warbler (Acar), house sparrow (Pado) and tree sparrow (Pamo) MHC class I alleles/allomorphs based on amino acid sequences and peptide-binding specificities, respectively. a Functional clustering of the passerine allomorphs and corresponding peptide-binding motifs. The relative height of the bars in the motifs relates to the frequency of a given amino acid at that position in the bound 9mer peptide. The MHCcluster method was used to predict the functional clustering and the NetMHCcons method was used to predict the peptide-binding motifs. b The tree based on passerine amino acid sequences is a simplified illustration of the ML tree found in Fig. 5 to allow for a better comparison between the trees based on peptide-binding and amino acid sequences
Two trees with different topologies
A tree based on the amino acid sequences from the α1 and α2 domains (exons 2 and 3) was estimated using ML (Fig. 5). Here the sparrow sequences formed a monophyletic cluster with high bootstrap support (bt = 95) and the tree sparrow and house sparrow sequences within this cluster were mixed across species. Alleles with a deletion corresponding to two amino acids at position 149 (Pamo_01, Pamo_02 and Pado_230) formed a significant monophyletic cluster (bt = 100) distant from the five other house sparrow and tree sparrow alleles. The origin of the two allelic lineages, with and without this insert coding for two amino acids, probably pre-dates the separation of house sparrows and tree sparrows (Fig. 1). When graphically comparing the tree based on amino acid sequences and the tree based on functional clustering, it is clear that their topologies are different (Fig. 3a, b). The tree based on amino acids clearly separates the house and tree sparrow alleles from the great reed warbler alleles (Fig. 5), whereas in the tree based on functional clustering, two of the house sparrow allomorphs (Pado_240 and Pado_241) are found among the great reed warbler allomorphs (Fig. 3a, b). However, two trees based on amino acid sequences from exon 2 and exon 3, respectively, give different phylogenetic signals. In a tree based on amino acid sequences from exon 2 the great reed warbler sequences form a single highly supported cluster (bt = 99), while they do not in the tree based on amino acid sequences from exon 3 (Online Resource 2). Two trees based on synonymous substitutions in exon 2 and exon 3, respectively, do place the great reed warbler sequences in a single cluster, though with low bootstrap support in the tree based on exon 3 (exon 2, bt = 89; exon 3 bt = 51; Online Resource 2). Our interpretation of the trees based on functional clustering and amino acid sequences is that the house sparrow and the great reed warbler studied here have MHC class I molecules with similar peptide-binding repertoire although these molecules are encoded by partly different amino acid sequences.
Phylogenetic relationships of great reed warbler (Acar), house sparrow (Pado) and tree sparrow (Pamo) MHC class I amino acid sequences (α1 and α2 regions) using Maximum Likelihood (JTT + G model) and bootstrap with 100 replicates in RAxML (Stamatakis et al. 2008)
Discussion
Previous studies of passerine MHC class I are mostly based on DNA or amino acid sequence analysis. Here we have included an additional feature (i.e., peptide-binding) of the MHC class I allomorphs and with the help of in silico prediction tools we have been able to study how amino acids in the peptide-binding cleft influence the peptide-binding. By studying the predicted peptide-binding properties of MHC class I molecules in passerines we take one step closer to resolving the phenotype, on which natural selection acts. We envision that peptide-binding prediction tools in combination with sequence data will provide a comprehensive framework for a better understanding of the enormous diversity found in the MHC class I genes of passerines.
The passerine MHC class I allomorphs differ in their anchor positions
The major aim of the present study was to model and evaluate peptide-binding motifs of passerine MHC class I allomorphs to better understand the function of the high number of MHC class I genes observed in passerines. The peptide-binding motifs of the 14 studied passerine MHC class I allomorphs showed differences in their preferred anchor positions (Figs. 3a and 4). This is in line with what has been observed for human HLA class I allomorphs that differ significantly in their peptide-binding motifs (Rammensee et al. 1995). However, in the present study we have investigated peptide-binding motifs not only within a single species but also across three different species; house sparrow, tree sparrow and great reed warbler. We find that the preferred anchor positions for the different allomorphs are shared across these species. The house sparrow and the tree sparrow that diverged 10MYA have overlapping peptide-binding motifs and so do the house sparrow and the great reed warbler that diverged considerably earlier (30 MYA) (Sibly et al. 2012).
Peptide-binding specificities
In the analyses of the peptide-binding motifs, we found that the preference for a hydrophobic amino acid at position nine in the C-terminal part of the bound peptide was shared across all allomorphs (Fig. 3a). However, in the N-terminal part the preferred anchor positions differed both within and between species, considering both number of positions and amino acid preference. The peptide-binding predictions also revealed that eight of the 14 passerine allomorphs had a preference for a positively charged amino acid at position five (Fig. 3a). Interestingly, these eight allomorphs showed a slight overlap in their peptide-binding specificity (Fig. 4) and they are subsequently found in the same part of the tree based on functional clustering (Fig. 3a). This finding suggests that position five is of high importance for determining the peptide-binding specificity of these allomorphs. Two of the house sparrow allomorphs and all four tree sparrow allomorphs formed two separate clusters wherein the allomorphs have considerable overlap in their peptide-binding specificities (Figs. 3a and 4). The same overlapping pattern in peptide-binding specificity was not found for the great reed warbler allomorphs. The great reed warbler alleles (Acar_3, Acar_5, Acar_15 and Acar_20) show a weak clustering pattern with low bootstrap support in the tree based on amino acid sequences (Fig. 5). However, the pattern in the tree based on functional clustering suggests that these four great reed warbler allomorphs are functionally different in regards to their peptide-binding specificities (Figs. 3a and 4).
The increased separation of the great reed warbler allomorphs observed in the tree based on functional clustering compared to the tree based on amino acid sequences (Fig. 3) can partly, but most likely not fully, be explained by how these trees were generated. The method that was used for the peptide-binding predictions assumes that the pseudo sequence of each MHC class I molecule defines its peptide-binding specificity and thus its function. In essence, this means that only the most important amino acids responsible for the peptide-binding will be taken into account, hence a single amino acid substitution within the pseudo sequence can generate a very different repertoire of bound peptides. In contrast, in the tree based on MHC class I exons 2 and 3 amino acid sequences a single amino acid substitutions will be less important. However, a larger difference on the functional level of MHC class I allomorphs compared to the amino acid level has also been observed in other vertebrates, for example, in humans (Axelsson-Robertson et al. 2011). Taken together our data suggests that the great reed warbler manage to bind a broader repertoire of peptides compared to both the tree sparrow and the house sparrow studied here. However, more allomorphs from a larger number of individuals need to be studied to further support these preliminary findings on a species level.
Pseudo sequences hold sites subject to positive selection
A key obstacle to the present analyses to determine peptide-binding repertoires was the encoding of the passerine MHC class I sequences into so-called pseudo sequences on which the peptide-binding predictions are based. The pseudo sequence consists of the residues in the peptide-binding cleft of the MHC class I molecule that are estimated to bind the peptide for presentation on the cell surface. Because of the relatively distant relationship with the HLA-A and -B structures, which are used to define the typical amino acid contacts between the bound peptide and the MHC class I molecule, and the passerine MHC class I structures it may be suspected that accurate pseudo sequences for passerine MHC class I allomorphs could be difficult to infer using this method. However, the significant overlap found between positively selected sites and the obtained pseudo sequences provide an independent verification that the obtained pseudo sequences for the passerines are reasonable (Fig. 1). Moreover, we find that the pseudo sequence positions, as well as the positively selected sites, are matched directly to the peptide-binding cleft in the homology models (Fig. 2). The peptide-binding prediction method, NetMHCconc, used here is a consensus of three previously described MHC class I peptide-binding prediction methods, the NetMHC (Lundegaard et al. 2008), the NetMHCpan (Hoof et al. 2009; Nielsen et al. 2007) and the PickPocket (Zhang et al. 2009). The NetMHCpan has been used previously with good accuracy in a number of non-human studies, for example, in chimpanzees (Pan trglodytes), rhesus macaques (Macaca mulatta), pigs (Sus scrofa), and cattle (Bos primigenius) (Erup Larsen et al. 2011; Hoof et al. 2009; Nene et al. 2012; Pedersen et al. 2011). Furthermore, it has recently been shown that a consensus of the NetMHCpan and the PickPocket outperforms the two methods when the alleles studied are more distantly related to the training data, as is the case in this study (Karosiene et al. 2012).
Pathogens shape the peptide-binding repertoire
Pathogens exert a strong selection pressure on their host. The selection pressure from pathogens is more pronounced in tropical regions were the parasite diversity and abundance is considerably higher than in temperate regions (Stearns and Koella 2007). The selection from pathogens plays a crucial role in shaping the MHC allelic composition within a population (Hill et al. 1991; Prugnolle et al. 2005). An individual with a broad peptide-binding repertoire of the MHC allomorphs is likely to be more successful combating various infections than an individual with a more narrow peptide-binding repertoire. Great reed warblers winter in the tropics and breed in temperate regions, and are thus exposed to more pathogens than house sparrows and tree sparrows that are sedentary and naturally distributed in temperate regions (Cramp 1992). However, individuals with the broadest peptide-binding repertoire are likely to be at an advantage in both tropical and temperate regions, although the actual selection pressure is likely to be stronger in the tropics.
The overlapping peptide-binding specificities, found in the tree based on functional clustering, between great reed warbler and house sparrow MHC class I allomorphs suggests that there has been selection for a similar peptide-binding repertoire in both species despite significantly different amino acid sequences (Figs. 3 and 5). This may be a result of exposure to similar pathogens. For example, both great reed warblers and house sparrows are known to be infected with the avian malaria parasite Plasmodium relictum (strain SGS1) (Loiseau et al. 2011; Zehtindjiev et al. 2008). We did not find any overlap in peptide-binding specificities between great reed warbler and tree sparrow MHC class I allomorphs. However, we believe that this lack of overlap could be an effect of sampling. The overlap in peptide-binding specificity found between great reed warbler and house sparrow suggests either convergent evolution on the level of protein function or trans-species evolution, where the age of an allele is greater than the species, in combination with frequent gene conversion events (Bos and Waldman 2006). The trees based on exon 2, amino acids or synonymous substitutions, suggest that convergent evolution is more likely, since here the gene trees are equal to the species tree for great reed warblers and house sparrows (Online Resource 2). A tree based on exon 4, the structural part of the MHC molecule, also results in a gene tree that is equal to the species tree (data not shown). Meanwhile, the tree based on amino acid sequences on exon 3 suggests trans-species evolution, since here the gene tree is different from the species tree and (e.g.) the great reed warbler sequences do not form a supported monophyletic cluster (online Resource 2). Finally, the tree based on synonymous substitutions in exon 3, results in a gene tree equal to the species tree for great reed warblers and house sparrows, again indicating convergent evolution as the most likely explanation for our findings (Online Resource 2).
MHC class I and the linkage to TAP
Koch et al. (2007) demonstrated that chicken MHC class I molecules have promiscuous peptide-binding (Koch et al. 2007), which could partly explain the low number of allomorphs per individual found in the chicken (Kaufman et al. 1995, 1999; Shaw et al. 2007; Wallny et al. 2006). The predicted functional differences in peptide-binding repertoires and the large number of great reed warbler alleles per individual (Westerdahl et al. 1999) compared to the chicken suggest that the great reed warbler MHC class I allomorphs do not have the promiscuous peptide-binding that has been proposed for chicken MHC class I molecules (Koch et al. 2007). However, without functional studies of expressed passerine MHC class I molecules, this cannot be conclusively determined as it has been in chickens (Koch et al. 2007). In chickens, co-evolution between MHC class I and TAP has been proposed to favour the dominant expression of one MHC class I allomorph (Walker et al. 2011). The large number of MHC class I genes, with different peptide-binding specificity, found in passerines in the present study suggest that the passerine TAP genes are likely to be less specific in what peptides they translocate, thus enabling a more diverse pool of peptides to be bound by different MHC class I molecules compared to in the chicken. In addition, there are indications that the MHC class I and TAP genes map to different chromosomes in the zebra finch, suggesting that these genes are not at all as closely linked as they are in chickens (Balakrishnan et al. 2010; Kaufman et al. 1999). This would indicate that co-evolution between the MHC class I and TAP genes is probably not as pronounced, or even absent, in passerines.
Importance of diversity in MHC class I alleles in passerines
Passerines have been shown to have many MHC class I genes, both functional and non-functional (Bonneaud et al. 2004; Promerova et al. 2009; Schut et al. 2011; Westerdahl 2007; Westerdahl et al. 1999). This large number of genes is intriguing as there are theoretical models and field studies suggesting that the best presentation of pathogenic peptides is accomplished by an optimal number of MHC class I allomorphs rather than a large one, as a high MHC class I diversity would limit the T cell repertoire through negative selection (Nowak et al. 1992; Wegner et al. 2003). In contrast, another model suggests that a higher MHC class I diversity increases the T cell repertoire by positive selection thus making a high number of MHC class I allomorphs beneficial (Borghans et al. 2003). Although passerines, in general, seem to have a larger number of MHC class I genes compared to most mammals, it is most likely not the number of MHC genes per se that determines the ability to fight of a large pool of pathogens but rather the diversity in peptide-binding specificity found within a single individual. An interesting observation, that ought to be further explored, is that the functional diversity that we have seen in the passerines in the present study is smaller than the functional diversity observed in humans. Perhaps a lower functional diversity in MHC class I allomorphs in passerines have been compensated by a larger number of allomorphs per individual.
Conclusions
We investigated selection, sequence divergence and peptide-binding properties of MHC class I alleles/allomorphs in passerines and found differences on the level of MHC class I alleles and allomorph characteristics rather than on a species level. Our interpretation is that convergent evolution on the level of the protein function, possibly driven by selection from shared pathogens, has resulted in allomorphs with similar peptide-binding repertoires, though trans-species evolution in combination with gene conversion cannot be excluded. The peptide-binding prediction data suggests that the large number of MHC class I genes found in passerines probably are of functional importance. However, this study only provides an indication to the MHC class I functional diversity in these passerines and experiments to determine protein expression as well as crystal structures are needed to more fully understand the MHC class I diversity.
References
Aguilar A, Edwards SV, Smith TB, Wayne RK (2006) Patterns of variation in MHC class II beta loci of the little greenbul (Andropadus virens) with comments on MHC evolution in birds. J Hered 97:133–142
Apanius V, Penn D, Slev PR, Ruff LR, Potts WK (1997) The nature of selection on the major histocompatibility complex. Crit Rev Immunol 17:179–224
Axelsson-Robertson R, Ahmed RK, Weichold FF, Ehlers MM, Kock MM, Sizemore D, Sadoff J, Maeurer M (2011) Human leukocyte antigens A*3001 and A*3002 show distinct peptide-binding patterns of the Mycobacterium tuberculosis protein TB10.4: consequences for immune recognition. Clin vaccine immunol CVI 18:125–134
Balakrishnan CN, Ekblom R, Volker M, Westerdahl H, Godinez R, Kotkiewicz H, Burt DW, Graves T, Griffin DK, Warren WC, Edwards SV (2010) Gene duplication and fragmentation in the zebra finch major histocompatibility complex. BMC Biol 8:29
Bjorkman PJ, Saper MA, Samraoui B, Bennett WS, Strominger JL, Wiley DC (1987) The foreign antigen binding site and T cell recognition regions of class I histocompatibility antigens. Nature 329:512–518
Bollmer JL, Dunn PO, Whittingham LA, Wimpee C (2010) Extensive MHC class II B gene duplication in a passerine, the common Yellowthroat (Geothlypis trichas). J Hered 101:448–460
Bonneaud C, Sorci G, Morin V, Westerdahl H, Zoorob R, Wittzell H (2004) Diversity of MHC class I and IIB genes in house sparrows (Passer domesticus). Immunogenetics 55:855–865
Borghans JA, Noest AJ, De Boer RJ (2003) Thymic selection does not limit the individual MHC diversity. Eur J Immunol 33:3353–3358
Bos DH, Waldman B (2006) Evolution by recombination and transspecies polymorphism in the MHC class I gene of Xenopus laevis. Mol Biol Evol 23:137–143
Brown JW, Rest JS, Garcia-Moreno J, Sorenson MD, Mindell DP (2008) Strong mitochondrial DNA support for a Cretaceous origin of modern avian lineages. BMC Biol 6:6
Chaves LD, Krueth SB, Reed KM (2009) Defining the turkey MHC: sequence and genes of the B locus. J Immunol 183:6530–6537
Cramp S (1992) Handbook of the birds of Europe, the Middle East and North Africa. Oxford University Press, Oxford
Ekblom R, Stapley J, Ball AD, Birkhead T, Burke T, Slate J (2011) Genetic mapping of the major histocompatibility complex in the zebra finch (Taeniopygia guttata). Immunogenetics 63:523–530
Erup Larsen M, Kloverpris H, Stryhn A, Koofhethile CK, Sims S, Ndung'u T, Goulder P, Buus S, Nielsen M (2011) HLA restrictor—a tool for patient-specific predictions of HLA restriction elements and optimal epitopes within peptides. Immunogenetics 63:43–55
Guillemot F, Billault A, Pourquie O, Behar G, Chausse AM, Zoorob R, Kreibich G, Auffray C (1988) A molecular map of the chicken major histocompatibility complex: the class II beta genes are closely linked to the class I genes and the nucleolar organizer. EMBO J 7:2775–2785
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Hee CS, Gao S, Loll B, Miller MM, Uchanska-Ziegler B, Daumke O, Ziegler A (2010) Structure of a classical MHC class I molecule that binds "non-classical" ligands. PLoS Biol 8:e1000557
Hill AV, Allsopp CE, Kwiatkowski D, Anstey NM, Twumasi P, Rowe PA, Bennett S, Brewster D, McMichael AJ, Greenwood BM (1991) Common west African HLA antigens are associated with protection from severe malaria. Nature 352:595–600
Hoof I, Peters B, Sidney J, Pedersen LE, Sette A, Lund O, Buus S, Nielsen M (2009) NetMHCpan, a method for MHC class I binding prediction beyond humans. Immunogenetics 61:1–13
Karosiene E, Lundegaard C, Lund O, Nielsen M (2012) NetMHCcons: a consensus method for the major histocompatibility complex class I predictions. Immunogenetics 64:177–186
Katoh K, Misawa K, Kuma K, Miyata T (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30:3059–3066
Kaufman J (1999) Co-evolving genes in MHC haplotypes: the "rule" for nonmammalian vertebrates? Immunogenetics 50:228–236
Kaufman J, Milne S, Gobel TW, Walker BA, Jacob JP, Auffray C, Zoorob R, Beck S (1999) The chicken B locus is a minimal essential major histocompatibility complex. Nature 401:923–925
Kaufman J, Volk H, Wallny HJ (1995) A "minimal essential MHC" and an "unrecognized MHC": two extremes in selection for polymorphism. Immunol Rev 143:63–88
Koch M, Camp S, Collen T, Avila D, Salomonsen J, Wallny HJ, van Hateren A, Hunt L, Jacob JP, Johnston F, Marston DA, Shaw I, Dunbar PR, Cerundolo V, Jones EY, Kaufman J (2007) Structures of an MHC class I molecule from B21 chickens illustrate promiscuous peptide binding. Immunity 27:885–899
Loiseau C, Zoorob R, Robert A, Chastel O, Julliard R, Sorci G (2011) Plasmodium relictum infection and MHC diversity in the house sparrow (Passer domesticus). Proc Biol Sci Roy Soc 278:1264–1272
Lundegaard C, Lamberth K, Harndahl M, Buus S, Lund O, Nielsen M (2008) NetMHC-3.0: accurate web accessible predictions of human, mouse and monkey MHC class I affinities for peptides of length 8–11. Nucleic Acids Res 36:W509–W512
Lundegaard C, Lund O, Buus S, Nielsen M (2010) Major histocompatibility complex class I binding predictions as a tool in epitope discovery. Immunology 130:309–318
Miller HC, Lambert DM (2004) Gene duplication and gene conversion in class II MHC genes of New Zealand robins (Petroicidae). Immunogenetics 56:178–191
Neefjes J, Jongsma ML, Paul P, Bakke O (2011) Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat Rev Immunol 11:823–836
Nene V, Svitek N, Toye P, Golde WT, Barlow J, Harndahl M, Buus S, Nielsen M (2012) Designing bovine T cell vaccines via reverse immunology. Ticks Tick-Borne Dis 3:188–192
Nielsen M, Lund O, Lundegaard C (2012) MHCcluster, a method for functional clustering of MHC molecules. ISCB-Latin
Nielsen M, Lundegaard C, Blicher T, Lamberth K, Harndahl M, Justesen S, Roder G, Peters B, Sette A, Lund O, Buus S (2007) NetMHCpan, a method for quantitative predictions of peptide binding to any HLA-A and -B locus protein of known sequence. PLoS One 2:e796
Nielsen M, Lundegaard C, Lund O, Petersen TN (2010) CPHmodels-3.0—remote homology modeling using structure-guided sequence profiles. Nucleic Acids Res 38:W576–W581
Nowak MA, Tarczyhornoch K, Austyn JM (1992) The optimal number of major histocompatibility complex—molecules in an individual. Proc Natl Acad Sci USA 89:10896–10899
Pedersen LE, Harndahl M, Rasmussen M, Lamberth K, Golde WT, Lund O, Nielsen M, Buus S (2011) Porcine major histocompatibility complex (MHC) class I molecules and analysis of their peptide-binding specificities. Immunogenetics 63:821–834
Promerova M, Albrecht T, Bryja J (2009) Extremely high MHC class I variation in a population of a long-distance migrant, the Scarlet Rosefinch (Carpodacus erythrinus). Immunogenetics 61:451–461
Promerova M, Babik W, Bryja J, Albrecht T, Stuglik M, Radwan J (2012) Evaluation of two approaches to genotyping major histocompatibility complex class I in a passerine-CE-SSCP and 454 pyrosequencing. Mol Ecol Resour 12:285–292
Prugnolle F, Manica A, Charpentier M, Guegan JF, Guernier V, Balloux F (2005) Pathogen-driven selection and worldwide HLA class I diversity. Curr Biol CB 15:1022–1027
Rammensee HG, Friede T, Stevanoviic S (1995) MHC ligands and peptide motifs: first listing. Immunogenetics 41:178–228
Reed KM, Bauer MM, Monson MS, Benoit B, Chaves LD, O'Hare TH, Delany ME (2011) Defining the turkey MHC: identification of expressed class I- and class IIB-like genes independent of the MHC-B. Immunogenetics 63:753–771
Sato A, Mayer WE, Tichy H, Grant PR, Grant BR, Klein J (2001) Evolution of MHC class II B genes in Darwin's finches and their closest relatives: birth of a new gene. Immunogenetics 53:792–801
Schut E, Aguilar JR, Merino S, Magrath MJ, Komdeur J, Westerdahl H (2011) Characterization of MHC-I in the blue tit (Cyanistes caeruleus) reveals low levels of genetic diversity and trans-population evolution across European populations. Immunogenetics 63:531–542
Sepil I, Moghadam HK, Huchard E, Sheldon BC (2012) Characterization and 454 pyrosequencing of Major Histocompatibility Complex class I genes in the great tit reveal complexity in a passerine system. BMC Evol Biol 12:68
Shaw I, Powell TJ, Marston DA, Baker K, van Hateren A, Riegert P, Wiles MV, Milne S, Beck S, Kaufman J (2007) Different evolutionary histories of the two classical class I genes BF1 and BF2 illustrate drift and selection within the stable MHC haplotypes of chickens. J Immunol 178:5744–5752
Shiina T, Shimizu S, Hosomichi K, Kohara S, Watanabe S, Hanzawa K, Beck S, Kulski JK, Inoko H (2004) Comparative genomic analysis of two avian (quail and chicken) MHC regions. J Immunol 172:6751–6763
Sibly RM, Witt CC, Wright NA, Venditti C, Jetz W, Brown JH (2012) Energetics, lifestyle, and reproduction in birds. Proc Natl Acad Sci USA 109:10937–10941
Stamatakis A, Hoover P, Rougemont J (2008) A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol 57:758–771
Stearns SC, Koella JC (2007) Evolution in health and disease. Oxford University Press, New York
Strandh M, Lannefors M, Bonadonna F, Westerdahl H (2011) Characterization of MHC class I and II genes in a subantarctic seabird, the blue petrel, Halobaena caerulea (Procellariiformes). Immunogenetics 63:653–666
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Thomsen MC, Nielsen M (2012) Seq2Logo: a method for construction and visualization of amino acid binding motifs and sequence profiles including sequence weighting, pseudo counts and two-sided representation of amino acid enrichment and depletion. Nucleic acids res
Walker BA, Hunt LG, Sowa AK, Skjodt K, Gobel TW, Lehner PJ, Kaufman J (2011) The dominantly expressed class I molecule of the chicken MHC is explained by coevolution with the polymorphic peptide transporter (TAP) genes. Proc Natl Acad Sci USA 108:8396–8401
Walker BA, van Hateren A, Milne S, Beck S, Kaufman J (2005) Chicken TAP genes differ from their human orthologues in locus organisation, size, sequence features and polymorphism. Immunogenetics 57:232–247
Wallny HJ, Avila D, Hunt LG, Powell TJ, Riegert P, Salomonsen J, Skjodt K, Vainio O, Vilbois F, Wiles MV, Kaufman J (2006) Peptide motifs of the single dominantly expressed class I molecule explain the striking MHC-determined response to Rous sarcoma virus in chickens. Proc Natl Acad Sci USA 103:1434–1439
Warren WC, Clayton DF, Ellegren H, Arnold AP, Hillier LW, Kunstner A, Searle S, White S, Vilella AJ, Fairley S, Heger A, Kong L, Ponting CP, Jarvis ED, Mello CV, Minx P, Lovell P, Velho TA, Ferris M, Balakrishnan CN, Sinha S, Blatti C, London SE, Li Y, Lin YC, George J, Sweedler J, Southey B, Gunaratne P, Watson M, Nam K, Backstrom N, Smeds L, Nabholz B, Itoh Y, Whitney O, Pfenning AR, Howard J, Volker M, Skinner BM, Griffin DK, Ye L, McLaren WM, Flicek P, Quesada V, Velasco G, Lopez-Otin C, Puente XS, Olender T, Lancet D, Smit AF, Hubley R, Konkel MK, Walker JA, Batzer MA, Gu W, Pollock DD, Chen L, Cheng Z, Eichler EE, Stapley J, Slate J, Ekblom R, Birkhead T, Burke T, Burt D, Scharff C, Adam I, Richard H, Sultan M, Soldatov A, Lehrach H, Edwards SV, Yang SP, Li X, Graves T, Fulton L, Nelson J, Chinwalla A, Hou S, Mardis ER, Wilson RK (2010) The genome of a songbird. Nature 464:757–762
Wegner KM, Kalbe M, Kurtz J, Reusch TBH, Milinski M (2003) Parasite selection for immunogenetic optimality. Science 301:1343
Westerdahl H (2007) Passerine MHC: genetic variation and disease resistance in the wild. J Ornithol 148:S469–S477
Westerdahl H, Wittzell H, von Schantz T (1999) Polymorphism and transcription of Mhc class I genes in a passerine bird, the great reed warbler. Immunogenetics 49:158–170
Westerdahl H, Wittzell H, von Schantz T (2000) Mhc diversity in two passerine birds: no evidence for a minimal essential Mhc. Immunogenetics 52:92–100
Wittzell H, Madsen T, Westerdahl H, Shine R, von Schantz T (1998) MHC variation in birds and reptiles. Genetica 104:301–309
Yang Z (1997) PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci CABIOS 13:555–556
Yang Z (2007) PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24:1586–1591
Yang Z, Bielawski JP (2000) Statistical methods for detecting molecular adaptation. Trends Ecol Evol 15:496–503
Yang Z, Wong WS, Nielsen R (2005) Bayes empirical bayes inference of amino acid sites under positive selection. Mol Biol Evol 22:1107–1118
Zagalska-Neubauer M, Babik W, Stuglik M, Gustafsson L, Cichon M, Radwan J (2010) 454 sequencing reveals extreme complexity of the class II Major Histocompatibility Complex in the collared flycatcher. BMC Evol Biol 10:395
Zehtindjiev P, Ilieva M, Westerdahl H, Hansson B, Valkiunas G, Bensch S (2008) Dynamics of parasitemia of malaria parasites in a naturally and experimentally infected migratory songbird, the great reed warbler Acrocephalus arundinaceus. Exp Parasitol 119:99–110
Zhang H, Lund O, Nielsen M (2009) The PickPocket method for predicting binding specificities for receptors based on receptor pocket similarities: application to MHC-peptide binding. Bioinformatics 25:1293–1299
Acknowledgments
This study was financed by grants from the Swedish Research Council, Crafoords foundation and Schybergs foundation (the Royal Physiographic foundation) to Helena Westerdahl, by grants from the Swedish Research Council to Stefan Wallin, by grants from the Swedish Research Council, Kocks foundation, Crafoords foundation, the Royal Physiographic foundation, Groschinskys foundation and Alfred Österlunds foundation to Kajsa Paulsson, and by grants from the foundation of ‘Regementsläkaren Dr Hartelii’ to Elna Follin. We would like to thank Anna Drews and Ester Arévalo Sureda for assistance in the lab and Emily O´Connor and three anonymous referees for giving feedback on the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
K. Paulsson and H. Westerdahl contributed equally to this work.
Rights and permissions
About this article
Cite this article
Follin, E., Karlsson, M., Lundegaard, C. et al. In silico peptide-binding predictions of passerine MHC class I reveal similarities across distantly related species, suggesting convergence on the level of protein function. Immunogenetics 65, 299–311 (2013). https://doi.org/10.1007/s00251-012-0676-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-012-0676-3