Abstract
The passerine major histocompatibility complex (MHC) class I and IIB genes are different from those of the avian model species the chicken because passerines have (1) a larger number of MHC genes, (2) MHC genes with longer introns, and (3) MHC genes that are pseudogenes. Most passerine MHC genes are transcribed (coding), extremely variable and subject to balancing selection. The high genetic diversity of the MHC genes of passerines is most likely maintained by selection from a large number of different pathogens. Association between MHC alleles and resistance to avian malaria infections have been reported in House Sparrows and Great Reed Warblers. Passerines are infected by a large number of different avian malaria infections. Therefore passerines and avian malaria is a study system that is well-suited to investigations of balancing selection and associations between MHC genes and disease resistance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The major histocompatibility complex (MHC) is a well-known gene complex that is found in all vertebrates and it is known to be of importance for survival, and potentially also in mate choice. The MHC genes are the most variable coding genes known in vertebrates and this polymorphism is most likely maintained by selection from pathogens (Klein 1986). In recent years particular MHC alleles have been shown to correlate with parasite burdens in a very divergent group of species from natural populations: Malagasy Mouse Lemur, Yellow-necked Mouse, Three-spined Stickleback, Great Reed Warbler, House Sparrow and Tropical Python (Meyer-Lucht and Sommer 2005; Kurtz et al. 2004; Schad et al. 2005; Westerdahl et al. 2005; Bonneaud et al. 2006b; Madsen and Ujvari 2006). These studies indicate that there are associations between particular MHC alleles in the hosts and the prevalence of certain parasites; hence, there are associations between the host’s genes and the host’s fitness that can be quantified, given that the parasites affect the fitness of their host.
The genetic component of an individual’s phenotype has been investigated using DNA sequences in a large number of molecular ecological studies during the last decade, but only a few investigations have found a good correlation between genotype and actual fitness of the phenotype in the natural selecting environment (Hansson et al. 2001). The MHC genes have the potential to bridge the gap between phenotype and genotype because these genes are related to fitness and survival. It is likely that the MHC provides the best system available in vertebrates for investigating how natural selection can promote local adaptation at the genetic level (Bernatchez and Landry 2003; Piertney and Oliver 2006).
An alternative or additional mechanism for maintaining genetic diversity in the MHC genes is an MHC-based mate choice. Female mate choice may be driven by genetic compatibility in such a way that females choose mates who are genetically compatible with them (reviewed in Tregenza and Wedell 2000). The MHC is a good candidate for such mate choice, since these genes may increase offspring viability and may also be the actual cue used during mate choice (reviewed in Penn and Potts 1999). Several mate choice experiments in mice and some in humans have found that females preferentially mate with males with dissimilar MHC genes, though other studies have failed to find a mating preference (Wedekind et al. 1995; reviewed in Penn and Potts 1999; Eklund et al. 2000). So far, there are a handful of studies that have investigated an MHC-based mate choice in passerines, and none of these studies have found any evidence of a female mate choice based on the male MHC, at least not in the female’s first and main social mate choice (Freeman-Gallant et al. 2003; Westerdahl et al. 2004; Richardson et al. 2005; Bonneaud et al. 2006a). However, two of these mate choice studies have detected an MHC-related mate choice when investigating extra pair paternity (Freeman-Gallant et al. 2003; Richardson et al. 2005).
This short review on current knowledge about passerine MHC genes will focus on three different MHC topics; (1) I will compare the characteristics of passerine MHC genes with those of the avian model species the domestic chicken Gallus gallus; (2) I will then discuss aspects of evolution and selection for the MHC genes in passerines, and; (3) I will finally compare two studies that have investigated associations between MHC alleles and resistance to avian malaria infections and discuss the associations that can be expected.
Characteristics of the passerine MHC genes
MHC genes and selection for polymorphism
The MHC genes encode cell-surface proteins (MHC molecules) that play a key role in adaptive immune defense. Each MHC molecule can bind a limited number of peptides, and if the bound peptide is from a pathogen there will be an appropriate immune reaction (Abbas 1994). The leading hypothesis for the diversity of the MHC is that there is an ongoing evolutionary arms race between the pathogens in the environment and the MHC genes of their hosts (Potts and Wakeland 1990; Borghans et al. 2004). Different MHC alleles will be at an advantage at different times, and therefore a large number of different MHC alleles will be maintained within populations over very long time intervals (Edwards and Hedrick 1998). It is advantageous for each individual to carry rare beneficial MHC alleles to which the pathogens have not adapted or to be heterozygous at MHC loci (Bodmer 1972; Hedrick 2002). The latter is advantageous either because heterozygote individuals are more likely than homozygote individuals to carry a rare beneficial MHC allele by chance, or because it is advantageous to be heterozygote at a locus per se—the so-called heterozygote advantage (Doherty and Zinkernagel 1975a, 1975b; Hughes and Hughes 1995).
MHC genes in birds
There are two classes of MHC genes—MHC class I and MHC class II—where the function is well understood. MHC class I deals predominantly with intracellular infections (e.g., viruses) and MHC class II mostly with extracellular infections (e.g., bacteria) (Abbas 1994; Hughes and Yeager 1998). There are also infections like malaria which most likely activate both MHC classes I and II. The domestic chicken has a “minimal essential MHC,” which means that it has few genes in the MHC region on chromosome 16, few copies of the MHC class I and II genes, and also that each gene is of a small size (i.e., it has short introns). The chicken has two class I and two class II genes at the classical MHC-B locus (Kaufman 1999; Kaufman et al. 1999). Furthermore, the chicken has a MHC-Y locus. The function and expression level of the genes at this locus is not known, and at this locus there are two or three additional class I and II genes, respectively (Miller et al. 2004). The Ring-necked Pheasant Phasianus colchicus possesses two MHC class I and two class II genes at the MHC-B locus and the Black Grouse Tetrao tetrix also has a low number of MHC-B and MHC-Y class II genes (Wittzell et al. 1999; Strand et al. 2007). These three galliforms, chicken, Ring-necked Pheasant and Black Grouse, all have a low number of MHC class II genes, and so do penguins (Bollmer et al. 2007). However, except for these two bird orders, all of the birds studied so far have presented a larger number of MHC class I and II genes (Hess and Edwards 2002; Ekblom 2003), and this finding is particularly pronounced for passerine birds (e.g., Bonneaud et al. 2004; Miller and Lambert 2004a; Richardson and Westerdahl 2003).
MHC genes in passerines
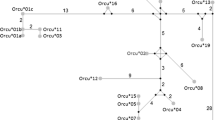
Passerines have a considerably higher number of MHC genes than the chicken, and they have class I and II genes that are much larger than chickens, hence the introns are longer in passerines (class I, Great Reed Warbler, Westerdahl et al. 2004b; class II, Red-winged Blackbird, Edwards et al. 1998; House Finch, Hess et al. 2000; Fig. 1). It is not clear whether there is an MHC-Y locus in passerines, although MHC alleles with low variability have been reported (Jarvi et al. 2004). Pseudogenes (nonfunctional genes) have been reported for MHC classes I and II in passerines, and MHC class I and II pseudogenes have not yet been reported in chicken (Table 1). In Great Reed Warblers the MHC class I pseudogenes have a 5 bp deletion in exon 3 that leads to a shift in the reading frame, and these pseudogenes are less variable than the transcribed class I genes (Westerdahl et al. 1999, 2004b). The House Sparrow has two groups of transcribed alleles for MHC class I, one group that is very variable and another group that is much less variable (Bonneaud et al. 2004). The general picture from the genetic organization of the MHC genes in passerines is that they are much more complex than in chicken due to the passerines having (1) a larger number of genes, (2) genes that are highly polymorphic and also genes that have low diversity within the classical MHC region, and (3) pseudogenes (Fig. 1; Table 1).
Comparison of the genomic organization (indicated as number of loci) of MHC for chicken and passerines. Available number of MHC loci in the Great Reed Warbler represent the number of passerine MHC loci here. MHC class I loci are indicated with filled squares, MHC class II with open squares, and Y-MHC loci that have not been identified as grey squares
MHC alleles are homogenized
In passerines it is difficult to determine the locus to which a certain MHC class I or II allele belongs. The high similarity of alleles between loci is most likely the result of gene conversion. Gene conversion is a process where a sequence fragment is transferred from one gene to another (segmental mutation), where the origin of the template sequence is outside the mutated gene. This process causes alleles from different loci to become similar to one another, at least when longer fragments are transferred, and it can therefore erase locus-specific patterns of alleles (Högstrand and Böhme 1999). Gene conversion has been observed both within and between loci in several species and, e.g., in the Ring-necked Pheasant identical MHC class II alleles have been reported at the two class IIB loci (Wittzell et al. 1999). When screening for MHC diversity in study populations we preferentially want to amplify alleles from a single locus at a time, primarily because we want to know the locus to which a certain allele belongs, but also for practical reasons when calculating, e.g., heterozygosity estimates. However, it is important to amplify alleles from several loci since it is not known which locus is the most informative or is subject to the strongest selection in most species. Furthermore, we need information from several loci to obtain more knowledge about gene conversion within and between loci. In passerines it seems difficult to amplify alleles from a single locus at a time. One approach that potentially could separate alleles on a locus basis is to study a noncoding region of the gene. The 3′-untranslated region of the mRNAs could potentially indicate the locus from which a certain allele comes. This has been suggested for both class II in South Island Robins and for class I in Great Reed Warblers (Miller and Lambert 2004a; Westerdahl et al. 1999). However, in general, a locus-specific amplification might be less critical in species where the locus-specific patterns of alleles have been erased by the gene conversion process, because in this case functional alleles (irrespective of level of expression) potentially look very similar. Locus-specific amplification, and hence separation of alleles per locus, has actually been reported for class II in Red-winged Blackbird (Gasper et al. 2001). Genome sequencing of the entire MHC region in several passerines is most likely the best way to find locus specificity in passerine MHC genes. The MHC sequences from cosmid libraries (House Finch, Little Green Bull and Red-winged Blackbird) and the Zebra Finch genome projects are a significant step in that direction (Edwards et al. 2000; Hess et al. 2000; Gasper et al. 2001; Aguilar et al. 2006).
Aspects of evolution and selection of the MHC genes
Trans-species evolution of MHC alleles
A large number of MHC alleles are maintained within a population over long time periods, and this is likely to be accomplished by balancing selection (Hedrick et al. 2002). Trans-species evolution of MHC alleles suggests that not only will MHC alleles be maintained in populations, but MHC alleles will even survive several speciation events [alleles are identical in the regions of the allele that encode the peptide binding region (PBR) of the MHC molecule], and a phylogenetic tree based on MHC genes from several species can therefore be in discordance with the true species tree (Edwards and Hedrick 1998). The most frequently mentioned explanatory forces behind balancing selection are negative frequency-dependent selection and heterozygote advantage (Bodmer et al. 1972; Doherty and Zinkernagel 1975a, 1975b). Negative frequency-dependent selection is when rare alleles are at an advantage while more common alleles are selected against (Bodmer et al. 1972). Heterozygote advantage is when individuals that carry two different alleles at a locus are at an advantage compared with individuals that carry a single allele (Doherty and Zinkernagel 1975a, 1975b; Parham and Ohta 1996). Great Reed Warblers and Seychelles Warblers are both members of the genus Acrocephalus and MHC class I alleles from these warblers do not form separate clusters on a species level in a phylogenetic tree. Aguilar et al. (2006) investigated how two different exons (exon 2 and exon 3) of the class IIB alleles from passerines clustered in phylogenetic trees. As expected, the exon 2 sequences (which encode the PBR for MHC class II and are subject to strong balancing selection) from related species, e.g., House Finch and Bengalese Finch, did not cluster in a species-specific manner. Less related species, e.g., finches and warblers, did form separate clusters in the phylogenetic tree, so species that have been diverged for longer time periods often have diverged MHC alleles even though the MHC sequence is subject to balancing selection. When Aguilar et al. (2006) analyzed class IIB sequences from exon 3 in a phylogenetic tree, these sequences did cluster in a species-specific way because exon 3 encodes a structural region of the MHC class II molecule which is not subject to balancing selection.
A selection pressure from a single pathogen that is shared between two diverged species, like Great Reed Warblers and House Sparrows, could potentially counteract divergence and even maintain identical MHC alleles in these two distantly related species. Great Reed Warblers and House Sparrows are, e.g., known to be infected by identical strains of avian malaria (Plasmodium relictum GRW4) and these bird species could therefore be subject to similar selection pressures from the avian malaria infection GRW4. However, identical MHC alleles (considering the PBR) have not yet been reported in Great Reed Warblers and House Sparrows.
Balancing selection
The classical way to investigate whether an MHC gene has been subject to balancing selection is to investigate the ratio between nonsynonymous and synonymous substitutions (dN/dS) in the PBR (Li 1997). This dN/dS measure gives an indication of whether the gene actually has been subject to balancing selection or not, but the dN/dS ratios does not tell us whether a gene is currently under balancing selection. This is because the dN/dS pattern was most likely generated a long time ago, and thus visualizes an ancient selection. Often it is an ongoing selection that we want to be able to detect and investigate. A few avian studies have recently tried to screen for ongoing balancing selection in the MHC genes. Westerdahl et al. (2004a) showed that the MHC class I alleles in Great Reed Warblers varied significantly in frequency between different cohorts of breeding adults. This result suggests that there is selection on the MHC genes and that selection is occurring in the Great Reed Warbler population at present. However, this study did not show that rare alleles are at an advantage and hence that negative frequency-dependent selection was actually acting. Miller and Lambert (2004b) compared the genetic diversity in large source populations of New Zealand Robins Petroicidae with those of bottlenecked island populations. They screened for diversity in both neutral markers and in selected MHC alleles 20 years after five individuals had been transferred from source to island populations. The expected result was that the genetic polymorphism should be maintained to a higher degree in the MHC alleles compared with the neutral markers, although their results did not show any difference between the neutral and selected genetic markers. Very few individuals actually founded the island populations (two and four), which could explain the limited amount of MHC genetic variation that was observed.
MHC alleles and resistance to avian malaria infections
Selection from pathogens
The high genetic diversity in the MHC genes is likely maintained by selection from a large number of different pathogens and not by a single pathogen, because a single pathogen would exert a uniform directional selection pressure resulting in few MHC alleles, and hence in low MHC diversity. As mentioned above, negative frequency-dependent selection could potentially maintain the MHC diversity, but then the prevalence of pathogens must vary in either time and/or space (Hedrick 2002). Individuals that are heterozygous at an MHC locus could possibly mount an immune response against more pathogens than an individual that is homozygous at that locus. Therefore, heterozygotes are beneficial and heterozygote advantage is, as mentioned earlier, a selective force that could maintain some of the MHC diversity. Different species of avian malaria (including parasites from the genera Plasmodium, Haemoproteus and Leucocytozoon) could be one set of pathogens that maintains the MHC genetic diversity in passerines because (a) these pathogens are common, (b) experiments have shown that they could be lethal, and (c) their prevalence varies in time and space (Atkinson and van Riper 1991; Bensch et al. 2000; Ricklefs and Fallon 2002; Bensch and Åkesson 2003).
Associations between prevalence of malaria and MHC alleles
Two recent passerine studies, one in House Sparrows and one in Great Reed Warblers, investigated associations between prevalence of malaria infections and presence of certain MHC class I alleles. Prevalence of avian malaria infections were screened by polymerase chain reaction (PCR) that amplified parts of the cytochrome b gene of avian malaria parasites (of the genera Plasmodium and Heamoproteus) from DNA extracted from avian blood according to Waldenström et al. (2004). Bonneaud et al. (2006b) investigated associations between malaria infection SGS1 and ten different MHC class I alleles in two populations of resident House Sparrows. The prevalence of the SGS1 infection was high, 68 and 56%, respectively, in the two populations, and it is therefore likely that this is a relatively mild form of avian malaria. The authors found a negative association between MHC allele a151 and the SGS1 infection in one population and between MHC allele a172 and SGS1 in the other study population.
Westerdahl et al. (2005) investigated associations between three different malaria infections (GRW1, GRW2 and GRW4) and 23 different MHC class I alleles in a long-distance migratory bird, the Great Reed Warbler. It should be noted that these three malaria infections are all transmitted at the African wintering grounds and that the Great Reed Warblers are only studied at their Swedish breeding grounds (Waldenström et al. 2002; Bensch et al. 2007). The authors found a positive association between MHC allele B4b and malaria infection GRW2. The prevalence of GRW2 is low in the study population—8% of the individuals carry the infection—and preliminary experimental data suggests that GRW2 is severe (even lethal) in Great Reed Warblers (S. Bensch, unpublished).
At first thought it is surprising that there is a negative association between prevalence of an avian malaria infection and presence of an MHC allele in the House Sparrow, while there is a positive association between avian malaria infection and an MHC allele in the Great Reed Warbler. However, if there is a difference in virulence between the two malaria infections, one being benign and the other lethal, then there is a reasonable explanation for these findings. The virulence of the malaria parasites affects its competitive ability and more virulent strains have been shown to have a competitive advantage in mixed-strain infections (de Roode et al. 2005; Råberg et al. 2006). According to studies of human malaria, the genetic resistance to malaria varies markedly from population to population, and the resistance genes can affect different parts of the immune defense, from parasite invasion to immune responses to the infection (Hill et al. 1997).
An MHC allele that provides resistance to an avian malaria infection could potentially either give full protection against the infection or it could make a potentially lethal malaria infection milder. Hill et al. (1991) found that children with certain MHC haplotypes were resistant to lethal cerebral malaria even though all children in the study were suffering from a milder clinical malaria infection. The virulence of an avian malaria infection probably affects the sign of the association that should be expected. A nonlethal malaria infection is likely to generate a negative association between prevalence of a malaria infection and presence of an MHC allele, provided that the MHC allele gives complete resistance against the malaria infection. This negative association comes from the idea that there are no individuals that are simultaneously infected and carrying the resistance allele, because these individuals never become infected (nonexistent, Fig. 2a). A lethal malaria infection is on the other hand likely to generate a positive association between prevalence of a malaria infection and presence of an MHC allele that gives resistance. In this scenario only the individuals that carry the resistance allele survive the malaria infection, while the individuals that do not carry the MHC allele die from the infection. The individuals that are infected and do not carry the resistance allele do not exist long enough to be sampled (infected, died, Fig. 2b). These two hypotheses should be testable in an experimental setup where wild birds are inoculated with either mild or severe malaria infections (Zehtindjiev et al., submitted to Exp Parasitol), although several factors like the genetic backgrounds of the birds and the natural selection in the original wild population preferably need to be taken into account.
a House Sparrow individuals that carried certain MHC alleles never became infected with malaria (nonexistent), and this results in a negative association between prevalence of malaria and certain MHC alleles (Bonneaud et al. 2006b). b In the Great Reed Warbler, only individuals that carried a specific identified MHC allele survived the severe tropical malaria infection GRW2, and this generates a positive association between prevalence of malaria and a certain MHC allele (Westerdahl et al. 2005)
The results from the House Sparrow study fit with Fig. 2a, where individuals with a certain resistance MHC allele do not become infected with the relatively mild avian malaria infection. The data from the Great Reed Warbler study fit with Fig. 2b, because here only individuals with a certain resistance allele survive when they become infected with lethal malaria. Individuals that do not carry the resistance allele die from the lethal malaria infection.
Detection of avian malaria infections in natural populations
In both the House Sparrow and the Great Reed Warbler studies, it would have been preferable to separate individuals that never were infected from those that completely cleared the infection, because this would have made the associations between malaria infection and the presence of MHC allele more linear and it would have also decreased the variance of the association. There are really five different groups of individuals that we preferably want to separate when studying avian malaria infections in natural populations: (a) individuals that become infected and later die from the malaria infection; (b) individuals that become infected, survive and then carry the infection chronically; (c) individuals that become infected, survive, and completely clear the infection; (d) individuals that were exposed to the infection but never became infected, and finally; (e) individuals that never were exposed to the infection. In migratory birds that become infected at their wintering site, and are only studied at their breeding site (like the Great Reed Warbler), group (a) will never be observed, because these birds die from the primary malaria infection at their wintering site. In resident birds (like the House Sparrow), it is theoretically possible to catch all five groups of birds, although (d) and (e) are more or less impossible to separate. However, all birds disperse to some degree, and a major problem when studying the prevalence of avian malaria in natural populations is that it is not possible to separate birds that have dispersed from those that have died from avian malaria. Furthermore, in order to separate birds that have cleared the infection (group c) from those that were never infected (groups d and e), each bird needs to be caught repeatedly during the period when the malaria infection is spread. An alternative strategy for separating groups (c) and (d and e) is to use a standard immune-blotting technique where plasma samples from the birds are tested for prevalence of antimalarial antibodies (Jarvi et al. 2002).
Study systems for avian malaria and disease resistance
One major difference between the Great Reed Warbler and House Sparrow studies is that House Sparrows get infected with avian malaria at their breeding site, while Great Reed Warblers, which are long-distance migrants, get infected at their wintering site in Africa (Waldentröm et al. 2002; Bonneaud et al. 2006b). In House Sparrows, the prevalence of a mild malaria infection was measured whether a bird was carrying the infection or not, and no consequences of fitness were reported (Bonneaud et al. 2006b). In the Great Reed Warbler study, only birds that actually returned to their breeding site were screened for prevalence of malaria. The authors suggest that Great Reed Warblers without advantageous MHC alleles did not return to breed if they had been infected with severe malaria because then they would have died at their wintering site (Westerdahl et al. 2005). There appears to be no fitness effect of carrying avian malaria infections chronically (GRW1, 2 or 4) at the breeding site in Great Reed Warblers (Bensch et al. 2007). Fitness costs of malaria infection are likely to be most severe during the primary infection (Atkinson et al. 2001), and for Great Reed Warblers the primary infection occurs in Africa where mortality cannot be observed. Birds that are sampled shortly after they become infected with malaria potentially offer a better study system than these Great Reed Warblers because then it would be possible to measure fitness effects that are directly related to the malaria infection. This means that resident birds (e.g., House Sparrows) or birds that are sampled when they become infected with malaria are more suitable study systems. Scientists tend to screen for the prevalence of avian malaria in surviving birds, because it is extremely rare to catch birds that are about to die from an avian malaria infection. This might explain why a limited number of studies have found fitness effects caused by avian malaria infections under natural conditions (Richner et al. 1995; Opplinger et al. 1996; Sol et al. 2003). Experiments, both in the laboratory and in the field, have on the other hand repeatedly detected severe and even lethal effects of avian malaria infections (Hayworth et al. 1987; Nordling et al. 1998; Marzal et al. 2005; Tomas et al. 2007). A potential way to improve the study system of avian malaria and resistance alleles is to use a closed but natural avian population. Data from a stationary island population where dispersing birds could be separated from birds that have died from malaria would be very informative.
Conclusions
All passerines studied so far have a larger number of MHC class I and IIB genes than the avian model species the chicken. Passerine MHC genes can be either coding genes or pseudogenes, and the coding genes are polymorphic and have been subject to balancing selection. The MHC alleles are similar between related species and this indicates trans-species evolution. An association between particular MHC alleles and resistance to an avian malaria infection has been reported in both House Sparrows and Great Reed Warblers. The MHC haplotype (or MHC alleles) has now been shown to determine the outcome of malaria infections in humans, mice and birds (Hill et al. 1991; Wedekind et al. 2005; Westerdahl et al. 2005; Bonneaud et al. 2006b). Passerines are infected by a large number of different avian malaria infections and are hence a well-suited species group for future studies on associations between MHC and disease resistance.
References
Abbas AK, Lichtman AH, Pober JS (1994) Cellular and molecular immunology. WB Saunders, Philadelphia, PA
Aguilar A, Edwards SV, Smith TB, Wayne RK (2006) Patterns of variation in MHC class II ß loci of the little greenbul (Andropadus virens) with comments on MHC evolution in birds. J Hered 97:133–142
Atkinson CT, van Riper C (1991) Pathogenicity and epizootiology of avian haematozoa: Plasmodium, Leucocytozoon and Haemoproteus. In: Loye JE, Zuk M (eds) Bird−parasite interactions. Oxford University Press, New York
Atkinson CT, Dusek RJ, Lease JK (2001) Serological responses and immunity to superinfection with avian malaria in experimentally-infected Hawaii amakihi. J Wildl Dis 37:20–27
Bensch S, Åkesson S (2003) Temporal and spatial variation of hematozoans in Scandinavian willow warblers. J Parasitol 89:388–391
Bensch S, Stjernman M, Hasselquist D, Ostman O, Hansson B, Westerdahl H, Pinheiro RT (2000) Host specificity in avian blood parasites: a study of Plasmodium and Haemoproteus mitochondrial DNA amplified from birds. Proc R Soc Lond B 267:1583–1589
Bensch S, Waldenström J, Jonzén N, Westerdahl H, Hansson B, Sejberg D, Haselquist D (2007) Temporal dynamics and diversity of avian malaria parasites in a single host species. J Anim Ecol 76:112–122
Bernatchez L, Landry C (2003) MHC studies in nonmodel vertebrates: what have we learned about natural selection in 15 years? J Evol Biol 16:363–377
Bodmer WF (1972) Evolutionary significance of the HLA system. Nature 237:139–145
Bollmer JL, Vargas FH, Parker RG (2007) Low MHC variation in the endangered Galápagos penguin (Spheniscus mendiculus). Immunogenetics 59:593–602
Bonneaud C, Sorci G, Morin V, Westerdahl H, Zoorob R, Wittzell H (2004) Diversity of MHC class I and IIB genes in house sparrows (Passer domesticus). Immunogenetics 55:855–865
Bonneaud C, Richard M, Faivre B, Westerdahl H, Sorci G (2006a) Complex MHC-based mate choice in a wild passerine. Proc R Soc Lond B 273:1111–1116
Bonneaud C, Perez-Tris J, Federici P, Chastel O, Sorci G. (2006b) MHC alleles confer local resistance to malaria in a wild passerine. Evolution 60:383–389
Borghans JAM, Beltman JB, De Boer RJ (2004) MHC polymorphism under host–pathogen coevolution. Immunogenetics 55:732–739
Doherty PC, Zinkernagel RM (1975a) A biological role for the major histocompatibility antigens. Lancet 1:1406
Doherty PC, Zinkernagel RM (1975b) Enhanced immunological surveillance in mice heterozygous at the H-2 gene complex. Nature 256:50–52
Edwards SV, Grahn M, Potts WK (1995) Dynamics of MHC evolution in birds and crocodilians: amplification of class II genes with degenerate primers. Mol Ecol 4:719–729
Edwards SV, Gasper J, March M (1998) Genomics and polymorphism of Agph-DAB1, an MHC class II B gene in red-winged blackbirds (Agelaius phoeniceus). Mol Biol Evol 15:236–50
Edwards SV, Gasper J, Garrigan D, Martindale D, Koop BF (2000) A 39-kb sequence around a blackbird MHC class II gene: ghost of selection past and songbird genome architecture. Mol Biol Evol 17:1384–95
Edwards SV, Hedrick PW (1998) Evolution and ecology of MHC molecules: from genomics to sexual selection. Trends Ecol Evol 13:305–311
Ekblom R, Grahn M, Höglund J (2003) Patterns of polymorphism in the MHC class II of a non-passerine bird, the great snipe (Gallinago media). Immunogenetics 54:734–741
Eklund AC, Belchak MM, Lapidos K, Raha-Chowdhury R, Ober C (2000) Polymorphisms in the HLA-linked olfactory receptor genes in the Hutterites. Hum Immunol 61:711–717
Freeman-Gallant CR, Johnson EM, Saponara F, Stanger M (2002) Variation at the MHC in Savannah sparrows. Mol Ecol 11:1125–1130
Freeman-Gallant CR, Meguerdichian M, Wheelwright NT, Sollecito SV (2003) Social pairing and female mating fidelity predicted by RFLP similarity at the major histocompatibility complex in a songbird. Mol Ecol 12:3077–3083
Gasper JS, Shiina T, Inoko H, Edwards SV (2001) Songbird genomics: analysis of 45 kb upstream of a polymorphic MHC class II gene in red-winged blackbirds (Agelaius phoeniceus). Genomics 75:26–34
Hansson B, Bensch S, Hasselquist D, Akesson M (2001) Microsatellite diversity predicts recruitment of sibling great reed warblers. Proc R Soc Lond B 268:1287–1291
Hayworth AM, van Riper C III, Weathers WW (1987) Effects of plasmodium relictum on the metabolic rate and body temperature in canaries (Serinus canarius). J Parasitol 73:850–853
Hedrick PW (2002) Pathogen resistance and genetic variation at MHC loci. Evolution 56:1902–1908
Hess CM, Edwards SV (2002) The evolution of the major histocompatibility complex in birds. Bioscience 52:423–431
Hess CM, Gasper J, Hoekstra HE, Hill CE, Edwards SV (2000) MHC class II pseudogene and genomic signature of a 32-kb cosmid in the house finch (Carpodacus mexicanus). Genome Res 10:613–23
Hill AVS, Allsopp CEM, Kwiatkowski D, Anstey NM, Twumasi P, Rowe PA, Bennett S, Brewster D, McMichael AJ, Greenwood BM (1991) Common West African HLA antigens are associated with protection from severe malaria. Nature 352:595–600
Hill AVS, Jepson A, Plebanski M, Gilbert SC (1997) Genetic analysis of host–parasite coevolution in human malaria. Philos Trans R Soc Lond B 352:1317–1325
Hughes AL, Hughes MK (1995) Natural selection on the peptide-binding regions of major histocompatibility complex molecules. Immunogenetics 42:233–43
Hughes AL, Yeager M (1998) Natural selection at major histocompatibility complex loci of vertebrates. Annu Rev Genet 32:415–35
Högstrand K, Böhme J (1999) Gene conversion can create new MHC alleles. Immunol Rev 167:305–317
Jarvi SI, Schultz JJ, Atkinson CT (2002) PCR diagnostics underestimate the prevalence of avian malaria (Plasmodium relictum) in experimentally-infected passerines. J Parasitol 88:153–8
Jarvi SI, Tarr CL, McIntosh CE, Atkinson CT, Fleischer RC (2004) Natural selection of the major histocompatibility complex (MHC) in Hawaiian honeycreepers (Drepanidinae). Mol Ecol 13:2157–2168
Kaufman J (1999) Co-evolving genes in MHC haplotypes: the “rule” for nonmammalian vertebrates? Immunogenetics 50:228–236
Kaufman J, Jacob J, Shaw I, Walker B, Milne S, Beck S, Salomonsen J (1999) Gene organisation determines evolution of function in the chicken MHC. Immunol Rev 167:101–117
Klein J (1986) Natural history of the major histocompatibility complex. Wiley, New York
Kurtz J, Kalbe M, Aeschlimann PB, Haberli MA, Wegner KM, Reusch TB, Milinski M (2004) Major histocompatibility complex diversity influences parasite resistance and innate immunity in sticklebacks. Proc R Soc Lond B 271:197–204
Li W-H (1997) Molecular evolution. Sinauer, Sunderland, MA
Madsen T, Ujvari B (2006) MHC class I variation associates with parasite resistance and longevity in tropical pythons. J Evol Biol 19:1973–1978
Marzal A, de Lope F, Navarro C, Pape Møller A (2005) Malarial parasites decrease reproductive success: an experimental study in a passerine bird. Oecologia 142:541–45
Meyer-Lucht Y, Sommer S (2005) MHC diversity and the association to nematode parasitism in the yellow-necked mouse (Apodemus flavicollis). Mol Ecol 14:2233–2243
Miller HC, Lambert DM (2004a) Gene duplication and gene conversion in class II MHC genes of New Zealand robins (Petroicidae). Immunogenetics 56:178–91
Miller HC, Lambert DM (2004b) Genetic drift outweighs balancing selection in shaping post-bottleneck major histocompatibility complex variation in New Zealand robins (Petroicidae). Mol Ecol 13:3709–3721
Miller MM, Bacon LD, Hala K, Hunt HD, Ewald SJ, Kaufman J, Zoorob R, Briles WE (2004) Nomenclature for the chicken major histocompatibility (B and Y) complex. Immunogenetics 56:261–279
Nordling D, Andersson M, Zohari S, Gustafsson L (1998) Reproductive effort reduces specific immune response and parasite resistance. Proc R Soc Lond B 265:1291–1298
Oppliger A, Christe P, Richner H (1996) Clutch size and malaria resistance. Nature 381:565
Parham P, Ohta T (1996) Population biology of antigen presentation by MHC class I molecules. Immunogenetics 272:67–74
Penn DJ, Potts WK (1999) The evolution of mating preferences and major histocompatibility complex genes. Am Nat 153:145–164
Piertney SB, Oliver MK (2006) The evolutionary ecology of the majotr histocompatibility complex. Heredity 96:7–21
Potts WK, Wakeland EK (1990) Evolution of diversity at the major histocompatibility complex. Trends Ecol Evol 5:181–187
Raberg L, de Roode JC, Bell AS, Stamou P, Gray D, Read AF (2006) The role of immune-mediated apparent competition in genetically diverse malaria infections. Am Nat 168:41–53
Richardson DS, Westerdahl H (2003) MHC diversity in two Acrocephalus species: the outbred great reed warbler and the inbred Seychelles warbler. Mol Ecol 12:3523–3529
Richner H, Christe P, Oppliger A (1995) Paternal investment affects prevalence of malaria. Proc Natl Acad Sci USA 92:1192–1194
Richardson DS, Komdeur J, Burke T, von Schantz T (2005) MHC-based patterns of social and extra-pair mate choice in the Seychelles warbler. Proc R Soc Lond B 272:759–767
Ricklefs RE, Fallon SM (2002) Diversification and host switching in avian malaria parasites. Proc R Soc Lond B 269:885–892
de Roode JC, Helinski ME, Anwar MA, Read AF (2005) Dynamics of multiple infection and within-host competition in genetically diverse malaria infections. Am Nat 166:531–542
Schad J, Ganzhorn JU, Sommer S (2005) Parasite burden and constitution of major histocompatibility complex in the Malagasy mouse lemur, Microcebus murinus. Evolution 59:439–450
Sol D, Jovani R, Torres J (2003) Parasite mediated mortality and host immune response explain age-related differences in blood parasitism in birds. Oecologia 135:542–547
Strand T, Westerdahl H, Höglund J, Alatalo R, Siitari H (2007) The Mhc class II of the black grouse (Tetrao tetrix) consists of low numbers of B and Y genes with variable diversity and expression. Immunogenetics 59:725–734
Tomas G, Merino S, Moreno J, Morales J, Martinez-De la Puente J (2007) Impact of blood parasites on immunoglobulin level and parental effort: a medication field experiment on a wild passerine. Funct Ecol 21:125–1323
Tregenza T, Wedell N (2000) Genetic compatibility, mate choice and patterns of parentage: invited review. Mol Ecol 9:1013–1027
Vincek V, Klein D, Graser RT, Figueroa F, O'Huigin C, Klein J (1995) Molecular cloning of major histocompatibility complex class II B gene cDNA from the Bengalese finch Lonchura striata. Immunogenetics 42:262–267
Vincek V, O’Huigin C, Satta Y, Takahata N, Boag PT, Grant PR, Grant BR, Klein J (1997) How large was the founding population of Darwin’s finches? Proc R Soc Lond B 264:111–118
Waldenström J, Bensch S, Kiboi S, Hasselquist D, Ottosson U (2002) Cross-species infection of blood parasites between resident and migratory songbirds in Africa. Mol Ecol 11:1545–1554
Waldenström J, Bensch S, Hasselquist D, Östman Ö (2004) A new nested PCR method very efficient in detecting Plasmodium and Haemoproteus infections from avian blood. J Parasitol 90:191–194
Wedekind C, Seebeck T, Bettens F, Paepke AJ (1995) MHC-dependent mate preferences in humans. Proc R Soc Lond B 260:245–249
Wedekind C, Walker M, Little TJ (2005) The course of malaria in mice: major histocompatibility complex (MHC) effects, but no general MHC heterozygote advantage in single-strain infections. Genetics 170:1427–1430
Westerdahl H (2004) No evidence of an MHC based mate choice in the great reed warbler. Mol Ecol 13:2465–2470
Westerdahl H, Wittzell H, von Schantz T (1999) Polymorphism and transcription of MHC class I genes in a passerine bird, the great reed warbler. Immunogenetics 49:158–170
Westerdahl H, Wittzell H, von Schantz T (2000) MHC diversity in two passerine birds: no evidence for a minimal essential MHC. Immunogenetics 52:92–100
Westerdahl H, Hanssson B, Bensch S, Hasselquist D (2004a) Between-year variation of MHC allele frequencies in great reed warblers: selection or drift? J Evol Biol 17:485–492
Westerdahl H, Wittzell H, von Schantz T, Bensch S (2004b) MHC class I typing in a songbird with numerous loci and high polymorphism using motif-specific PCR and DGGE. Heredity 92:534–542
Westerdahl H, Waldenström J, Hansson B, Hasselquist D, Bensch S (2005) An association between malaria infection and MHC in great reed warblers. Proc R Soc Lond B 272:1511–1518
Wittzell H, Bernot A, Auffray C, Zoorob R (1999) Concerted evolution of two MHC class II B loci in pheasants and domestic chickens. Mol Biol Evol 16:479–490
Acknowledgements
The present study was financed by grants from the Swedish Research Council, The Royal Swedish Academy of Sciences and Lunds Djurskyddsfond to Helena Westerdahl. I would like to thank Matt Hale, Staffan Bensch and Lars Råberg for reading and improving earlier versions of this manuscript. I also want to thank Terry Burke (my PostDoc Host) and Jarrod Hadfield for helping me to improve the IOC talk on which this manuscript is based.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by F. Bairlein.
Rights and permissions
About this article
Cite this article
Westerdahl, H. Passerine MHC: genetic variation and disease resistance in the wild. J Ornithol 148 (Suppl 2), 469–477 (2007). https://doi.org/10.1007/s10336-007-0230-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10336-007-0230-5