Abstract
Binding of peptides to major histocompatibility complex (MHC) molecules is the single most selective step in the recognition of pathogens by the cellular immune system. The human MHC genomic region (called HLA) is extremely polymorphic comprising several thousand alleles, each encoding a distinct MHC molecule. The potentially unique specificity of the majority of HLA alleles that have been identified to date remains uncharacterized. Likewise, only a limited number of chimpanzee and rhesus macaque MHC class I molecules have been characterized experimentally. Here, we present NetMHCpan-2.0, a method that generates quantitative predictions of the affinity of any peptide–MHC class I interaction. NetMHCpan-2.0 has been trained on the hitherto largest set of quantitative MHC binding data available, covering HLA-A and HLA-B, as well as chimpanzee, rhesus macaque, gorilla, and mouse MHC class I molecules. We show that the NetMHCpan-2.0 method can accurately predict binding to uncharacterized HLA molecules, including HLA-C and HLA-G. Moreover, NetMHCpan-2.0 is demonstrated to accurately predict peptide binding to chimpanzee and macaque MHC class I molecules. The power of NetMHCpan-2.0 to guide immunologists in interpreting cellular immune responses in large out-bred populations is demonstrated. Further, we used NetMHCpan-2.0 to predict potential binding peptides for the pig MHC class I molecule SLA-1*0401. Ninety-three percent of the predicted peptides were demonstrated to bind stronger than 500 nM. The high performance of NetMHCpan-2.0 for non-human primates documents the method’s ability to provide broad allelic coverage also beyond human MHC molecules. The method is available at http://www.cbs.dtu.dk/services/NetMHCpan.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In the majority of higher vertebrates, major histocompatibility complex (MHC) molecules select and present antigenic peptides to T cells, thereby controlling the specificity of cellular immune reactions (Thompson 1995). Indeed, peptide binding to MHC is the most selective of the events involved in antigen presentation (Yewdell and Bennink 1999). Each MHC molecule potentially has a unique binding specificity presenting a distinct set of antigenic peptides to the immune system (Falk et al. 1991).
The MHC genomic region (called HLA, in short for human leukocyte antigen) is the most polymorphic in humans. More than three thousand allelic variants have been discovered so far (Robinson et al. 2001). Most MHC molecules have uncharacterized binding specificity. Out of the more than 1,500 known HLA class I molecules, for example, less than 5% have their binding specificity characterized experimentally (Rammensee et al. 1999; Sette et al. 2005a). In particular, the binding specificity of the classical HLA-C molecules and the non-classical HLA-E, HLA-F, and HLA-G molecules remains unsolved due to lack of experimental data.
There is an even greater lack of experimental data for non-human species. In the case of non-human primates, which frequently serve as models for the study of the human immune response to pathogenic infections, less than 15 alleles have been characterized experimentally (Sette et al. 2005a). This limited knowledge of the epitope-binding specificity of relevant species such as chimpanzee (Pan troglodytes, Patr; Sidney et al. 2006) and rhesus macaque (Macaca mulatta, Mamu) MHC class I molecules (Sette et al. 2005b) often compromises a detailed understanding correlating immunity between humans and non-human primates. Moreover, additional primate species, whose MHC class I binding specificities remain uncharacterized, are emerging as new model organisms for viral infections like HIV-1 (Pendley et al. 2008).
In light of the degree of MHC polymorphism, identifying a subset of peptides mediating cellular immunity, which at the same time provides broad allelic coverage, is an essential yet daunting task for vaccine discovery (Moutaftsi et al. 2006; Watkins et al. 2008). Characterizing the binding motif of a given MHC molecule requires a significant amount of experimental work. As a result, development of in silico methods aimed at predicting the binding motif for uncharacterized MHC molecules is important. Conventional allele-specific MHC class I binding prediction methods are limited to alleles that are characterized by peptide-binding data (Brusic et al. 1994; Buus et al. 2003; Donnes and Elofsson 2002; Lundegaard et al. 2008; Mamitsuka 1998; Nielsen et al. 2003; Segal et al. 2001; Tenzer et al. 2005). Recently, several groups have developed prediction methods designed to provide broad allelic coverage of the MHC polymorphism (Jacob and Vert 2008; Jojic et al. 2006; Nielsen et al. 2007; Zhang et al. 2005). These methods all focus on the human MHC class I A and B (and to some extent C) loci. No publicly available method covers the HLA-E and HLA-G loci or offers binding prediction for a broad set of non-human class I MHC alleles.
For the analysis and interpretation of immune responses in out-bred populations, detailed knowledge of the peptide-binding capacity of each individual is necessary (Frahm et al. 2007; Perez et al. 2008). To date, such analyses have been greatly hindered due to limited knowledge of the specificities of the majority of MHC molecules.
In this work, we present an updated pan-specific MHC class I binding prediction method. NetMHCpan-2.0 has been trained on the hitherto largest set of quantitative MHC binding data available, covering HLA-A and HLA-B, as well as chimpanzee, rhesus macaque, gorilla (Gorilla gorilla), and mouse (Mus musculus) MHC class I molecules. By this, we expand the original NetMHCpan method (Nielsen et al. 2007) to cover a diverse set of non-human alleles, including chimpanzee, rhesus macaque, and even pig (Sus scrofa) MHC class I alleles. Further, we extend the coverage of the human class I loci and demonstrate that a pan-specific method trained on quantitative human, non-human primate, and mouse data can predict the binding specificities of HLA-C, HLA-E, and HLA-G molecules. Also, the method allows the user to upload any full-length MHC class I protein sequence together with a query source protein sequence and search for putative peptide binders to the class I molecule in question. The NetMHCpan-2.0 method, together with the benchmark evaluation data, is available at http://www.cbs.dtu.dk/services/NetMHCpan.
Materials and methods
Source data
Quantitative nonameric peptide–MHC class I binding data were obtained from the IEDB database (Sette et al. 2005a) and an in-house database of quantitative peptide–MHC binding data. In total, the data set consisted of 79,137 unique peptide–MHC class I interactions covering 34 HLA-A, 32 HLA-B, eight chimpanzee (Patr), seven rhesus macaque (Mamu), one gorilla (Gogo), and six mouse MHC class I alleles. See Supplementary Table S1 for a list of the number of data points per allele. The data are highly diverse containing a total number of 25,525 unique peptides. Only a minor fraction of the peptides (1,112 or 4%) share more than seven amino acid identity to any other peptide in the data set. The data set contains a large fraction of non-binding data for each allele (on average 70%). The low data redundancy and the large amount of non-binding data make this an ideal data set for machine learning data mining.
Qualitative nonameric MHC ligand data for HLA-A, HLA-B, HLA-C, and HLA-G were obtained from the SYFPEITHI database (Rammensee et al. 1999), and MHC ligand data for HLA-E*0101 were obtained from the IEDB (Sette et al. 2005a).
Quantitative data for the swine MHC molecule SLA-1*0401 was obtained as described in the “Materials and methods”.
The evaluation data for HLA ligands and quantitative non-human primate peptide binding are available online at http://www.cbs.dtu.dk/suppl/immunology/NetMHCpan-2.0.php.
MHC class I pseudo-sequence
The MHC class I molecule was represented by a pseudo-sequence consisting of amino acid residues in contact with the peptide. The contact residues are defined as being within 4.0 Å of the peptide in any of a representative set of HLA-A and HLA-B structures binding a nonameric peptide. Of all contact residues, only those that were polymorphic in any known HLA-A, HLA-B, and HLA-C protein sequence were included, giving rise to a pseudo-sequence consisting of 34 amino acid residues (Nielsen et al. 2007). This pseudo-sequence mapping was applied to all MHC molecules in this study. This could lead us to discard essential peptide–MHC interactions for non-classical and non-human MHC molecules. However, no quantitative peptide-binding data are available for non-classical HLA molecules, and only very limited data are available for non-human primates. The pan-specific approach relies on the ability of the neural networks to capture general features of the relationship between peptides and HLA pseudo-sequences and interpret these in terms of binding affinity. Only interactions that are polymorphic in the training data can aid the neural network learning. It would hence not be possible for the NetMHCpan method to learn from such extended pseudo-sequence mappings due to the lack of polymorphism at the extended MHC positions in the training data.
Neural network training
Artificial neural networks were trained in a fivefold cross-validation manner as described in Nielsen et al. (2007). For each data point, both the peptide sequence and MHC class I pseudo-sequence served as input to the networks. The input sequences were presented to the neural network in three distinct manners: (a) conventional sparse encoding (i.e., encoded by 19 zeros and a one); (b) Blosum encoding, where each amino acid was encoded by the BLOSUM50 matrix score vector (Henikoff and Henikoff 1992); and (c) a mixture of the two, where the peptide was sparse-encoded, and the HLA pseudo-sequence was Blosum encoded. The log-transformed experimentally determined affinity data served as the output value to train the networks (Nielsen et al. 2003).
Pseudo-distance and nearest neighbor
The pseudo-distance between two alleles is calculated from the similarity score of the HLA pseudo-sequences using the relation \(d = 1 - \frac{{s\left( {A,B} \right)}}{{\sqrt {s\left( {A,A} \right) \times s\left( {B,B} \right)} }}\), where s(A,B) is the BLOSUM50 similarity score vector (Henikoff and Henikoff 1992) between the pseudo-sequences A and B, respectively.
The nearest neighbor of a specific MHC molecule is defined as being the molecule in the training set with the smallest pseudo-distance to this molecule.
Peptide affinity assays
The SLA-1*0401 (haplotype 4/NIH d; sequence gi: 158253220), positions 22–297, was generated with a biotinylated fusion tag and purified as previously described (Ferre et al. 2003; Leisner et al. 2008). The peptide-binding affinity was determined largely as previously described (Sylvester-Hvid et al. 2002) except that the sandwich ELISA capture step was effected by streptavidin coated onto Maxisorb microtiter plates (Nunc).
Results
The NetMHCpan-2.0 method was trained on a large set of quantitative peptide–MHC class I binding data (see “Materials and methods”). This updated method extends the original method (Nielsen et al. 2007), in the following referred to as NetMHCpan-1.0, in two dimensions: First of all, the training data set for NetMHCpan-2.0 had broader allelic coverage, including data for a larger set of HLA molecules as well as non-human MHC binding data for rhesus macaque (Mamu), chimpanzee (Patr), gorilla (Gogo), and mouse (H-2) MHC class I molecules. In the second dimension, the NetMHCpan-2.0 training data included more binding data for HLA molecules that were also part of the original method (see “Materials and methods” for details).
Leave-one-out (LOO) evaluation
The performance of NetMHCpan-2.0 was evaluated by following a leave-one-out training approach, meaning that the data for the molecule in question was not included into the training. The leave-one-out experiment simulated the situation where the MHC molecule in question had an uncharacterized binding specificity, thus providing an ideal benchmark approach for investigating the performance of the pan-specific binding prediction algorithm for a broad range of MHC specificities. If not indicated differently, the performance for the omitted MHC molecule was then calculated as Pearson’s correlation coefficient (PCC; Press et al. 1992) between the predicted and known log-transformed binding-affinity values. One-tailed paired t tests were used to compare the performances of different methods.
Performance for HLA-A and HLA-B
In the original publication, it was shown that the performance of the NetMHCpan method for a given MHC molecule depended strongly on the coverage of its neighborhood (Nielsen et al. 2007). As described in the original NetMHCpan publication, the neighborhood of an MHC molecule can be characterized in terms of the distance to and peptide coverage of the nearest MHC molecule with an experimentally characterized binding specificity. Here, the distance is measured in terms of the amino acid similarity distance between the pseudo-sequences of the two MHC molecules (see “Materials and methods”). The NetMHCpan method performs well for MHC molecules, where the close neighborhood is populated with well-characterized MHC molecules, and likewise, the method performs less accurate if the close neighborhood is either empty or populated with poorly characterized MHC molecules. We compared the performance of NetMHCpan-1.0 and NetMHCpan-2.0 for HLA-A and HLA-B molecules in terms of Pearson’s correlation coefficient and the area under the ROC curve (AUC; Swets 1988; see Supplementary Table S2). Only alleles characterized by ten or more peptide-binding data points and at least one peptide binder (affinity stronger than 500 nM) were included in the analysis. Since no data for the allele in question were included in the training, the LOO experiment allowed for a direct comparison of the predictive performance of the two methods even though their training data sets differ in size. With an average performance of 0.67, NetMHCpan-2.0 performed significantly better than NetMHCpan-1.0 (0.62, p = 0.0002, n = 61). This, however, was largely due to the improved performance on HLA-B. While on average the performance for HLA-A molecules increased only slightly, the average performance for the HLA-B molecules increased significantly (p = 0.0001, n = 29) from 0.5 to 0.6 (see Fig. 1). This improvement is most likely due to the fact that the number of HLA-B molecules included in the training was increased from 18 to 32 and that more training data were available for those HLA-B molecules that were already part of NetMHCpan-1.0. Thus, overall, the amount of HLA-B data were increased by approximately 2.5-fold. For a given molecule, we estimate its distance to any molecule in the training set in terms of the pseudo-sequence distance (for details see “Materials and methods”). Overall, there is a larger decrease in the average distance of the HLA-B molecules to their nearest neighbor (0.139 for NetMHCpan-1.0 to 0.115 for NetMHCpan-2.0), as compared to the HLA-A molecules (0.078 for NetMHCpan-1.0 to 0.073 for NetMHCpan-2.0). On average, HLA-A molecules have closer neighbors and, as expected, show a higher predictive performance than HLA-B molecules (see Fig. 1).
For some HLA-B molecules, including additional HLA affinity data into the training helped to populate their close neighborhood (see Supplementary Table S2). This was the case for HLA-B*1501, which gained the two close neighbors B*1502 and B*1503, thereby reducing the pseudo-distance to the nearest neighbor from 0.19 to 0.09. To investigate if these two molecules were responsible for the increase in performance from 0.41 to 0.7, we trained a network based on the original NetMHCpan-1.0 dataset and including the data for B*1502 and B*1503. This resulted in a performance of 0.72, confirming our assumption and showing that enriching the immediate neighborhood boosts the performance for an MHC molecule.
In addition to the distance to the closest neighbor, the peptide-binding data provided by the closest neighbor seem to be an important factor in determining prediction performance. In the case of B*2705, for example, adding data for B*2702 to the training reduced the distance to the closest neighbor dramatically from 0.31 to 0.08. With only 14 data points, however, B*2702 did not add sufficient data to the training to dramatically improve the leave-one-out performance for B*2705 (0.05 versus 0.23 for NetMHCpan-1.0 and NetMHCpan-2.0, respectively).
The only HLA molecule that gained a non-human molecule as closest neighbor was HLA-B*0702, for which adding more data increased the performance from 0.54 to 0.6. We tested if this improvement could be specifically attributed to adding Patr-B*1301 data to the training by training the method exclusively on HLA and Patr-B*1301 data. This training increased the performance for HLA-B*0702 from 0.54 to 0.61 and, thus, confirmed our assumption.
Performance on non-human primate MHC class I molecules
The leave-one-out performance values of NetMHCpan-1.0 and NetMHCpan-2.0 applied to non-human primate alleles are listed in Table 1. Five out of six rhesus macaque (Mamu) alleles reached a higher PCC for NetMHCpan-2.0. The average performance increased by 44% (from 0.39 to 0.56). The performance of six out of eight chimpanzee (Patr) alleles was improved, and, in total, the average performance for the Patr alleles increased from 0.42 to 0.57. Taken together, NetMHCpan-2.0 performed significantly better than NetMHCpan-1.0 on Patr and Mamu alleles (p = 0.02, n = 14). These results confirm that we also for non-human primates improve the prediction accuracy by adding data for MHC molecules that form their neighborhood.
The motif logos in Fig. 2 illustrate the predicted and known binding specificity for several Patr and Mamu molecules. The logos were generated using the leave-one-out networks, meaning that NetMHCpan-2.0 succeeded in establishing the specificity of these Mamu and Patr molecules without having encountered data for these molecules during the training process. This illustrates clearly how NetMHCpan-2.0 is able to infer the specificity also for unknown non-human MHC molecules.
Binding motifs of Mamu-A*02, Mamu-A*11, Patr-A*0101, and Patr-B*1301 generated from reported and predicted binders. The predicted binders were generated as the top scoring 1% best binders of 100,000 randomly selected natural 9mer peptides. Position specific scoring matrices (PSSM) were calculated from the set of binding peptides using sequence weighting and correction for low counts (Altschul et al. 1997; Nielsen et al. 2004). The binding motifs were visualized using the logo-plot method by Schneider and Stephens (1990). In a sequence logo, the height of a column of letters is equal to the information content at that position, and the height of each letter within a column is proportional to the frequency of the corresponding amino acid at that position
Potential pitfalls of leave-one-out performances
We were surprised by the significant improvement for Patr-B*2401 and Mamu-B*01 (performance gains from −0.32 to 0.38 and from −0.30 to 0.47, respectively). In both cases, adding more data to the training did not strongly change their close neighborhood, as measured by the pseudo-distances between the sequences of the molecules (see Table 1). A closer investigation, however, revealed that these two molecules, despite of their large mutual pseudo-distance of 0.453, showed marked binding motif similarities (see Supplementary Figure S1). It also turned out that the data sets of these two alleles overlap by 68 peptides, which was assumed likely to have largely influenced the prediction outcome for the other allele. These observations gave rise to the concern that the method might be merely learning the binding-affinity values of the individual peptides by heart independent of the context of the MHC molecule. To investigate if this was indeed the case, we followed a cross-validation approach, in turn leaving out a third of the overlapping peptide data from the training and then evaluating on the left-out peptides. This approach reestablished most of the improved performance values (0.33 for Mamu-B*01 and 0.34 for Patr-B*2401), thus, demonstrating that these alleles indeed learned their binding specificity from the alleles in the training data (most likely from each other) without merely learning the individual peptide affinity values by heart.
We investigated the effect of peptide-overlap between training and evaluation set for a large number of alleles by performing the cross-validated leave-one-out training, which avoids any peptide-overlap between training and evaluation. This analysis showed that the overall performance decreased only slightly (from 0.65 to 0.63) when compared to the original leave-one-out experiment. The result, thus, demonstrates that the training data are of sufficient size and diversity so that peptide-overlap between training and evaluation set only poses a minor problem, and we can assume the leave-one-out performance values to be proper estimates of the predictive performance of uncharacterized MHC molecules.
In the final NetMHCpan-2.0 version, we have taken care of peptide overlaps between MHC molecules by assigning affinity data for the same peptide to the same subset in the fivefold cross-validation. In this way, a peptide is prevented from occurring in training and evaluation set at the same time.
Estimation of the prediction accuracy
NetMHCpan-2.0 is intended to find its application in predicting the binding specificity of uncharacterized MHC class I molecules for which there is no binding data available. The obvious downside of this is that we are not able to make any statements on the performance for a particular MHC molecule unless affinity data for this allele exists. While NetMHCpan-2.0 achieves a high performance for some alleles, it shows a low performance for others. Our aim is to estimate the prediction performance for any given MHC molecule based on its protein sequence.
We have earlier shown that filling the immediate neighborhood of an uncharacterized MHC class I molecule with binding data contributes largely to the prediction performance (Nielsen et al. 2007). In general, a training set of at least 50 data points is needed to train a network with acceptable performance (Lundegaard et al. 2008; Nielsen et al. 2003; Yu et al. 2002). We therefore selected those alleles from our training set for which we had more than 50 data points and, among these, at least ten binding peptides, reducing the set of possible neighbor MHC molecules to 69. From this set, we identified the closest neighbor for each of the 82 alleles in the LOO experiment. The strong correlation between the distance to the closest nearest neighbor and the predictive accuracy is apparent from the plot shown in the insert of Fig. 3. Next, we transformed the nearest neighbor distance into a direct interpretation in terms of prediction accuracy. Following a fivefold cross-validation approach, we calculated the best linear fit between the performance of an allele and the distance to its closest neighbor and used this linear relationship to predict the performance. This resulted in a Pearson’s correlation of 0.67 (R 2 = 0.45) between the expected performance and the actual performance (see Fig. 3). The performance estimation is offered as part of the output of the NetMHCpan-2.0 server.
The NetMHCpan method
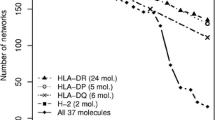
The final NetMHCpan-2.0 method was trained in a fivefold cross-validation manner as described by Nielsen et al. (2007). In short, the complete pool of unique peptides was randomly split into five groups with all MHC binding data for a given peptide placed in the same group (in this way, no peptide can belong to more than one group). Artificial neural networks were next trained as described in the “Materials and methods”. The cross-validation performances for all alleles in the training set are listed in Supplementary Table S3. The average performance per species is given in Fig. 4.
Scatter plots showing the relation between the predicted and experimentally measured binding affinities for an MHC molecule can provide an informative illustration of the predictive performance of the NetMHCpan method. Such plots can demonstrate to what extent a prediction method is indeed capable of reproducing the measured binding-affinity values. Figure 5 and Supplementary figure S2 give examples of such scatter plots for three HLA alleles. Figure 5 gives one particular example illustrating the power of the NetMHCpan method to successfully leverage information from neighboring MHC molecules to boost performance in cases where the training data for that particular allele are scarce. In the figure, the relation between the predicted and measured IC50 values is shown for the HLA-A*0302 allele for the pan-specific NetMHCpan-2.0 and single-allele NetMHC (Lundegaard et al. 2008) methods, respectively. The HLA-A*0302 allele is characterized with limited number of binding data but has a close neighborhood populated with a MHC molecule (HLA*0301) for which many binding data are available. For this allele, the predictive performance of the NetMHCpan method in terms of the Pearson’s CC is 0.77, and the slope of the best linear fit is 0.71. For the NetMHC method, on the other hand, the corresponding numbers are 0.29 (Pearson’s CC) and 4.04 (slope of best linear fit), respectively. Similar plots are shown in Supplementary figure S2 for two other alleles. The first case is an allele (HLA-A*0301) for which a large number of peptide-binding data are available, and the second, an allele (HLA-B*7301) where the close neighborhood is empty, i.e., no similar MHC molecule exists for which binding data are available. The relative poor performance of the NetMHCpan method for the “lonely” HLA-B*7301 manifests the essential need of a well-characterized neighborhood in order for the pan-specific prediction approach to succeed.
Scatter plots of the predicted versus experimental IC50 values for the HLA-A*0302 alleles. NetMHCpan refers to the method developed in this paper, and NetMHC refers to the single-allele neural-network-based method developed by Lundegaard et al. (2008). The lines in the plots are least square fits for NetMHCpan (solid line) and NetMHC (dashed line), respectively. The HLA-A*0302 is characterized with 21 peptide data points. The Pearson’s correlation between the prediction and experimental log(IC50) values is 0.77 and 0.29 for the NetMHCpan and NetMHC methods, respectively, and the slope of the best linear fit is 0.71 and 4.04
The NetMHCpan-2.0 method was made available as a web server, which provides affinity predictions for any peptide and MHC class I molecule. The MHC molecule can either be chosen from a list of more than 1,600 different alleles or the user can provide the full-length protein sequence of any MHC class I molecule. This makes the method especially attractive because it can provide suggestions on how the binding motif might look, even if there is no prior information on the specificity available. NetMHCpan-2.0 is available at http://www.cbs.dtu.dk/services/NetMHCpan.
We used NetMHCpan-2.0 to predict the binding specificities of HLA, macaque, chimpanzee, and mouse alleles and made the sequence logos of these motifs available online. The motif logos can be viewed at http://www.cbs.dtu.dk/biotools/MHCMotifViewer/Home.html (Rapin et al. 2008).
Identifying endogenously presented peptides
The NetMHCpan method was validated using a large set of data from the SYFPEITHI database (Rammensee et al. 1999), which were not included in the training data of the method. This set consists of 596 HLA ligands restricted to 34 different HLA-A and HLA-B alleles. For each of the reported ligands, we identified its source protein in the UniProt database (UniProt 2008) and predicted the affinity of all overlapping nonamers contained in the source protein sequence for the HLA allele in question. In each case, all peptides, with the exception of the reported HLA ligand, were assumed to be non-binders. Performing this experiment, we found an average rank of the HLA ligands of 2% and an average rank per HLA allele of 3.3%. This means that on average for a protein with 200 amino acids, a number of peptides less than five are needed to be tested in order to identify the ligand. In particular, for ligands restricted to HLA alleles not included in the training of the NetMHCpan method, we found that the average rank was 2.3%, whereas the average rank was 3.4% for ligands restricted to HLA alleles included in the training. This result clearly demonstrates the power of the NetMHCpan method to do accurate extrapolations to specificity-wise unknown MHC molecules. The details of this analysis are shown in Supplementary Table S4.
Performance for HLA-C
Quantitative binding data for HLA-C molecules are sparse. There are, however, qualitative data available from the SYFPEITHI database (Rammensee et al. 1999). In total, we obtained 77 ligands covering eight HLA-C alleles (see Supplementary Table S5). For each of the reported ligands, we identified its source protein and predicted the affinity of all nonamers contained in the source protein like we did for the HLA-A and HLA-B ligands earlier. In doing this, we were able to calculate the relative rank of each ligand and the average relative rank per HLA-C allele (see Supplementary Tables S5 and S6). With an average relative rank of 3.6% and 1.9%, respectively, the best performance was achieved for HLA-Cw*0102 and HLA-Cw*0304. Figure 6 shows sequence logos of the predicted specificities of these two molecules as well as the sequences of the reported ligands. The anchor positions of both molecules are assumed to be P2 and P9 (Rammensee et al. 1999) with a preference for alanine (A) at P2 and leucine (L) at P9. This conforms very well with our predicted motifs.
These results suggest that NetMHCpan-2.0 may be useful for predicting binders for HLA-C alleles. Based on their pseudo-sequence, HLA-C alleles are closest to HLA-B alleles. The predicted performances in terms of Pearson’s correlation coefficient, estimated from the pseudo-distance to the closest neighbor, range from 0.34 to 0.68. We expect that adding quantitative HLA-C binding data to the training of NetMHCpan-2.0 would largely improve the performance for HLA-C binding prediction.
Performance for the non-classical HLA molecules HLA-E and HLA-G
In contrast to the other HLA class I alleles, the HLA-E locus is not very polymorphic. The IMGT/HLA database reports only two full-length HLA-E molecules (Robinson et al. 2001). The closest neighbor of HLA-E*0101 is the mouse allele H-2-Kk with a distance of 0.42. The closest among the HLA alleles is B*4001 with a distance of 0.56. This indicates that HLA-E*0101 is extremely isolated, and based on the large distance to the closest neighbor, we estimate a relative low prediction performance of 0.21. We obtained qualitative binding data for HLA-E*0101 from the IEDB (Sette et al. 2005a), which consisted of seven ligands stemming from the same source protein and performed an analysis similar to the one described above for HLA-A and HLA-B. The details of this analysis are shown in Supplementary Table S7. On average, the known binders rank among the top 4% of all peptides.
For HLA-G, we obtained the sequences of 11 HLA-G ligands from the SYFPEITHI database (Rammensee et al. 1999). The closest neighbor of HLA-G*0101 is HLA-A*2403 with a pseudo-distance of 0.35. Based on this, we would predict a performance of 0.32. Predicting the binding affinity for the ligands and all the peptides in their respective source protein resulted in an average rank of 3.4% of the ligands (see Supplementary Table S8). Figure 7 shows the predicted motif logo for HLA-G*0101 and the list of known ligands. HLA-G was reported to have P2, P3, and P9 as anchor positions, with a preference for hydrophic residues at P2 and P9 and a preference for proline at P3 (Diehl et al. 1996). In addition, a preference for basic residues at the N-terminus of the peptides has been reported, which is also reflected by the predicted motif (Clements et al. 2007). Moreover, HLA-G was suggested to share binding specificities with HLA-A2, which is in agreement with the predicted preference for hydrophobic residues at P9. The predicted motif shows a slight preference for proline at P3 (12.3% of the predicted binders have a proline at P3) but does not put forth P3 as an anchor position. It should be noted that for both HLA-E and HLA-G, the number of experimentally verified HLA ligands is very limited, making a detailed comparison of the predicted and experimental sequence logos difficult. For instance, does the predicted sequence motif for HLA-G show a preference for Y and F at position P2 that is not present in the limited set of known ligands. Further experimental validation is needed to clarify if the HLA-G motif indeed does support binding of peptides with Y and F at P2.
In spite of not being able to predict binding for HLA-E*0101 and G*0101 as accurately as for HLA-A and HLA-B molecules, NetMHCpan-2.0 appears to be a useful tool to gain insight into the binding motifs also for non-classical HLA molecules.
Performance for a pig MHC class I molecule
We used NetMHCpan-2.0 to predict potential binding peptides for the pig MHC class I molecule SLA-1*0401. We then experimentally tested the affinity of 14 high scoring peptides. Thirteen of these 14 peptides turned out to be binders, five of them being strong binders with an IC50 value of less than 50 nM. Figure 8 shows the predicted binding specificity of SLA-1*0401 and the motif that was generated from the 13 verified binders.
Sequence motifs generated from the 13 verified binders and predicted binders for the swine MHC class I molecule SLA-1*0401. The predicted binders and motif sequence logos were generated as described in Fig. 3
Interpretation of cellular immune responses
Interpretation of cellular immune responses in large out-bred populations has often been greatly hindered by the limited knowledge of the peptide-binding specificities of the majority of the populations’ MHC molecules. In a large HIV cohort study, where 184 peptides were tested for recognition in 31 HIV-1-infected patients, NetMHCpan has earlier proven powerful in identifying the HLA allele most likely responsible for the observed CTL responses. Eighty-five percent of the 225 CTL responses could be explained based on the HLA phenotype of the patients (Perez et al. 2008).
A similar study, in which 242 well-defined viral HIV and EBV epitopes were tested for CTL responses in 100 patients, came to the conclusion that HLA class I epitopes are surprisingly promiscuous (Frahm et al. 2007). Frahm et al. reported that half of the responses in their study were seen in the absence of the originally reported restricting HLA class I allele, indicating that the epitopes must be restricted also by other HLA alleles. Based on two different statistical approaches, they suggested alternative restricting HLA alleles for 303 epitope–HLA pairs, 33 of them significant using both approaches. Interestingly, these alternative HLA alleles were picked solely based on statistics, disregarding any knowledge of the HLA binding specificity of possible alternative alleles. Since a large set of probable associations between original and alternative restrictions occurred between alleles that fell into different supertypes and in some cases even different loci, the authors suggested that in some cases, features other than readily apparent MHC binding similarities may contribute to epitope promiscuity.
Given this data, we were interested in whether we could explain these results based on predicted HLA class I binding. For 283 of the 303 epitopes, we were able to identify the source proteins. For each of these epitopes, we then applied NetMHCpan-2.0 to all peptides contained in its source protein, predicting the binding affinity for both the original restricting HLA molecule and the suggested alternative HLA molecule. Since most restrictions were only specified with two digits, we applied NetMHCpan-2.0 for all known four-digit alleles that fit the denoted two-digit specification. AUC values were calculated assuming that the reported epitope is the only binder and all other peptides contained in the source protein are non-binders. Epitopes with an AUC value of at least 0.9 were interpreted as predicted binders. With this approach, we were able to explain 97.3% of the original restrictions and 90.9% of the alternative restrictions in the set of 33 most significant epitope associations. If applied to the complete list of 283 HLA-associations, we were able to explain 96.9% of the original restrictions and 75% of the alternative restrictions, of which 28.8% could only be explained by peptides embedded in the reported epitopes. We repeated this analysis limiting the set of HLA alleles to the ones that have been reported to exist in North America (Middleton et al. 2003; since the study subjects stemmed from the Boston area). Still, we were able to explain 78.8% of the 33 significant alternative restrictions, 34.1% of these as embedded peptides. Our results support the suggested alternative restrictions reported by Frahm et al. and indicate that the epitope promiscuity is defined by the binding specificity to the HLA molecules.
Discussion
Binding of peptides to the MHC molecule is the single most selective step in distinguishing immunogens from non-immunogens of the cellular immune system in most animals (Yewdell and Bennink 1999). The MHC genomic region is highly polymorphic, and each MHC molecule has a unique binding motif presenting a distinct set of peptides to the immune system (Falk et al. 1991). Characterizing which peptides will bind a given MHC molecule is hence of pivotal importance for the understanding of cellular immune responses. Only a very limited set of the currently known MHC molecules in humans and animals has been characterized experimentally (Rammensee et al. 1999; Sette et al. 2005a). This lack of specificity characterization has made it difficult to correlate immunity to antigenic presentation due to the fact that little is known about the peptide-binding specificity in the host (Sidney et al. 2006).
An essential step in peptide vaccine research is the testing of potential immunogenic peptides in animal models. To date, there are no tools available that offer peptide-binding prediction for a wide array of non-human primate MHC class I molecules. Likewise, binding prediction with broad allelic coverage for HLA loci other than HLA-A and HLA-B has not been publicly available thus far.
With NetMHCpan-2.0, we have taken MHC class I binding prediction beyond HLA-A and HLA-B. We demonstrated that adding peptide-binding information for non-human MHC class I molecules to the training extends the method’s applicability to HLA-C and the non-classical HLA loci HLA-E and HLA-G. Furthermore, we showed that NetMHCpan-2.0 is able to accurately predict the binding specificity of chimpanzee and Indian rhesus macaque alleles. This shows the potential of the method to assist in the characterization of MHC class I molecules of upcoming model organisms like rhesus macaques of Chinese origin and cynomolgus macaques whose MHC specificities are still largely unknown (Karl et al. 2008; Pendley et al. 2008). We have earlier demonstrated how such pan-specific MHC binding predictions can provide novel insights to the co-evolution of the host immune system and infectious pathogens in, for instance, HIV-infected humans and chimpanzees (Hoof et al. 2008). It is thus likely that NetMHCpan-2.0 may assist in vaccine research by providing insights into the binding repertoire of the model organism and thereby enabling a direct correlation between the immune responses in animal models and the human natural host.
NetMHCpan-2.0 was able to predict the binding motif of the pig MHC class I molecule SLA-1*0401 with high accuracy. This result shows the potential of the method to predict binding beyond HLA and non-human primate alleles. It remains to be seen how far NetMHCpan will be able to reach in predicting peptide binding for animal MHC molecules.
In the process of reaching further into the world of animal MHC molecules, NetMHCpan-2.0 can be of great value to limit the number of peptides to be tested in affinity experiments because potential binders are likely to rank among the top-scoring peptides. This allows for a more efficient characterization of the binding motif of unknown MHC class I alleles because a smaller set of peptides has to be tested to establish the binding specificity.
The unique ability of the method to characterize binding specificities for any MHC class I molecule can rationalize the selection of peptides for vaccine studies and guide interpretation of immune responses in large out-bred populations where a detailed characterization of each individual is essential (Frahm et al. 2007). In their work, Frahm et al. observed epitope promiscuity between HLA supertypes and HLA loci and suggested that in some cases, features other than binding motif similarities may contribute to epitope promiscuity. The data presented here indicates, however, that the majority of the observed epitope promiscuity can be explained by peptide-binding specificities alone.
The pan-specific approach relies on the ability of the algorithm to capture general features of the relationship between peptides and MHC sequences, and it is therefore apparent that the method should perform better when the query MHC molecule is represented by closely related MHC molecules with characterized binding specificity (Nielsen et al. 2007). We illustrated this importance of a well-defined neighborhood for the prediction performance both in a large-scale leave-one out experiment and for a set of alleles not included in the training of the NetMHCpan-2.0 method. We demonstrated how the correlation between the performance for a molecule and its distance (as measured in terms of amino acid similarity) to a well-defined neighbor enabled us to estimate the prediction performance of NetMHCpan-2.0 for uncharacterized MHC molecules.
The comparison of the NetMHCpan-1.0 and the NetMHCpan-2.0 performances for HLA and non-human alleles illustrates that adding quantitative data for additional alleles to the training improves the performance for alleles in their immediate pseudo-sequence neighborhood. We aim to complete the MHC specificity space by identifying informative alleles that would bring the largest gain in performance by filling the holes in the specificity space. An approach to this has already been suggested for the MHC class II specificity space, which would involve the identification of informative alleles, the development of immunoassays for these alleles, and the retraining of the method on the extended data set (Nielsen et al. 2008). This approach will further extend the prediction capacity of NetMHCpan-2.0 into the animal world and will improve the prediction performance for the today sparsely covered HLA loci C, E, and G.
References
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402 doi:10.1093/nar/25.17.3389
Brusic V, Rudy G, Harrison LC (1994) Prediction of MHC binding peptides using artificial neural networks. In: a. Y. X. Stonier RJ (ed) Complex systems: mechanism of adaptation. IOS, Amsterdam, pp 253–260
Buus S, Lauemoller SL, Worning P, Kesmir C, Frimurer T, Corbet S, Fomsgaard A, Hilden J, Holm A, Brunak S (2003) Sensitive quantitative predictions of peptide–MHC binding by a ‘Query by Committee’ artificial neural network approach. Tissue Antigens 62:378–384 doi:10.1034/j.1399-0039.2003.00112.x
Clements CS, Kjer-Nielsen L, McCluskey J, Rossjohn J (2007) Structural studies on HLA-G: implications for ligand and receptor binding. Hum Immunol 68:220–226 doi:10.1016/j.humimm.2006.09.003
Diehl M, Munz C, Keilholz W, Stevanovic S, Holmes N, Loke YW, Rammensee HG (1996) Nonclassical HLA-G molecules are classical peptide presenters. Curr Biol 6:305–314 doi:10.1016/S0960-9822(02)00481-5
Donnes P, Elofsson A (2002) Prediction of MHC class I binding peptides, using SVMHC. BMC Bioinformatics 3:25 doi:10.1186/1471-2105-3-25
Falk K, Rotzschke O, Stevanovic S, Jung G, Rammensee HG (1991) Allele-specific motifs revealed by sequencing of self-peptides eluted from MHC molecules. Nature 351:290–296 doi:10.1038/351290a0
Ferre H, Ruffet E, Blicher T, Sylvester-Hvid C, Nielsen LL, Hobley TJ, Thomas OR, Buus S (2003) Purification of correctly oxidized MHC class I heavy-chain molecules under denaturing conditions: a novel strategy exploiting disulfide assisted protein folding. Protein Sci 12:551–559 doi:10.1110/ps.0233003
Frahm N, Yusim K, Suscovich TJ, Adams S, Sidney J, Hraber P, Hewitt HS, Linde CH, Kavanagh DG, Woodberry T, Henry LM, Faircloth K, Listgarten J, Kadie C, Jojic N, Sango K, Brown NV, Pae E, Zaman MT, Bihl F, Khatri A, John M, Mallal S, Marincola FM, Walker BD, Sette A, Heckerman D, Korber BT, Brander C (2007) Extensive HLA class I allele promiscuity among viral CTL epitopes. Eur J Immunol 37:2419–2433 doi:10.1002/eji.200737365
Henikoff S, Henikoff JG (1992) Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci USA 89:10915–10919 doi:10.1073/pnas.89.22.10915
Hoof I, Kesmir C, Lund O, Nielsen M (2008) Humans with chimpanzee-like major histocompatibility complex-specificities control HIV-1 infection. AIDS 22:1299–1303
Jacob L, Vert JP (2008) Efficient peptide–MHC-I binding prediction for alleles with few known binders. Bioinformatics 24:358–366 doi:10.1093/bioinformatics/btm611
Jojic N, Reyes-Gomez M, Heckerman D, Kadie C, Schueler-Furman O (2006) Learning MHC I-peptide binding. Bioinformatics 22:e227–e235 doi:10.1093/bioinformatics/btl255
Karl JA, Wiseman RW, Campbell KJ, Blasky AJ, Hughes AL, Ferguson B, Read DS, O’Connor DH (2008) Identification of MHC class I sequences in Chinese-origin rhesus macaques. Immunogenetics 60:37–46 doi:10.1007/s00251-007-0267-x
Leisner C, Loeth N, Lamberth K, Justesen S, Sylvester-Hvid C, Schmidt EG, Claesson M, Buus S, Stryhn A (2008) One-pot, mix-and-read peptide–MHC tetramers. PLoS ONE 3:e1678
Lundegaard C, Lamberth K, Harndahl M, Buus S, Lund O, Nielsen M (2008) NetMHC-3.0: accurate web accessible predictions of human, mouse and monkey MHC class I affinities for peptides of length 8–11. Nucleic Acids Res 36:W509–W512
Mamitsuka H (1998) Predicting peptides that bind to MHC molecules using supervised learning of hidden Markov models. Proteins 33:460–474 doi:10.1002/(SICI)1097-0134(19981201)33:4<460::AID-PROT2>3.0.CO;2-M
Middleton D, Menchaca L, Rood H, Komerofsky R (2003) New allele frequency database: http://www.allelefrequencies.net. Tissue Antigens 61:403–407
Moutaftsi M, Peters B, Pasquetto V, Tscharke DC, Sidney J, Bui HH, Grey H, Sette A (2006) A consensus epitope prediction approach identifies the breadth of murine T(CD8+)-cell responses to vaccinia virus. Nat Biotechnol 24:817–819 doi:10.1038/nbt1215
Nielsen M, Lundegaard C, Worning P, Lauemoller SL, Lamberth K, Buus S, Brunak S, Lund O (2003) Reliable prediction of T-cell epitopes using neural networks with novel sequence representations. Protein Sci 12:1007–1017 doi:10.1110/ps.0239403
Nielsen M, Lundegaard C, Worning P, Hvid CS, Lamberth K, Buus S, Brunak S, Lund O (2004) Improved prediction of MHC class I and class II epitopes using a novel Gibbs sampling approach. Bioinformatics 20:1388–1397 doi:10.1093/bioinformatics/bth100
Nielsen M, Lundegaard C, Blicher T, Lamberth K, Harndahl M, Justesen S, Roder G, Peters B, Sette A, Lund O, Buus S (2007) NetMHCpan, a method for quantitative predictions of peptide binding to any HLA-A and -B locus protein of known sequence. PLoS One 2:e796 doi:10.1371/journal.pone.0000796
Nielsen M, Lundegaard C, Blicher T, Peters B, Sette A, Justesen S, Buus S, Lund O (2008) Quantitative predictions of peptide binding to any HLA-DR molecule of known sequence: NetMHCIIpan. PLOS Comput Biol 4:e1000107 doi:10.1371/journal.pcbi.1000107
Pendley CJ, Becker EA, Karl JA, Blasky AJ, Wiseman RW, Hughes AL, O’Connor SL, O’Connor DH (2008) MHC class I characterization of Indonesian cynomolgus macaques. Immunogenetics 60:339–351 doi:10.1007/s00251-008-0292-4
Perez CL, Larsen MV, Gustafsson R, Norstrom MM, Atlas A, Nixon DF, Nielsen M, Lund O, Karlsson AC (2008) Broadly immunogenic HLA class I supertype-restricted elite CTL epitopes recognized in a diverse population infected with different HIV-1 subtypes. J Immunol 180:5092–5100
Press WH, Flannery BP, Teukolsky SA, Vetterling WT (1992) Numerical recipes in C: the art of scientific computing. Cambridge University Press, Cambridge
Rammensee H, Bachmann J, Emmerich NP, Bachor OA, Stevanovic S (1999) SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics 50:213–219 doi:10.1007/s002510050595
Rapin N, Hoof I, Lund O, Nielsen M (2008) MHC motif viewer. Immunogenetics, Sep 3 [Epub ahead of print]
Robinson J, Waller MJ, Parham P, Bodmer JG, Marsh SGE (2001) IMGT/HLA Database—a sequence database for the human major histocompatibility complex. Nucleic Acids Res 29:210–213 doi:10.1093/nar/29.1.210
Schneider TD, Stephens RM (1990) Sequence logos: a new way to display consensus sequences. Nucleic Acids Res 18:6097–6100 doi:10.1093/nar/18.20.6097
Segal MR, Cummings MP, Hubbard AE (2001) Relating amino acid sequence to phenotype: analysis of peptide-binding data. Biometrics 57:632–642 doi:10.1111/j.0006-341X.2001.00632.x
Sette A, Fleri W, Peters B, Sathiamurthy M, Bui HH, Wilson S (2005a) A roadmap for the immunomics of category A–C pathogens. Immunity 22:155–161 doi:10.1016/j.immuni.2005.01.009
Sette A, Sidney J, Bui HH, del Guercio MF, Alexander J, Loffredo J, Watkins DI, Mothe BR (2005b) Characterization of the peptide-binding specificity of Mamu-A*11 results in the identification of SIV-derived epitopes and interspecies cross-reactivity. Immunogenetics 57:53–68 doi:10.1007/s00251-004-0749-z
Sidney J, Asabe S, Peters B, Purton KA, Chung J, Pencille TJ, Purcell R, Walker CM, Chisari FV, Sette A (2006) Detailed characterization of the peptide binding specificity of five common Patr class I MHC molecules. Immunogenetics 58:559–570 doi:10.1007/s00251-006-0131-4
Swets JA (1988) Measuring the accuracy of diagnostic systems. Science 240:1285–1293 doi:10.1126/science.3287615
Sylvester-Hvid C, Kristensen N, Blicher T, Ferre H, Lauemoller SL, Wolf XA, Lamberth K, Nissen MH, Pedersen LO, Buus S (2002) Establishment of a quantitative ELISA capable of determining peptide–MHC class I interaction. Tissue Antigens 59:251–258 doi:10.1034/j.1399-0039.2002.590402.x
Tenzer S, Peters B, Bulik S, Schoor O, Lemmel C, Schatz MM, Kloetzel PM, Rammensee HG, Schild H, Holzhutter HG (2005) Modeling the MHC class I pathway by combining predictions of proteasomal cleavage, TAP transport and MHC class I binding. Cell Mol Life Sci 62:1025–1037 doi:10.1007/s00018-005-4528-2
Thompson CB (1995) New insights into V(D)J recombination and its role in the evolution of the immune system. Immunity 3:531–539 doi:10.1016/1074-7613(95)90124-8
UniProt (2008) The universal protein resource (UniProt). Nucleic Acids Res 36:D190–D195 doi:10.1093/nar/gkn141
Watkins DI, Burton DR, Kallas EG, Moore JP, Koff WC (2008) Nonhuman primate models and the failure of the Merck HIV-1 vaccine in humans. Nat Med 14:617–621 doi:10.1038/nm.f.1759
Yewdell JW, Bennink JR (1999) Immunodominance in major histocompatibility complex class I-restricted T lymphocyte responses. Annu Rev Immunol 17:51–88 doi:10.1146/annurev.immunol.17.1.51
Yu K, Petrovsky N, Schonbach C, Koh JY, Brusic V (2002) Methods for prediction of peptide binding to MHC molecules: a comparative study. Mol Med 8:137–148
Zhang GL, Khan AM, Srinivasan KN, August JT, Brusic V (2005) MULTIPRED: a computational system for prediction of promiscuous HLA binding peptides. Nucleic Acids Res 33:W172–W179
Acknowledgment
This work was supported by the NIH (contracts HHSN266200400025C, HHSN266200400083C, and HHSN26620040006C).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Figure S1
(DOC 285 KB)
Supplementary Figure S2
(DOC 223 KB)
Supplemental tables
(XLS 109 KB)
Rights and permissions
About this article
Cite this article
Hoof, I., Peters, B., Sidney, J. et al. NetMHCpan, a method for MHC class I binding prediction beyond humans. Immunogenetics 61, 1–13 (2009). https://doi.org/10.1007/s00251-008-0341-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-008-0341-z