Abstract
Purpose
To establish the population pharmacokinetic (PPK) model of cyclosporine (CsA) in Chinese renal transplant recipients and evaluate the influence of various indexes including CYP3A5 and MDR1 genetic polymorphism on pharmacokinetic parameters.
Methods
Trough (C0) and peak (C2) CsA concentration were monitored conventionally after renal transplantation. C0 and C2 were collected for 5 months in 146 patients. The CYP3A5*3 genotype and MDR1 haplotype were determined by methods based on amplification refractory mutant PCR. The data were analyzed by nonlinear mixed-effect modeling (NONMEM). The model was evaluated using goodness of fit plots and relative error measurements. Physiological and pathological factors including CYP3A5 and MDR1 genotypes were evaluated as covariates of CsA pharmacokinetic parameters.
Results
Pharmacokinetics of CsA was best described by a one-compartment disposition model followed a first-order absorption process. The estimated clearance (CL/F) was 49.5 l·h−1, the volume of distribution (Vd/F) was 226 l. Ka was fixed as 1.25 h−1. Post-transplant data, body weight, total bilirubin, and MDR1 genotype were covariates of CL/F (P < 0.005). Gender and MDR1 haplotype were covariates of Vd/F (P < 0.005). The AUC estimated based on the Bayesian method was 7,465 ± 1,708 ng·h·ml−1 (2,946 ∼13,926 ng·h·ml-1).
Conclusion
The PPK model developed in this study could be used to optimize CsA dose for Chinese renal transplant recipients by using conventional therapeutic drug monitoring (TDM) data.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Cyclosporine (CsA) is a member of the calcineurin inhibitor family, which is deemed to be a cornerstone of immunosuppressive therapy. CsA is widely used as a basic component of immunosuppressive regimens to prevent allograft rejection in solid organ transplantation [1, 2]. In the past decades, with the introduction of CsA, 1-year survival rate after renal grafts increased dramatically [3], although the long-term outcome is not optimal. There are great inter-individual and intra-individual differences in the response: with similar dosage, some patients experienced acute rejection, while others displayed acute nephrotoxicity [4, 5].
The difference in drug response of CsA is partly attributed to variations in CsA pharmacokinetics, which appear to be particularly significant in patients administered oral CsA in the first few weeks after transplantation [6, 7]. There are almost six-fold inter-individual differences in CsA absorption, which cause bioavailability of CsA ranging from 3∼50% [8, 9]. It has been recommended that dosing schedules be guided by therapeutic drug monitoring (TDM). Indexes such as trough levels (C0), concentration 2 h post dosage (C2), and the area under the curve (AUC) have been used [10–12]. The AUC is preferred because it is more informative [13].
Many pharmacokinetic studies on CsA have been conducted in patients after solid-organ transplantation [14, 15]. Previous studies proved that besides age, hepatic and renal functions or co-administered drugs, genetic polymorphism of drug metabolism enzymes and drug transporters are also important causes of pharmacokinetic variation. CYP3A family is responsible for CsA metabolism in the liver and intestine. The importance of CYP3A4 alleles (*4, *5, *6, *18A, and *19) is limited by their low frequency (<1% in Chinese subjects) [16, 17]. The frequency of CYP3A5*3 allele is about 75% [18, 19]. There is evidence that the CYP3A5*3 allele has an impact on the pharmacokinetics of CsA. P-glycoprotein (Pgp) is an ATP-dependent efflux transporter, which is encoded by multidrug resistance gene (MDR1/ABCB1) [20]. Genetic polymorphism of MDR1 has been studied in various populations. More than 48 single nucleotide polymorphisms (SNPs) have been found and C1236T, G2677T/A, C3435T are the most frequently studied [21]. Studies suggested that the MDR1 haplotype might be a better index of pharmacokinetics of Pgp substrates [22]. Our previous study found that CYP3A5 and MDR1 genotypes have an impact on CsA level in a group Chinese renal transplant recipients [23].
Population methods are particularly suitable in modeling pharmacokinetic responses in a relatively large group of subjects in which there are only limited observations for each subject. More importantly, population pharmacokinetics (PPK) has the additional benefit of being able to provide quantitative estimation of the inter-patient variability, the intra-patient variability, and the influence of demographic, clinical, and genetic factors on the pharmacokinetics. The PPK datasets of many studies on CsA include both full pharmacokinetic profiles (full-PK) and sparse pharmacokinetic profiles (sparse-PK). Some other studies used conventional TDM data, including C0 or C2 to establish a CsA PPK model.
In this study, we investigated PPK parameters in Chinese adult patients who were administered oral CsA for immunosuppressive therapy after renal transplantation. We also elucidated the influence of various factors including CYP3A5*3 and MDR1 genotypes on CsA pharmacokinetic parameters in Chinese renal transplant recipients.
Materials and methods
Patients
One hundred and forty-six renal transplants recipients (87 male and 59 female) were enrolled. All patients were of Han nationality. The study was approved by the Ethics Committee of Shanghai Ruijin Hospital. Informed consent was obtained from all patients. The demographic and clinical data of these patients are shown in Table 1. All patients received combined immunosuppressive therapy consisting of CsA, mycophenolate mofetil (MMF), and steroids. The initial dosage of CsA (Neoral, Novatis Pharma, Basel, Switzerland) was 6∼9 mg·(kg·day)−1, which was divided into two doses. The dosage was adjusted according to the blood level of CsA. MMF was given according to the body weight (BW) of the patients (2 g·day−1 for patients with BW over 50 kg, or 1.5 g·day−1 for those with BW under 50 kg). 6 mg·(kg·day)−1 of methylprednisolone was injected for 3 days. This schedule was followed by 80 mg·day−1 of oral prednisolone; the dose was reduced to 20 mg·day−1 after 1 month of therapy.
The demographic data, pathophysiological index were recorded on the day of CsA monitoring. Clearance of creatinine (CLcr) of the patients was calculated according to previous studies [24, 25]. Post-operation date (POD) was expressed as the days between the operation date and the TDM date. Medication known to affect CsA blood levels taken by patients was also recorded. To guarantee the accuracy, all data were double checked by different researchers.
Determination of CsA whole blood concentrations
Blood samples of CsA C0 and C2 were monitored. C0 were drawn at 8:00 a.m., just prior to the morning dose. Samples of C2 were drawn at 10:00 a.m., 2 h after the oral dose of CsA. All samples were anti-coagulated with ethylene diamine tetraacetic acid (EDTA). Whole blood CsA level was measured using a fluorescence polarization immunoassay (FPIA) on an Axsym analyzer (Abbott Diagnostic, Chicago, IL, USA) using the cyclosporine II Axsym reagent kit. The functional sensitivity of the assay was 25 ng·ml−1.
Genotyping
Leukocyte DNA was extracted by the phenol-chloroform method. A tetra-primers amplification method was used to detect the CYP3A5*3 allele [23]. A 25-μl reaction system contained 15∼50 ng DNA, 0.5 U Heat start Taq DNA polymerase (Bio Basic, Ontario, Canada), 1 × PCR buffer, 0.2 mmol·l−1 dNTP, 1.5 mmol·l−1 MgCl2, 0.3 μmol·l−1 of each flanking primers (3A5P1: 5′GCC CTT GCA GCA TTT AGT CCT T3′ and 3A5P2: 5′ CCT GCC TTC AAT TTT TCA CTG 3′) and 0.45 μmol·l−1 of each allele-specific primer (3A5wt: 5′ CCA AAC AGG GAA GAG ATA T 3′ and 3A5mu: 5′ GAG CTC TTT TGT CTT TCA G 3′). The reaction was carried out according to the following program: 15 min at 94°C; followed by 35 cycles of 94°C for 30 s, 57°C for 60 s and 72°C for 60 s with a final extension at 72°C for 7 min. PCR products were analyzed by electrophoresis with 2% agarose gels.
MDR1 C1236T, G2677T/A, and C3435T polymorphisms were detected by assays based on the amplification refractory mutation system established in our laboratory [26]. C1236T and C3435T genotypes were detected by single-tube tetra-primer amplification methods. G2677T/A genotypes were detected by a two-step allele-specific amplification (ASA) assay. LD between different pairs of MDR1 SNPs was determined by the absolute standardized LD coefficient |D’|. Haplotype assembly was performed using a Haploview 3.2 program (Broad Institute of Harvard and MIT, Cambridge, MA, USA), based on a standard expectation maximization algorithm to reconstruct individual haplotypes from population genotype data [27].
PPK modeling
One-compartment, two-compartment and Michaelis–Menten (M–M) models were evaluated in model construction. Modeling was performed with NONMEM (Version 6; GloboMax, Hanover, MD, USA). Log-transformed concentration data were used to ensure that the random effects are sufficiently distributed around zero. The first order conditional estimation method (FOCE) was applied for the modeling. Model selection was based on the objective function value (OFV), parameter estimates and standard errors. OFV is proportional to −2 log likelihood of the relevant model, and a lower value indicates a better model. The distribution of empirical Bayesian estimates was also an important factor for model selection. The primary pharmacokinetic parameters were clearance (CL/F) and volume of distribution (Vd/F). Ka was fixed based on previously reported literature values [28, 29].
Inter-individual and residual error model
The inter-individual variability of the parameters was assessed using an exponential function:
where P i was the individual value, TV(P i ) was the population value for the parameters described in the equation, ηi was the random deviation of P i from TV(P i ). The values of ηi were assumed to be independently normally distributed with a mean of 0 and a variance of ω2.
As the concentration data were log transformed, an additive model was used for residual error analysis:
where Cobs is the observed concentration, Cpred is the predicted concentration, and ε is a residual error with a mean of 0 and a variance of σ2.
Covariates
Patients’ physiological and pathological characteristics and genetic polymorphisms were evaluated as the possible covariates of the CsA pharmacokinetic model. The influence of continuous covariates on the pharmacokinetic parameter TV(P) was modeled according to the following equations:
For categorical covariates such as gender, and the CYP3A5*3 and MDR1 genotypes, discrete numbers were given to each index: 0 and 1 for male and female patients. 0, 1, 2 for CYP3A5 *1/*1, *1/*3, and *3/*3 patients; wild-type CGC haplotype is assumed to have no effect on Pgp activity, where the mutant haplotype TTT has the most significant influence on Pgp activity. The impact of the other 10 haplotypes on Pgp activity is assumed to be between CGC and TTT, and they are considered as one group. 0 to 5 were given to the MDR1 diplotypes CGC/CGC, CGC/Other, Other/Other, CGC/TTT, Other/TTT, and TTT/TTT respectively. The pharmacokinetic parameter TV(P) was modeled according to the following equations:
where TV(P) is the typical value of the pharmacokinetic parameters, θ P is the population estimation of the parameter. θ c is the factor contributed by the covariate.
A forward inclusion and backward elimination technique was used for the final regression model. Each candidate covariate was screened in turn by adding into the base model. Weighted residuals and the change in the OFV were noted in the model building process. Changes in the OFV approximate the χ2 distribution with the degrees of freedom (df) equal to the number of covariates introduced. A covariate was considered statistically significant when the OFV decreased by 6.63 or greater (P < 0.01, df = 1) when added to the base model (forward inclusion). The full model included all covariates that show a significant decrease in OFV. Hence, each covariate remaining in the model was removed in turn by fixing its value as zero. This procedure was repeated until the value of the objective function failed to increase by more than the critical value of 7.88 (P < 0.005, df = 1; backward elimination). Individual pharmacokinetic parameters, arithmetic means, and standard deviations were calculated using the NONMEM Bayesian estimates from POSTHOC output.
Model evaluation
The stability and performance of the final model was assessed through an internal validation method that involved a non-parametric bootstrap with resampling and replacement. In this study, 600 bootstrap samples were generated, and the PPK parameters were estimated for each of the 600 samples by using the final model. The mean and standard error of parameter estimates from the bootstrap analyses were then compared with the NONMEM estimates from the final model.
Results
Patient population
The demographic and pathophysiological data of 146 patients were shown in Table 1. A total of 1,577 CsA blood samples were collected from various stages post-transplant. The C0 and C2 were 229 ± 89 and 1,035 ± 388 ng·ml−1. Samples were collected from 3 days to 146 days after transplantation. Two hundred and nine samples were obtained within 1 week; 432 samples were obtained from 1 week to 2 weeks; 622 samples were obtained from 2 weeks to 1 month; 267 samples were obtained from 1 month to 3 months; 46 samples were obtained over 3 months (Table 2). Thirty-eight patients received medications that potentially interact with CsA, including cimetidine (31 patients), bifendate (2 patients), diltiazem (2 patients), fluconazole (3 patients), nifedipine (2 patients), amlodipine (5 patients), and sirolimus (1 patient). Twenty-nine patients experienced acute rejection. Most of the rejections occurred within 1 month of transplantation.
MDR1 and CYP3A5 genotypes
The allele frequency of the CYP3A5*1 and *3 allele in 146 patients was 25.1% (18.1∼32.1%) and 74.9% (67.9∼81.9%) respectively. There were 7, 55, and 84 patients with *1/*1, *1/*3, and *3/*3 genotype. This result was consistent with the Hardy–Weinberg equilibrium (χ2 = 0.004, df = 2, P > 1.05).
The allele frequencies of MDR1 1236C and 1236T were 38.7% (30.8–46.6%) and 61.3% (53.4–69.2%); those of 2677G, 2677T, and 2677A were 46.2% (38.1–54.3%), 37.2% (29.4–45.0%), and 16.6% (10.6–22.6%); and those of 3435C and 3435T were 64.5% (56.8–72.4%) and 35.5% (27.8–43.3%) respectively. The distribution of various genotypes was consistent with the Hardy–Weinberg equilibrium (χ2 = 0.675, df = 2, P = 0.452 for C1236T; χ2 = 1.855, df = 5, P = 0.632 for G2677T/A; χ2 = 0.972, df = 2, P = 0.636 for C3435T).
D’ between G2677T/A or C1236T and C3435T were 0.85 and 0.77 respectively, whereas |D’| between G2677T/A and C1236T was 0.33. This indicated that G2677T/A and C1236T are in linkage disequilibrium with C3435T. There were 12 MDR1 haplotypes in renal transplant recipients, comparable with a previous study on Chinese subjects [30, 31]. Among these haplotypes, TTT (24.7%), CGC (22.2%), TGC (20.0%) were most frequently observed. For patients who were homozygous at three variants (such as CC/GG/CC or TT/TT/TT) or who were heterozygous at only one variant (such as CT/GG/CC), the haplotypes could be assigned unambiguously. However, for patients who were heterozygous at more than one variant, there may be ambiguous results. For example, a patient with the CT/GT/CT genotype, the haplotype pair might be CGC/TTT, TGT/CTC, TGC/CTT or TTC/CGT. As frequencies of CGC and TTT were much higher than those of TGT (4.2%), CTC (1.5%), CTT (1.0%), TTC (3.5%) or CGT (1.2%), the diplotype CGC/TTT was assigned. There were 8, 32, 52, 18, 26, and 10 patients with the CGC/CGC, CGC/other, other/other, CGC/TTT, other/TTT, TTT/TTT diplotypes.
Compartmental pharmacokinetic analysis
A one-compartment model with a single first-absorption process without lag time is the most appropriate structural model. Since CsA was administered orally, the clearance (CL/F) and distribution volume (Vd/F) included bioavailability (F).
After the forward inclusion and backward elimination step, POD (Fig. 1, Table 2), body weight (BW), TBIL, and the MDR1 genotype (Fig. 2) reduced the OFV by >7.88 (P < 0.005) when tested as the covariates of CL/F individually against the base model. Gender and MDR1 reduced the OFV by >7.88 (P < 0.005) when tested as the covariates of Vd/F individually against the base model (Table 3). After all covariates had been included in the model, inter-individual variability of CL/F was decreased from 22.4% to 16.1%; inter-individual variability of Vd/F was decreased from 32.1% to 28.6% (Table 4). The PPK parameters of CsA are presented in Table 4. CL/F (mean ± SE) was estimated to be 49.5 ± 5.2 l·h−1; the Vd/F was 226 ± 30 l; Ka was fixed as 1.25 h−1.
a Clearance (CL/F) and b volume of distribution (Vd/F) in various MDR1 diplotypes in 146 Chinese renal transplant recipients. The bold line in the box indicates medians; the lower and upper border of the box indicates 25% percentiles and 75% percentiles; the whisker of the box means the minimum and maximum range of data. Circles indicate outliers; asterisks indicate extreme values
To assess the effect of including 38 patients using combined medication, which might potentially interfere with CsA PPK parameters, we deleted these data from the population dataset after the final model was estimated. The PPK parameters were estimated again, and no significant changes were observed. It seemed that co-administration of these drugs has little effect on CsA pharmacokinetics.
The assessment of the predictive performance of the final model is presented in scatter plots of observed concentration versus population-predicted (Fig. 3a) and individual-predicted CsA concentrations (Fig. 3b); weighted residual vs population-predicted CsA concentration (Fig. 3c) and vs time (Fig. 3d) are also presented in Table 4. Residuals of most concentration data were randomly distributed within 2 standard deviations (SD), which indicates good agreement. The average bias of CsA was 33.6% (95% CI: 14.0% to 53.2%).
Goodness of fit of the population pharmacokinetics (PPK) model of cyclosporine (CsA). a Population-predicted concentration vs true concentration; b individual-predicted concentration vs true concentration; c Weighted residual error vs population-predicted concentration; d Weighted residual error vs time. PRED predicted, CONC concentration, WRES weighted residual error
Most of the 600 bootstraps ran successfully, and the results indicated even smaller 95% CI than those calculated by NONMEM (Table 4). The results of CL/F and Vd/F calculated by NONMEM are in the center of the histogram of 588 bootstrap runs (Fig. 4).
The AUC of CsA was estimated by a limited sampling strategy (LSS), and the equation established by Keown et al. [32, 33] was used: \( {\hbox{AU}}{{\hbox{C}}_{{0 - 12}}} = 312.66 + 1.84 \times {{\hbox{C}}_0} + 4.39 \times {{\hbox{C}}_2}\,{\hbox{ng}} \times {\hbox{h}} \times {\hbox{m}}{{\hbox{l}}^{{ - 1}}} \). At the same time, the AUC was calculated through Bayesian assay. The AUC was 7,465 ± 1,708 ng·h·ml−1 (2,946∼13,926 ng·h·ml-1). Figure 5 shows the relationships between C0, C2, and AUC estimated by LSS and the AUC estimated through the PPK model respectively. The figures showed that the AUC estimated by the PPK model correlated well with C2 and the AUC estimated by LSS. On the basis of the PPK model and the Bayesian method, conventional TDM data can be used in the rational usage of CsA.
Discussion
In this study, we established a one-compartment PPK model of CsA using conventional TDM data. Various demographic, pathophysiological, and genetic factors were tested as covariates of the PPK model.
Different PPK models have been used to describe CsA pharmacokinetics: one-compartment [28, 34], two-compartment [29], three-compartment [35], and M-M models [36]. Models with a lag time of 0, 1 or 2 and gamma distribution for the absorption phase have been used by various studies to describe the CsA absorption. In the present study, all 1,577 samples were collected at 0 h (trough level) and at 2 h (near peak level) after administration of CsA. We tested different models and found that the one-compartment model is satisfactory in simulating CsA pharmacokinetics. Ka was fixed because limited blood time points in the CsA absorption phase caused difficulty in estimation. We tested different values of Ka based on the work of Wu et al. and Schädeli et al. [28, 29], and found that Ka = 1.25 h−1 is most suitable. The CL/F and Vd/F of the basic model of the present study were 23.9 l·h−1 and 163 l respectively. The results are comparable with those of Rosenbaum et al. (23.1 l·h−1 and 202 l) [34] and Wu et al. (28.5 l·h−1 and 133 l) [28].
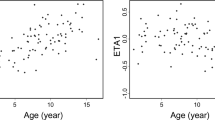
The impacts of various factors on the PPK parameters were evaluated. CL/F has been reported to decrease with POD, especially in the early stage [15, 28, 37, 38]. In the present study, we found significant differences in C0 and C2 among various groups classified according to POD (Table 2). In the basic model, weighted residuals (WRES) of the estimated concentrations increased with POD when POD ≤ 30 R2 = 0.21, P < 0.001), and there is no significant relationship when POD is over 30 R2 = 0.04, P > 0.05; Fig. 1). The results suggested that the concentration might be overestimated in the first month. We used POD as a covariate and fixed the POD at 30 for those samples collected after more than 1 month of therapy. The average decreasing CL/F in the first 4 weeks was 14.3%, 24.4%, 29.8%, and 33.4% respectively. The addition of POD caused an ΔOFV of over −400 points. The relationship between WRES and time was decreased in the final model (Fig. 3c). The role of POD may be partly attributed to the patients’ abnormal gastrointestinal function in the early stage post-transplantation, which impairs absorption of CsA and decreases the CsA bioavailability (F); hence, a higher CL/F is obtained. With the recovery of gastrointestinal motility during therapy the absorption of CsA is improved and the CL/F gradually stabilizes. On the other hand, prednisolone has been reported to activate steroid X receptor (SXR), which is an important regulator of CYP3A and MDR1 gene expression. The subsequent induction of CYP3A and Pgp can result in reduced absorption and accelerated metabolism of CsA. Tapering of steroid doses during immunosuppressive therapy may cause the reduction of expression of CYP3A and MDR1, and hence increase F and decrease CL/F of CsA. This effect may be reflected by the POD.
Consistent with a previous study [37], we found that BW had a significant influence on CsA clearance. There is a wide range of BW in patients in the present study (39–78 kg). BW changed dramatically even in the same patient during therapy. We found a 5.8∼8.0% increase in CL/F with every 10-kg increase in BW. Given a similar dosage, patients with higher BW had lower CsA concentrations. BW-adjusted dosage is applicable to maintain the CsA concentration within therapeutic range. On the other hand, in the forward inclusion step, we found that gender had a significant effect on both CL/F and Vd/F (ΔOFV was −6.2 and −7.9 respectively). Female patients have a 9% higher CL/F and 20% higher Vd/F. Previous studies have suggested that CYP3A in the intestine wall might be higher in female patients, which may decrease the F and further increase the Vd/F [39].Various biochemical indexes were tested as covariates. Patients with poor liver function are suggested to have impaired clearance of CsA. Patients with a high plasma total bilirubin level (TBIL) have higher blood concentrations of CsA [28, 37]. We also found the addition of TBIL caused ΔOFV to decrease by 32.7 points. For patients with one-fold higher TBIL over the average level, there will be a 15% decrease in CL/F.
Genetic polymorphism of drug metabolism enzymes and drug transporters is considered an important cause of pharmacokinetic variation [40]. The most important SNP of CYP3A5*3 is 6896A > G mutation in CYP3A5 intron 3, which results in a splice defect of the mRNA and produces an unstable and nonfunctional protein [41, 42]. Min et al. [43] studied 16 healthy subjects and found CL/F of *1/*1 was significantly higher than that of the *3/*3 group (P = 0.04). Haufroid et al. [44] showed that the CYP3A5 genotype could explain 9% of the total variance in CsA dose-adjusted C0 in 50 stable renal transplant recipients by using a stepwise multiple regression analysis. In the present study, we screened the CYP3A5*3 allele as a covariate. Compared with homozygous CYP3A5*1 patients, those with the mutant allele had a reduced CL/F for about 3.8% with each CYP3A5*3 allele (Fig. 2). However, CYP3A5*3 was excluded from the model in the backward exclusion stage as the increasing OFV is not sufficient (ΔOFV = 5.9). As previous studies have pointed out, CYP3A5 plays a minor role in the metabolism of CsA. Besides, the gene frequency of CYP3A5*3 is as high as 74.9%; there were only 7 patients with *1/*1 genotypes. It should also be noted that we constructed the PPK model using the conventional monitoring data, as limited data in the elimination phase further decrease the importance of the CYP3A5 genotype in CsA pharmacokinetics.
Recent studies suggest that the MDR1 haplotype is a good index of the genotype–phenotype relationship of Pgp. The study of Fredericks et al. [45] revealed significant lower dose-normalized CsA concentrations in patients with the CGC haplotype than those with the TTT haplotype (P = 0.024 for C0 and P = 0.048 for C2). The study classified patients as carriers or non-carriers of certain haplotypes. It should be noted that the diplotype effect can only be observed directly in vivo, and haplotype outcome is embodied in diplotypes. Our previous study [23] estimated MDR1 diplotypes on the basis of individual haplotypes reconstructed through a standard expectation maximization algorithm; patients were classified according to the diplotypes. We found that there is a significant difference in dose-adjusted C2 among various MDR1 diplotypes after 1 month of therapy. The CL/F and Vd/F of TTT/TTT subjects is about 29% and 31% lower than those of the CGC/CGC group. Pgp is found to be distributed widely in the brush border surface of the intestine, in the canalicular surface of hepatocytes, and in the apical surface of proximal tubular cells in the kidneys [46, 47]. Theoretically, both the absorption and the elimination of CsA can be affected by MDR1 polymorphism. However, CsA is extensively metabolized in vivo and less than 1% is excreted as the parent drug [48]. The role of MDR1 haplotypes in CsA absorption is more important. For CGC/CGC patients, more CsA was excreted back to the intestinal lumen. The F of CsA is lower for CGC/CGC patients than for TTT/TTT patients, causing higher CL/F and Vd/F for CGC/CGC patients.
Cyclosporine is the substrate of CYP3A and Pgp, and potential drug interactions have been reported in previous studies [49]. In this study, 38 patients using combined drugs may have altered absorption and metabolism of CsA in a certain period during therapy. At the time of administration, the duration and dosage of the drugs introduced were variable; thus, it is hard to test the coadministration of these drugs as a covariate. Thus, we tested the model with or without the patients who had coadministration of a certain drug to see if there is a difference in the pharmacokinetic parameters of the different models. We found that there is no significant change in the parameters between different models, so we did not consider drug interaction as a covariate in this model. The reason may be that these drugs are occasionally given, but the number of patients and duration of drug used are limited. We also tested the dosage of steroids as a covariate, and did not find any significant improvement. The reason may be that the dosage regimen of prednisolone for each patient was similar, the influence of SXR on CYP3A and Pgp for each patient is comparable, and hence the variation in CsA level caused by SXR is limited. It should also be noted that MPA has the adverse effect of causing diarrhea in some patients. This may be another reason for the variation in CsA absorption. As there are no complete data on the patients with abnormal gastrointestinal function, we did not test the compound use of MMF as a covariate. All these factors should be evaluated in personalized CsA therapy.
It is accepted that AUC is a sensitive predictor of outcomes such as acute rejection episodes and chronic rejection [6, 7]. A full time point pharmacokinetic profile is impractical because of the increased inconvenience and costs to patients. C0, C2 or other blood sample-collecting profiles are used to approximate the full time point AUC [33]. In this study, we divided dosage by CL/F and obtained AUC adjusted by F. We correlated AUC obtained through Bayesian assay with C0, C2, and AUC estimated through LSS, and found good correlation between AUC and C2 or estimated AUC0–12. By using the Bayesian method, we think that the AUC is more reliable and useful in CsA TDM.
The major limitation of this study was the fixed sampling time for CsA concentration. PPK studies ideally should consist of concentration–time data obtained at randomized times across a dosing interval. The data collected in the present study are all C0 and C2 after the dose. Unfortunately, because of the paucity of information in the early period after administration, it was not possible to model the absorption characteristics of CsA. Nevertheless, the use of fixed Ka enabled us to model the data and obtain good estimates for CL/F and Vd/F, which is valuable in the regulation of CsA therapy. Besides, CsA was monitored by FPIA in the present study, which is the most frequently used method in the conventional TDM assay of CsA because it is rapid, convenient, and less laborious. However, it should be noted that FPIA and other immunoassays are rather nonspecific compared with HPLC-UV or LC-MS because of the possibility of cross-reacting with CsA metabolites. This may be a cause of the relatively high residual error in this study.
In conclusion, in this study we established the PPK model of CsA in patients after renal transplantation from the early stage until 5 months after transplantation. We found that besides POD, body weight, gender, and total bilirubin, the MDR1 haplotype is also a significant covariate for describing the CL/F and Vd/F of CsA.
References
The Canadian Multicentre Transplant Study Group (1986) A randomized clinical trial of cyclosporine in cadaveric renal transplantation. Analysis at three years. N Engl J Med 314(19):1219–1225
Ponticelli C, Minetti L, Di Palo FQ, Vegeto A, Belli L, Corbetta G, Tarantino A, Civati G (1988) The Milan clinical trial with cyclosporine in cadaveric renal transplantation. A three year follow up. Transplantation 45(5):908–913
Kahan BD (2004) Therapeutic drug monitoring of cyclosporine: 20 years of progress. Transplant Proc 36(2 Suppl):378S–391S
Nankivell BJ, Hibbins M, Chapman JR (1994) Diagnostic utility of whole blood cyclosporine measurements in renal transplantation using triple therapy. Transplantation 58(9):989–996
Oellerich M, Armstrong VW, Kahan B, Shaw L, Holt DW, Yatscoff R, Lindholm A, Halloran P, Gallicano K, Wonigeit K (1995) Lake Louise Consensus Conference on cyclosporin monitoring in organ transplantation: report of the consensus panel. Ther Drug Monit 17(6):642–654
Lindholm A, Kahan BD (1993) Influence of cyclosporine pharmacokinetics, trough concentrations, and AUC monitoring on outcome after kidney transplantation. Clin Pharmacol Ther 54(2):205–218
Schroeder TJ, Hariharan S, First MR (1994) Relationship between cyclosporine bioavailability and clinical outcome in renal transplant recipients. Transplant Proc 26(5):2787–2790
Kahan BD, Grevel J (1988) Optimization of cyclosporine therapy in renal transplantation by a pharmacokinetic strategy. Transplantation 46(5):631–644
International Neoral Renal Transplantation Study Group (2002) Cyclosporine microemulsion (Neoral) absorption profiling and sparse-sample predictors during the first 3 months after renal transplantation. Am J Transplant 2(2):148–156
Lindholm A, Säwe J (1995) Pharmacokinetics and therapeutic drug monitoring of immunosuppressants. Ther Drug Monit 17(6):570–573
Cantarovich M, Barkun JS, Tchervenkov JI, Besner JG, Aspeslet L, Metrakos P (1998) Comparison of neoral dose monitoring with cyclosporine through levels versus 2-hr postdose levels in stable liver transplant patients. Transplantation 66(12):1621–1627
Morris RG, Russ GR, Cervelli MJ, Juneja R, McDonald SP, Mathew TH (2002) Comparison of trough, 2-hour, and limited AUC blood sampling for monitoring cyclosporin (Neoral) at day 7 post-renal transplantation and incidence of rejection in the first month. Ther Drug Monit 24(4):479–486
Marquet P (2005) Clinical application of population pharmacokinetic methods developed for immunosuppressive drugs. Ther Drug Monit 27(6):727–732
Awni WM (1992) Pharmacodynamic monitoring of cyclosporin. Clin Pharmacokinet 23(6):428–448
Saint-Marcoux F, Marquet P, Jacqz-Aigrain E, Bernard N, Thiry P, Le Meur Y, Rousseau A (2006) Patient characteristics influencing ciclosporin pharmacokinetics and accurate Bayesian estimation of ciclosporin exposure in heart, lung and kidney transplant patients. Clin Pharmacokinet 45(9):905–922
Dai D, Tang J, Rose R, Hodgson E, Bienstock RJ, Mohrenweiser HW, Goldstein JA (1992) Identification of variants of CYP3A4 and characterization of their abilities to metabolize testosterone and chlorpyrifos. Biochem Pharmacol 43(10):2201–2208
Hsieh KP, Lin YY, Cheng CL, Lai ML, Lin MS, Siest JP, Huang JD (2001) Novel mutations of CYP3A4 in Chinese. Drug Metab Dispos 29(3):268–273
Balram C, Zhou Q, Cheung YB, Lee EJ (2003) CYP3A5*3 and *6 single nucleotide polymorphisms in three distinct Asian populations. Eur J Clin Pharmacol 59(2):123–126
Miao LY, Huang CR, Hou JQ, Qian MY (2008) Association study of ABCB1 and CYP3A5 gene polymorphisms with sirolimus trough concentration and dose requirements in Chinese renal transplant recipients. Biopharm Drug Dispos 29(1):1–5
Ambudkar SV, Kimchi-Sarfaty C, Sauna ZE, Gottesman MM (2003) P-glycoprotein: from genomics to mechanism. Oncogene 22(47):7468–7485
Marzolini C, Paus E, Buclin T, Kim RB (2004) Polymorphisms in human MDR1 (P-glycoprotein): recent advances and clinical relevance. Clin Pharmacol Ther 75:13–33
Sakaeda T, Nakamura T, Okumura K (2002) MDR1 genotype-related pharmacokinetics and pharmacodynamics. Biol Pharm Bull 25:1391–1400
Chen B, Zhang W, Fang J, Jin Z, Li J, Yu Z, Cai W (2009) Influence of the MDR1 haplotype and CYP3A5 genotypes on cyclosporine blood level in Chinese renal transplant recipients. Xenobiotica 39(12):931–938
Cockcroft DW, Gault MH (1976) Prediction of creatinine clearance from serum creatinine. Nepharon 16:31–41
Traub SL, Johnson CE (1980) Comparison of methods of estimating creatinine clearance in children. Am J Hosp Pharm 37:195–201
Chen B, Fang J, Zhang W, Jin Z, Yu Z, Cai W (2009) Detection of C1236T, G2677T/A, and C3435T polymorphism of MDR1 by amplification refractory mutation system PCR. J Clin Lab Anal 23(2):110–116
Barrett JC, Fry B, Maller J, Daly MJ (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21(2):263–265
Wu KH, Cui YM, Guo JF, Zhou Y, Zhai SD, Cui FD, Lu W (2005) Population pharmacokinetics of cyclosporine in clinical renal transplant patients. Drug Metab Dispos 33(9):1268–1275
Schädeli F, Marti HP, Frey FJ, Uehlinger DE (2002) Population pharmacokinetic model to predict steady-state exposure to once-daily cyclosporin microemulsion in renal transplant recipients. Clin Pharmacokinet 41(1):59–69
Chowbay B, Cumaraswamy S, Cheung YB, Zhou Q, Lee EJ (2003) Genetic polymorphisms in MDR1 and CYP3A4 genes in Asians and the influence of MDR1 haplotypes on cyclosporin disposition in heart transplant recipients. Pharmacogenetics 13(2):89–95
Tang K, Ngoi SM, Gwee PC, Chua JM, Lee EJ, Chong SS, Lee CG (2002) Distinct haplotype profiles and strong linkage disequilibrium at the MDR1 multidrug transporter gene locus in three ethnic Asian populations. Pharmacogenetics 12:425–427
Keown P, Landsberg D, Halloran P, Shoker A, Rush D, Jeffery J, Russell D, Stiller C, Muirhead N, Cole E, Paul L, Zaltzman J, Loertscher R, Daloze P, Dandavino R, Boucher A, Handa P, Lawen J, Belitsky P, Parfrey P (1996) A randomized, prospective multicenter pharmacoepidemiologic study of cyclosporine microemulsion in stable renal graft recipients. Report of the Canadian Neoral Renal Transplantation Study Group. Transplantation 62(12):1744–1752
David OJ, Johnston A (2001) Limited sampling strategies for estimating cyclosporin area under the concentration-time curve: review of current algorithms. Ther Drug Monit 23(2):100–114
Rosenbaum SE, Baheti G, Trull AK, Akhlaghi F (2005) Population pharmacokinetics of cyclosporine in cardiopulmonary transplant recipients. Ther Drug Monit 27(2):116–122
Fanta S, Jönsson S, Backman JT, Karlsson MO, Hoppu K (2007) Developmental pharmacokinetics of ciclosporin—a population pharmacokinetic study in paediatric renal transplant candidates. Br J Clin Pharmacol 64(6):772–784
Grevel J, Post BK, Kahan BD (1993) Michaelis-Menten kinetics determine cyclosporine steady-state concentrations: a population analysis in kidney transplant patients. Clin Pharmacol Ther 53(6):651–660
Parke J, Charles BG (2000) Factors affecting oral cyclosporin disposition after heart transplantation: bootstrap validation of a population pharmacokinetic model. Eur J Clin Pharmacol 56(6–7):481–487
Irtan S, Saint-Marcoux F, Rousseau A, Zhang D, Leroy V, Marquet P, Jacqz-Aigrain E (2007) Population pharmacokinetics and bayesian estimator of cyclosporine in pediatric renal transplant patients. Ther Drug Monit 29(1):96–102
Lampen A, Christians U, Bader A, Hackbarth I, Sewing KF (1996) Drug interactions and interindividual variability of ciclosporin metabolism in the small intestine. Pharmacology 52(3):159–168
Lown KS, Kolars JC, Thummel KE, Barnett JL, Kunze KL, Wrighton SA, Watkins PB (1994) Interpatient heterogeneity in expression of CYP3A4 and CYP3A5 in small bowel. Lack of prediction by the erythromycin breath test. Drug Metab Dispos 22(6):947–955
Hustert E, Haberl M, Burk O, Wolbold R, He YQ, Klein K, Nuessler AC, Neuhaus P, Klattig J, Eiselt R, Koch I, Zibat A, Brockmöller J, Halpert JR, Zanger UM, Wojnowski L (2001) The genetic determinants of the CYP3A5 polymorphism. Pharmacogenetics 11:773–779
Kuehl P, Zhang J, Lin Y, Lamba J, Assem M, Schuetz J, Watkins PB, Daly A, Wrighton SA, Hall SD, Maurel P, Relling M, Brimer C, Yasuda K, Venkataramanan R, Strom S, Thummel K, Boguski MS, Schuetz E (2001) Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nat Genet 27:383–391
Min DI, Ellingrod VL, Marsh S, McLeod H (2004) CYP3A5 polymorphism and the ethnic differences in cyclosporine pharmacokinetics in healthy subjects. Ther Drug Monit 26(5):524–528
Haufroid V, Mourad M, Van Kerckhove V, Wawrzyniak J, De Meyer M, Eddour DC, Malaise J, Lison D, Squifflet JP, Wallemacq P (2004) The effect of CYP3A5 and MDR1 (ABCB1) polymorphisms on cyclosporine and tacrolimus dose requirements and trough blood levels in stable renal transplant patients. Pharmacogenetics 14(3):147–154
Fredericks S, Jorga A, MacPhee IA, Reboux S, Shiferaw E, Moreton M, Carter ND, Holt DW, Johnston A (2007) Multi-drug resistance gene-1 (MDR1) haplotypes and the CYP3A5*1 genotype have no influence on ciclosporin dose requirements as assessed by C0 or C2 measurements. Clin Transplant 21(2):252–257
Thiebaut F, Tsuruo T, Hamada H, Gottesman MM, Pastan I, Willingham MC (1987) Cellular localization of the multidrug-resistance gene product P-glycoprotein in normal human tissues. Proc Natl Acad Sci USA 84:7735–7738
Schinkel AH, Jonker JW (2003) Mammalian drug efflux transporters of the ATP binding cassette (ABC) family: an overview. Adv Drug Deliv Rev 55(1):3–29
Christians U, Sewing KF (1993) Cyclosporin metabolism in transplant patients. Pharmacol Ther 57(2–3):291–345
Lindholm A (1991) Factors influencing the pharmacokinetics of cyclosporine in man. Ther Drug Monit 13:464–477
Acknowledgement
Supported by the National Natural Science Foundation of China (No. 30500626).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Chen, B., Zhang, W., Gu, Z. et al. Population pharmacokinetic study of cyclosporine in Chinese renal transplant recipients. Eur J Clin Pharmacol 67, 601–612 (2011). https://doi.org/10.1007/s00228-010-0959-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00228-010-0959-2