Abstract
Runx2, a transcription factor, is essential for osteoblastic differentiation, bone formation, and maintenance. We examined the effect of Runx2 on transdifferentiation of 3T3-L1 preadipocytes into functional, mature osteoblasts. Forced expression of exogenous Runx2 using a retroviral gene-delivery system showed increases of alkaline phosphatase (ALP) activity and expression of the osteoblastic marker genes osteocalcin (OC), bone sialoprotein (BSP), and osterix (Osx), accompanied by low-level matrix mineralization. In contrast, adipocytic differentiation was completely blocked with downregulation of adipogenic transcription factors PPARγ2, C/EBPα, and C/EBPδ. Treatment of dexamethasone (Dex), a synthetic glucocorticoid, stimulated the formation of mineralized nodules in Runx2-overexpressing 3T3-L1 cells with increases of ALP, OC, BSP, and Osx expression. Here, we focused on a dual specific phosphatase, mitogen-activated protein kinase (MKP-1), since Dex significantly increased MKP-1 expression in Runx2-overexpressing 3T3-L1 cells. Forced expression of exogenous MKP-1 resulted in accumulation of robust matrix mineralization in parallel with induction of ALP activity and expression of OC, BSP, and Osx in Runx2-overexpressing 3T3-L1 cells. These results suggest that simultaneous overexpression of Runx2 and MKP-1 is effective for transdifferentiation of preadipocytes into fully differentiated bone-forming osteoblasts and provide a novel strategy for cell-based therapeutic applications requiring significant numbers of osteogenic cells to synthesize mineralized constructs for the treatment of large bone defects.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Osteoblasts and adipocytes are originated from common mesenchymal progenitor cells, and several transcription factors control the differentiation of the two lineages. Runx2 is a transcription factor that belongs to the runt domain gene family and has a DNA-binding domain that is homologous to the Drosophila pair-rule gene runt [1]. Runx2 was found to control bone formation during both skeletal development and postnatal life [2]. In the differentiation process of precursor cells into osteoblasts, Runx2 is activated and regulates the expression of osteoblastic marker genes such as osteocalcin (OC) [3], osteopontin (OPN) [4], and bone sialoprotein (BSP) [5]. This regulation occurs when Runx2 interacts with a DNA sequence termed “osteoblast-specific cis-acting element 2” (OSE2) [3, 5, 6]. Several studies have revealed that forced expression of exogenous Runx2 in nonosteoblastic cells or osteogenic cells upregulates the expression of osteoblastic makers and induces in vitro matrix mineralization [6–9]. In addition, gene-alteration studies have shown that heterozygous mutation of the Runx2 gene is implicated in the human disorder cleidocranial dysplasia [10], while homozygous mutation in mice results in a complete lack of mineralization and immediate postnatal death [6]. However, transgenic mice overexpressing exogenous Runx2 lead to osteopenia with a decrease of bone mineral density and multiple fractures [11, 12], although Runx2 is essential for bone formation.
Adipogenic differentiation requires the sequential expression of two major groups of transcription factors, including hormone receptor peroxisome proliferator-activated receptor γ (PPARγ) and CCAAT/enhancer-binding proteins (C/EBPs) [13]. PPARγ is expressed as two isoforms, PPARγ1 and PPARγ2, which are generated from alternative promoter usage and splicing [14]. The C/EBP families belong to the basic leucine zipper class of transcription factor and bind to specific DNA sequences as dimers with other C/EBPs [15]. When adipocytic differentiation is induced in preadipocytic cells, C/EBPβ and δ are rapidly and transiently induced [16, 17]. These regulators play important roles in the induction of C/EBPα and PPARγ2 via interaction with C/EBP regulatory elements present in the proximal promoter of these genes [18, 19]. Subsequently, C/EBPα and PPARγ2 involve the expression of adipocytic markers such as adipocyte P2 (aP2), which is necessary for the development of functional, mature adipocytes [20, 21]. Early markers of adipogenic differentiation are evaluated to increase the expression of PPARγ2 and aP2, whereas the formation of lipid droplets in the cytoplasm of adipocytes is a late marker of adipogenesis [22].
Transdifferentiation of bone marrow adipocytes into osteoblasts has been reported previously. In particular, a relatively pure population of adipocytes from bone marrow cells appears to have the ability to revert to a more proliferative phase and subsequently to differentiate in the osteogenic direction, which requires an intermediate step to induce morphological change into preadipocytes [23]. Another report has also shown that subcutaneous preadipocytes have the potential to differentiate into bone-forming osteoblasts [24]. These findings suggest that conversion of adipocytes to osteoblasts goes through a preadipocytic stage with enhanced proliferation potential, although lipid-filled mature adipocytes are not directly differentiated to osteoblasts [25]. However, the molecular mechanism for the preadipocyte conversion into osteoblasts is not fully understood. A recent study has reported that overexpression of Runx2 in adipose-derived stem cells stimulates the induction of osteoblastic differentiation accompanied by detectable levels of mineralized nodules and decreased PPARγ mRNA [26]. A preadipocytic cell line originated from mouse subcutaneous fat tissues also induces differentiation into mature osteoblasts with increased Runx2 mRNA [27]. These findings raise the possibility that Runx2 is a key factor promoting the transdifferentiation of adipocytes into functional osteoblasts.
3T3-L1 cells, an immortalized preadipocyte cell line, are established from subclones of 3T3 mouse embryonic fibroblasts [27]. Several days after reaching confluence, 3T3-L1 cells lose their fibroblastic morphology, round up, and differentiate into mature adipocytes, accumulating large amounts of cytoplasmic triglyceride [27, 28]. 3T3-L1 cells also differentiate and develop into tissue indistinguishable from normal adipose when transplanted subcutaneously into nude mice [28]. Here, we examined the effect of Runx2 on the transdifferentiation potential of 3T3-L1 cells into osteoblasts using a retroviral gene-delivery system. As will be shown, Runx2-overexpressing 3T3-L1 cells increased alkaline phosphatase (ALP) activity and expression of OC, BSP, and Osx with the formation of mineralized matrix. Osteoblastic differentiation was synergistically enhanced when Runx2-overexpressing 3T3-L1 cells were introduced with the expression vector encoding a full-length mitogen-activated protein kinase phosphatase-1 (MKP-1) gene, which modulates glucocorticoid-dependent osteoblast differentiation involving Runx2 activity [9]. These results demonstrated an important implication for our understanding of the role of Runx2 and MKP-1 in the transdifferentiation of preadipocytes into fully differentiated bone-forming osteoblasts.
Materials and Methods
Antibodies and Reagents
Polyclonal rabbit anti-mouse Runx2 antibody and HRP-conjugated donkey anti-goat IgG were obtained from Abcam (Cambridge, MA). Polyclonal rabbit anti-mouse PPARγ, polyclonal rabbit anti-human MKP-1, goat anti-rabbit actin, and HRP-conjugated anti-rabbit IgG antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Alexa Fluor 546-conjugated goat ant-mouse IgG and Alexa Fluor 488-conjugated goat anti-rabbit IgG were obtained from Invitrogen (Carlsbad, CA). Lipofectamine 2000, geneticin, penicillin-streptomycin, and reverse-transcription reagents including SuperScript IITM reverse transcriptase and oligonucleotide primers were also purchased from Invitrogen. Reagents for RNA isolation, standard reverse transcription (RT)-polymerase chain reaction (PCR) were acquired from TaKaRa Bio (Otsu, Shiga, Japan). Alizarin-Red S, Oil-Red O, water-soluble dexamethasone (Dex), and puromycin were obtained from Sigma (St. Louis, MO). Tablet containing NBT/BCIP or proteinase inhibitors was purchased from Roche Diagnostic (Mannheim, Germany). The calcium C-test kit and BCA protein assay kit were obtained from Wako Chemical (Tokyo Japan) and Pierce Biotechnology (Rockford, IL), respectively.
Plasmids
A full-length mouse Runx2 cDNA (Osf2-pCMV5) [6] and six-repeated osteoblast-specific cis-acting element 2 inserted upstream of the luciferase gene in pGL3-Basic (6× OSE2/pGL3) [3] were kindly provided by Professor Gérard Karsenty (College of Physicians and Surgeons, Columbia University, New York, NY). An expression vector, which was constructed by subcloning full-length mouse MKP-1 cDNA into pcDNA3.1(+) (MKP-1/pcDNA), was provided by Dr. Yoshikazu Mikami (Department of Anatomy, Nihon University School of Dentistry) [29].
Cell Culture
A mouse 3T3-L1 preadipocytic cell line was obtained from Health Science Research Resources Bank (JCRB9014, Osaka, Japan) and maintained in growth medium consisting of Dulbecco’s modified Eagle medium (DMEM, Wako Chemical) and 10% fetal bovine serum (FBS), 100 U/mL penicillin, and 100 μg/mL streptomycin. 3T3-L1 cells overexpressing Runx2 or/and MKP-1 were cultured with growth medium in the presence or absence 10 mM β-glycerophosphate and 50 μg/mL l-ascorbic acid to mineralize. A synthetic glucocorticoid, Dex, was also used. Cells at a density 1 × 105 or 1 × 104/mL were seeded on a 24-well plate or an eight-well culture slide, respectively, and maintained with growth medium overnight. Subsequently, medium was replaced and further cultured for 3, 6, 7, 9, 10, 14, 16, 18, and 21 days. Medium was changed every 3 days until the terminal assay. All culture plates and culture slides were obtained from BD Falcon (Franklin Lakes, NJ).
Construction of Runx2-Retroviral Vector and Retroviral Infection
A retroviral gene-transfer and expression system (Clontech, Mountain View, CA) was used for production of recombinant retrovirus for Runx2. A 112-bp fragment at the end of the 3′-untranslated region of Osf2-pCMV5 was amplified by PCR using primers 5′-CCA GAA TGA TGG TGT TGA CG-3′ and 5′-CTC GAG TCA ATA TGG CCG CCA AAC AG-3′, which contain restriction sites of BstXI and XhoI, respectively; ligated with a 1.71-kb fragment of Osf2-pCMV5 digested with EcoRI and BstXI; and then subcloned into EcoRI and XhoI sites in pBluescript KS (Osf2-pBS). Next, a 300-bp fragment of the 5′ region of Osf2 encoding the amino acid sequence of MSHLS of type II Runx2 [30] was amplified by PCR using primers 5′-AGA TCT ATG GCG TCA AAC AG-3′ and 5′-CCA TGG TGC GGT TGT CGT G-3′, containing restriction sites of BglII and NcoI, respectively. This product was ligated with a 1.3-kb fragment of Osf2-pBS digested with NcoI and XhoI and subcloned into BglII and XhoI sites of pMSCV puro.
Retroviral stocks were produced by transient transfection of PT67 cells with recombinant plasmid according to the manufacturer’s instruction. Briefly, cells were plated at a density of 5 × 106/mL on a 100-mm culture dish (BD Falcon) with growth medium for 24 hours and then transfected with 10 μg of pMSCVpuro containing Runx2cDNA or empty vector (no insert Runx2), using Lipofectamine 2000. Forty-eight hours after transfection, supernatants were collected and filtered through a 0.45-μm cellulose acetate membrane (Millipore, Bedford, MA) and aliquots were stored at −80°C until use.
For retroviral infection, 3T3-L1 cells were seeded on a 100-mm culture plate at a density 5 × 105/mL. Cells at 80% confluence were infected by adding 5 mL of retroviral supernatants and incubated for 24 h. Medium was changed to fresh growth medium containing 1 ng/mL puromycin, and cells were further cultured for 2 weeks. Some colonies were collected and analyzed.
Transfection of MKP-1 in Runx2-Overexpressing 3T3-L1 Cells
Runx2-overexpressing 3T3-L1 cells seeded on a 100-mm culture plate at a density of 1 × 105/mL were incubated overnight, and 10 μg of MKP-1/pcDNA was added with Lipofectamine 2000 in the growth medium. Five hours after incubation, medium was replaced with fresh growth medium and cells were cultured for 16 h and then expanded at a density of 1 × 104/mL with fresh growth medium containing 400 μg/mL geneticin. Three weeks after incubation, a positive clone, which simultaneously overexpresses Runx2 and MKP-1, was collected.
Histochemical Analyses
Cells in a 24-well plate were fixed with 10% buffered formalin, and ALP staining using NBT/BCIP tablet and Alizarin-Red S staining were performed to examine osteoblastic activity, respectively. After washing with distilled water to reduce nonspecific stain, plates were scanned using an Epson GT-X800 scanner (Seiko Epson, Tokyo, Japan).
Formation of lipid droplets was detected by Oil-Red O. Cells in eight-well culture slides were fixed and stained with Oil-Red O solution for 30 minutes at room temperature, washed with distilled water, and then mounted for inspection using light microscopy (Olympus, Tokyo, Japan).
Cell Growth Assay
Confluent cells in a 96-well culture plate were cultured for 1, 4, 7, 9, 14, and 21 days. A cell-growth assay was performed with a cell-counting kit (Dojindo, Kumamoto, Japan) according to the manufacturer’s instructions. After incubation with 100 μL solution containing WST reagent at 37°C for 45 min, supernatants were collected and cell-growth activity was detected by spectrophotometry at OD450.
Calcium Content
Calcium incorporation into mineralized matrix was determined as described previously [31]. Briefly, 300 μL of 0.5 M HCl was added to cells cultured in 24-well plates and incubated overnight at room temperature to decalcify the mineralized nodules. The calcium content was determined quantitatively using a calcium C-test kit, and the protein content was determined using a BCA protein assay kit after evaporation of HCl from the samples.
RT-PCR
Standard RT-PCR was performed using 1 μg of total RNA as a template. Double-stranded cDNA was synthesized from 1 μg of DNase I-treated total RNA in 20 μL of buffer containing 10 ng random primer, 10 mM dNTP mixture, 1 mM DTT, and 0.5 units SuperScript IITM reverse transcriptase at 50°C for 30 minutes and 94°C for 2 minutes. Subsequently, 1 μL of reaction solution was subjected to RT-PCR in 10 μL of buffer containing 10 mM NTPs, gene-specific primers, and 0.1 U Taq DNA polymerase (Finnzymes, Keilaranta, Finland). Forward and reverse primers are listed in Table 1. A reaction condition at 35 cycles was as follows: denaturing at 94°C for 15 s, annealing and extension at 64°C for 30 s, and final extension at 72°C for 10 min. PCR products were subjected to 2% agarose gel electrophoresis and visualized by ethidium bromide staining.
Luciferase Assay
Cells in 24-well plates were transfected with 2 μg of 6× OSE2/pGL3 using Lipofectamine 2000. Twenty-four hours after transfection, cells were washed with PBS(−) and scraped with 200 μL of lysis buffer. Luciferase assay was performed using the Dual Luciferase Assay Kit (Promega, Madison, WI) according to the manufacturer’s instructions. Experiments were performed in triplicate, and values of luciferase activity were normalized by that obtained by renilla luciferase activity as an internal control.
Western Blotting
Cells were washed with ice-cold PBS(−); resuspended with 200 μL of lysis buffer containing 0.2 M Tris–HCl (pH 7.5), 1% Triton X-100, 0.1% SDS, 150 mM NaCl, and a tablet of proteinase inhibitors; disrupted by gentle rocking for 30 min; and centrifuged at 15,000×g for 15 min at 4°C. Supernatants were collected and protein concentrations were determined with the BCA protein assay kit using bovine serum albumin (BSA) as a standard. Ten micrograms of samples were fractionated in a gradient SDS-PAGE gel (ATTO, Tokyo, Japan) and transferred to Imobilon-P membrane (Millipore). Membranes were blocked with 5% nonfat powdered milk dissolved with Tris-buffer saline (TBS) containing 20 mM Tris-HCl (pH 7.6), 137 mM NaCl for 1 h at room temperature and washed with TBS containing 0.1% Tween 20 (TBS-T). Rabbit polyclonal antibody to Runx2 or MKP-1 was added at a dilution of 1:250 and incubated for 2 hours. After washing with TBS-T, HRP-conjugated goat anti-rabbit IgG was added at a dilution of 1:500 for 1 h, a final wash was performed with TBS-T for 30 minutes three times, and then protein bands were visualized by ECL detection kit (GE Healthcare, Buckinghamshire, England, UK). Semiquantitative analysis was performed using the Image software package (version 1.63; NIH, Bethesda, MD).
Immunofluorescence Study
Cells in eight-well culture slides were fixed with 4% paraformaldehyde for 5 min, washed with PBS(−), and blocked with PBS(−) containing 1% BSA for 1 h at room temperature. After washing with PBS(−), polyclonal rabbit antibody to Runx2 or mouse antibody to PPARγ was added at a dilution of 1:250, incubated for 2 h, and then washed. Alexa Fluo 546-conjugated goat anti-rabbit IgG or Alexa Fluo 488-conjugated anti-mouse IgG was added at a dilution of 1:500 and incubated for 1 hour at room temperature. After a final wash with PBS(−), cells were mounted and visualized with a fluorescence microscope (Eclipse E600; Nikon, Tokyo, Japan).
Data Analysis
Values are expressed as mean ± SE. Statistical analysis was performed using Student’s t-test, and P < 0.05 was considered significant.
Results
Overexpression of Exogenous Runx2 in 3T3-L1 Cells
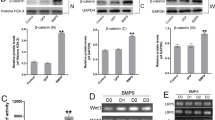
In order to generate Runx2-overexpressing 3T3-L1 cells, recombinant retrovirus encoding mouse type II Runx2 cDNA [30] was used as a transducer of the gene-delivery system. An immunofluorescence study using polyclonal anti-Runx2 antibody showed large amounts of Runx2 protein localized in nuclei of Runx2-overexpressing clones 5, 9, and 11 (Fig. 1a, left), whereas PPARγ were detected in clone 4 and controls including parental 3T3-L1 (L1) and a clone with empty vector (Co) (Fig. 1a, right). Western blotting also showed increased Runx2 expression in clones 5, 9, and 11 but low expression in clones 4 and Co (Fig. 1b). A molecular size reacting to anti-Runx2 antibody is approximately 65 kDa, which corresponds to that of type II Runx2 reported previously [32]. Semiquantitative analysis of these signals using NIH Image software detected the highest level of Runx2 expression in clone 9 among four clones examined (Fig. 1c). In contrast, PPARγ protein levels were lower in clones 5, 9, and 11 compared to that in clone 4 and Co (Fig. 1b, d). Taken together, we selected clone 9 for Runx2-overexpressing 3T3-L1 cells and further characterized it.
Overexpression of Runx2 in 3T3-L1 cells. Cells were infected with recombinant retrovirus encoding Runx2. By adding puromycin, Runx2-overexpressing clones 4, 5, 9, and 11 were selected and analyzed. 3T3-L1 cells (L1) and a clone with empty vector (Co) were used for negative controls. a Immunofluorescence analysis of Runx2 (left) and PPARγ (right) in Runx2-overexpressing clones was performed (original magnification of all images ×200). b Western blotting detected the expression level of Runx2 and PPARγ proteins in Runx2-overexpressing clones. c Semiquantitative analysis of Runx2 (left) and PPARγ (right) expression levels shown in b was performed using NIH image software
Effect of Runx2 on Osteogenic and Adipogenic Differentiation of 3T3-L1 Cells
At first, the effect of Runx2-overexpressing clone 9 on osteogenic differentiation was examined. Confluent cells of clone 9, L1, and Co were maintained for 7, 14, and 21 days; and the mRNA level of osteoblastic markers was detected by RT-PCR. As expected, Runx2-overexpressing clone 9 showed inductive ALP activity and mRNA levels of ALP, OC, and BSP. An increase of Osx mRNA level was also shown at 14 and 21 days, but that of OC, BSP, and Osx was undetectable in L1 and Co at all culture times (Fig. 2a, b). Basal mRNA levels of two osteoblastic transcription factors, Dlx5 and Msx2, did not change in clone 9, L1, and Co, even though they were cultured for 21 days (Fig. 2a). Alizarin-Red S staining in clone 9 showed small amounts of mineralized matrix at 21 days with the increase of calcium deposition (Fig. 2c, d). However, no mineralization was detected in clones 4, 5, and 11 (data not shown) as well as L1 and Co at 21 days of culture (Fig. 2c).
Osteogenic potential of Runx2-overexpressing 3T3-L1 cells. Confluent cells of Runx2-overexpressing clone 9, 3T3-L1 (L1), and a clone with empty vector (Co) were cultured at 3, 6, 7, 9, 14, and 21 days. a RT-PCR was performed with osteogenic primers OC, BSP, Osx, Dlx5, and Msx2. b ALP activity and ALP mRNA level were detected. As an internal control, β-actin was used. Alizarin-Red S staining (c) and amount of calcium deposition (d) in mineralized matrix shown in cells cultured for 21 days. * P < 0.05 vs. parental 3T3-L1
Next, we examined the expression pattern of adipocytic markers in Runx2-overexpressing clone 9 at 7, 14, and 21 days of culture. As shown in Fig. 3a, mRNA levels of PPARγ2, C/EBPα, and aP2 were increased in L1 and Co in a time-dependent manner. However, their levels were decreased in clone 9, PPARγ2 and C/EBPα were at 14 and 21 days, and aP2 was at 7, 14, and 21 days. A steady level of C/EBPβ and δ mRNAs was detected in L1, Co, and clone 9 at all culture times. Oil-Red O staining showed lipid droplets in L1 and Co at 14 and 21 days, but its accumulation was attenuated in clone 9 (Fig. 3b).
Adipogenic potential of Runx2-overexpressing 3T3-L1 cells. Confluent cells of Runx2-overexpressing clone 9, 3T3-L1 (L1), and a clone with empty vector (Co) were cultured for 7, 14, and 21 days. a RT-PCR was performed with adipogenic primers PPARγ2; C/EBPα, -β, and -γ; and aP2. As an internal control, β-actin was used. b Oil-Red O staining was performed in cells cultured for 21 days (original magnification of all images ×200)
Dex Increases Osteoblastic Differentiation in Runx2-Overexpressing 3T3-L1 Cells
Since previous studies have demonstrated that Dex stimulates the induction of osteoblastic differentiation in exogenous Runx2-overexpressing marrow stromal cells and primary fibroblasts [8, 9], the effect of Dex in Runx2-overexpressing clone 9 was examined. As shown in Fig. 4a, Dex-treated clone 9 drastically increased ALP activity from 3 to 6 days higher than that in Dex-untreated clone 9. RT-PCR also detected inductive mRNA levels of OC, BSP, and Osx in Dex-treated clone 9 at all culture times, while no significant changes in Dlx5 and Msx2 mRNA levels were observed in Dex-treated or untreated clone 9 (Fig. 4b). Furthermore, Dex increased the mineralized matrix and calcium deposition in Runx2-overexpressing clone 9 in a time-dependent manner (Fig. 4c, d).
Dex stimulates osteoblastic differentiation of Runx2-overexpressing 3T3-L1 cells. Confluent cells of Runx2-overexpressing clone 9 were cultured with or without Dex (10−7 M) for the indicated days. ALP activity and ALP mRNA level (a) and expression levels of OC, BSP, Osx, Dlx5, and Msx2 mRNAs (b) were detected. As an internal control, β-actin was used. Alizarin-Red S staining (c) and amount of calcium deposition (d) were determined in Dex-treated (black bars) and untreated (white bars) clone 9. * P < 0.05 vs. Dex-untreated clone 9
Dex Increases MKP-1 Expression Level in Runx2-Overexpressing 3T3-L1 Cells
Phillips and coworkers [9] reported that MKP-1, a dual specific phosphatase, stimulates osteoblast differentiation, increasing the dephosphorylation level of a serine residue (Ser125) of Runx2; but the exact mechanisms how MKP-1 is induced and Runx2 is activated are not well understood. In order to examine if endogenous MKP-1 modulates Dex-induced upregulation of osteoblast differentiation of clone 9, the MKP-1 expression level was determined by Western blotting. As shown in Fig. 5a, b, Dex significantly increased the expression level of endogenous MKP-1 compared to that of untreated cells, suggesting that Dex is closely related to the osteoblastic differentiation of Runx2-overexpressing clone 9 through MKP-1 expression.
Dex increased the expression level of MKP-1 in Runx2-overexpressing 3T3-L1 cells. Confluent cells of 3T3-L1 and clone 9 were cultured for 14 days in the presence (+) or absence (−) of Dex (10−7 M) for 1 week. a Cell lysates were prepared, then subjected to Western blotting. b Semiquantitative analysis of MKP-1 expression levels shown in a was performed using NIH Image software
Overexpression of Exogenous MKP-1 in Runx2-Overexpressing 3T3-L1 Cells
To determine the effect of MKP-1 in Runx2-overexpressing clone 9, exogenous MKP-1 was overexpressed using expression vector encoding full-length mouse MKP-1 cDNA. Several clones were collected, and the expression levels of Runx2 and MKP-1 were determined by Western blotting, resulting in a clone strongly expressing MKP-1 and/or Runx2, shown in Fig. 6a (lane 6). A luciferase assay was performed to examine the binding activity of Runx2 to 6× OSE2/pGL3. As shown in Fig. 6b, a higher level of Runx2-mediated luciferase activity was determined in MKP-1-overexpressing clone 9 than in clone 9 with or without empty vector but there was no significant change in 3T3-L1 cells, demonstrating that MKP-1 is a candidate for the increase of Runx2-binding activity to the 6× OSE2 sequence.
Forced expression of MKP-1 in Runx2-overexpressing 3T3-L1 cells. 3T3-L1 (L1) and Runx2-overexpressing clone 9 were transfected with an expression vector encoding MKP-1 cDNA, and then a clone overexpressing MKP-1 was chosen. a Western blotting was performed to examine the expression level of MKP-1 and Runx2 (lane 1, 3T3-L1; lane 2, 3T3-L1 with pcDNA; lane 3, 3T3-L1 with MKP-1; lane 4, clone 9; lane 5, clone 9 with pcDNA; lane 6, clone 9 with MKP-1). Actin was used as an internal control. b Runx2-mediated luciferase activity to 6 × OSE2/pGL3 was measured in 3T3-L1 (L1) and Runx2-overexpressing clone 9 (#9) (white bars, cells without vector; gray bars, cells with empty vector; black bars, cells with MKP-1). * P < 0.05 vs. clone 9 with or without empty vector
MKP-1 Stimulates Osteoblastic Conversion of Runx2-Overexpressing 3T3-L1 Cells
The effect of MKP-1 on osteoblastic differentiation of Runx2-overexpressing clone 9 was examined. As shown in Fig. 7a (lanes 4–6), MKP-1 overexpression in clone 9 increased ALP activity as well as mRNA expression levels of ALP, OC, BSP, and Osx compared to clone 9 with or without empty vector but not induction of Dlx5 and Msx2. However, no significant expression level of osteoblastic markers was detected in MKP-1-overexpressing 3T3-L1 (Fig. 7a, lanes 1–3). Alizarin-Red S staining showed that MKP-1 overexpression drastically increased matrix mineralization, which first appeared at 3 days, increased from 7 days, and then reached a plateau at 14 days. Compared with Dex-treated clone 9, exogenous MKP-1 quickly induced accumulation of a large amount of mineralized matrix at 10, 14, and 21 days, whereas a small amount was observed at 14 and 21 days in clone 9 with or without empty vector (Fig. 7b). Calcium deposition in MKP-1-overexpressing clone 9 also increased from 7 days, higher than that in Dex-treated clone 9 (Fig. 7c). To rule out cell death, cell-growth activity was further measured in these cells cultured for 1, 4, 7, 10, 14, and 21 days in the presence or absence of β-glycerophosphate and ascorbic acid, resulting in no significant change among four growth curves obtained by clone 9, clone 9 with pcDNA, Dex-treated clone 9/pcDNA, and clone 9 overexpressed by MKP-1 (Fig. 7d).
MKP-1 overexpression stimulates osteoblastic differentiation of Runx2-overexpressing 3T3-L1 cells. Osteogenic potential was compared between MKP-1-overexpressing clone 9 (#9) and 3T3-L1 (L1). a RT-PCR was performed with primers of OC, BSP, Dlx5, Msx2, and Osx (lane 1, L1; lane 2, L1 with pcDNA; lane 3, L1 with MKP-1; lane 4, clone 9; lane 5, clone 9 with pcDNA; lane 6, clone 9 with MKP-1). Actin was used as an internal control. b Alizarin-Red S staining was subjected in clone 9 (#9), clone 9 with pcDNA3.1 (#9/pcDNA), Dex (10−7 M)-treated clone 9 with pcDNA3.1 (#9/pcDNA+Dex), and clone 9 with MKP-1 (#9/MKP-1) at 3, 7, 10, 14, and 21 days. c Amounts of calcium deposition were measured in clone 9, Dex (10−7 M)-treated clone 9, untreated clone 9, and MKP-1-overexpressing clone 9 at 3, 7, 10, 14, and 21 days. d Cell-growth activity was detected in clone 9, Dex (10−7 M)-treated clone 9, untreated clone 9, and MKP-1-overexpressing clone 9 at 1, 4, 7, 10, 14, and 21 days. * P < 0.05 vs. clone 9
Discussion
This is the first report that overexpression of Runx2 promotes transdifferentiation of 3T3-L1 preadipocytes into bone-forming osteoblasts with inhibition of adipogenesis. In vitro studies suggested a reciprocal relationship between the differentiation process into osteoblasts and adipocytes under several experimental conditions. For example, increased PPARγ2 expression in bone marrow cells inhibits differentiation into osteoblast lineages [33, 34]. Overexpression of PPARγ2 in MC3T3-E1 preosteoblastic cells downregulates ALP activity and mRNA levels of osteoblastic markers including OC and BSP, resulting in stimulation of adipocytic differentiation [35]. Conversely, osteoblastic differentiation is initiated with decreased PPARγ mRNA level when Runx2 predominantly expresses in mature adipocyte-derived preadipocytic cells or adipose-derived stem cells [25, 26]. These findings confirm that Runx2 or PPARγ2 expression is a milestone for cell fate toward the osteoblast or adipocyte lineage. Interestingly, a decrease of C/EBPα mRNA level was detected in Runx2-overexpressing 3T3-L1 cells (Fig. 3a). C/EBPα is induced at later stages and active in mature adipocytes [36]. Forced expression of C/EBPα and PPARγ2 in NIH3T3 fibroblasts has synergistic effects on adipogenetic conversion [21], and coexpression of C/EBPα and -δ increases transcriptional activity from PPARγ2 promoter [19]. These findings suggest that Runx2 expression inhibits the cooperation of PPARγ with C/EBPα or their signal pathways necessary for optimal differentiation into mature adipocytes. On the other hand, no significant change in the mRNA level of osteoblastic transcription factors Dlx5 and Msx2 was shown during all culture times (Figs. 2a, 4b), suggesting that Dlx5 and Msx2 expression is independent of Runx2-mediated osteoblast differentiation of 3T3-L1 cells.
Several studies have reported cell type-dependent upregulation of osteoblastic differentiation and mineralization after overexpression of exogenous Runx2. In C3H10T1/2 fibroblasts and MC3T3-L1 preosteoblasts, Runx2 overexpression increases the formation of mineralized matrix with inductive expression of ALP, OC, and BSP [6–9]. In contrast, Runx2-overexpressing NIH3T3 and IMR-90 fibroblasts fail to mineralize, but ALP activity and OC mRNA levels were increased in Runx2-overexpressing NIH3T3 cells [7]. These findings suggest that, although functional Runx2 is crucial to the expression of osteoblastic marker genes, matrix mineralization may require additional tissue-specific activators, which supplement Runx2 activity. Here, we focus on the effect of MKP-1, a dual specific phosphatase, since Dex increased endogenous MKP-1 expression in Runx2-overexpressing 3T3-L1 cells (Fig. 5a, b). Notably, MKP-1 is thought to stimulate Dex-induced osteoblastic differentiation in bone marrow cells and primary fibroblasts through an increase of Runx2 activity [9]. Indeed, our previous study has shown that Dex stimulates MKP-1 expression with an increase of Runx2-mediated luciferase activity of 6× OSE2/pGL3 in the rat mesenchymal progenitor cell line ROB-C26 [29]. In this study, forced expression of exogenous MKP-1 in Runx2-overexpressing 3T3-L1 cells synergistically increased Runx2-mediated luciferase activity to 6× OSE2 (Fig. 6b) and expression of osteoblastic marker with a high mineralized efficiency (Fig. 7a–c), suggesting the importance of MKP-1 expression and Runx2-binding activity to 6× OSE2 for transdifferentiation of 3T3-L1 cells into the osteoblastic lineage. However, MKP-1-overexpressing 3T3-L1 cells did not show osteoblastic differentiation, although they have low Runx2 expression (Fig. 6a, lane 3). This evidence suggests that high-level expression of Runx2 and MKP-1 involves the conversion of preadipocytes into functional and mature osteoblasts.
The present study clearly demonstrates that overexpression of MKP-1 stimulates Runx2-dependent osteoblastic differentiation in 3T3-L1 cells (Fig. 7a–c). The ability of MKP-1 to increase expression of osteoblastic markers and accumulation of matrix mineralization suggests that MKP-1 is an important regulator that supports Runx2 activity. However, the physiological association of other transcriptional activators involving Runx2-mediated osteoblast differentiation cannot be ignored. It is well known that Runx2 activity is tightly controlled by protein–protein interactions of transcriptional activators. For example, Runx2 is controlled by TAZ, a WW domain-containing molecule of a transcriptional activator that directly interacts with Runx2 and activates Runx2-dependent gene transcription [37]. Cbfβ1 is the most important transcriptional coactivator of Runx2, which is required for Runx2-dependent bone formation [38, 39]. Signal transducers of transforming growth factor β superfamily receptors, Smad1 and Smad5, interact with Runx2 to regulate bone-specific genes [40, 41]. Runx2 is also negatively controlled by transcriptional factors such as Stat1 [42], Sox 9 [43], AJ18 [44], Nrf2 [45], and Hoxa2 [46]. In particular, antisense MKP-1 oligonucleotide prolonged the accumulation of phosphorylated transcription factor Stat1 [47], indicating a novel activation pathway of Runx2 in which MKP-1 attenuates Stat1 activity and promotes a Runx2-dependent osteoblast differentiation program. Indeed, Stat1−/− mice reveal increasing nuclear localization and transcriptional activity of Runx2 [48].
A number of mesenchymal cells or bone marrow stromal cells genetically modified using viral delivery systems have been implanted into recipient animal models to regenerate bone and heal bone defect [25, 47]. The present study demonstrated that overexpression of Runx2 and MKP-1 promoted the transdifferentiation of 3T3-L1 cells into osteoblasts with large amounts of accumulation of mineralized nodules, suggesting that Runx2 and MKP-1 are a good choice for the application of genetic engineering in bone regeneration. Similar to the present study, overexpression of BMP receptors suppresses adipogenesis of 3T3-F442 preadipocytes and stimulates osteoblastic differentiation in response to retinoic acid, resulting in the deposition of large amounts of mineralized nodules [49]. However, our results clearly demonstrate that Runx2 and MKP-1 in 3T3-L1 cells stably induced the conversion into fully differentiated osteoblasts in the absence of other growth factors and chemicals. Therefore, combined expression of Runx2 and MKP-1 is a powerful tool for osteogenic induction of preadipocytes in bone marrow. Whether the present results can be applied to the management of bone disease will be a challenge for future research.
References
Ogawa E, Maruyama M, Kagoshima H et al (1993) PEBP2/PEA2 represents a family of transcription factors homologous to the products of the Drosophila runt gene and the human AML1 gene. Proc Natl Acad Sci USA 90:6859–6863
Komori T, Yagi H, Nomura S et al (1997) Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell 89:755–764
Ducy P, Karsenty G (1995) Two distinct osteoblast-specific cis-acting elements control expression of a mouse osteocalcin gene. Mol Cell Biol 15:1858–1869
Sato M, Morii E, Komori T et al (1998) Transcriptional regulation of osteopontin gene in vivo by PEBP2αA/CBFA1 and ETS1 in the skeletal tissues. Oncogene 17:1517–1525
Benson MD, Aubin JE, Xiao G et al (1999) Cloning of a 2.5 kb murine bone sialoprotein promoter fragment and functional analysis of putative Osf2 binding sites. J Bone Miner Res 14:396–405
Ducy P, Zhang R, Geoffroy V et al (1997) Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell 89:747–754
Byers BA, Pavlath GK, Murphy TJ et al (2002) Cell-type-dependent up-regulation of in vitro mineralization after overexpression of the osteoblast-specific transcription factor Runx2/Cbfal. J Bone Miner Res 17:1931–1944
Byers BA, García AJ (2004) Exogenous Runx2 expression enhances in vitro osteoblastic differentiation and mineralization in primary bone marrow stromal cells. Tissue Eng 10:1623–1632
Phillips JE, Gersbach CA, Wojtowicz AM et al (2006) Glucocorticoid-induced osteogenesis is negatively regulated by Runx2/Cbfa1 serine phosphorylation. J Cell Sci 119:581–591
Otto F, Thornell AP, Crompton T et al (1997) Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell 89:765–771
Liu W, Toyosawa S, Furuichi T et al (2001) Overexpression of Cbfa1 in osteoblasts inhibits osteoblast maturation and causes osteopenia with multiple fractures. J Cell Biol 155:157–166
Geoffroy V, Kneissel M, Fournier B et al (2002) High bone resorption in adult aging transgenic mice overexpressing Cbfa1/Runx2 in cells of the osteoblastic lineage. Mol Cell Biol 22:6222–6233
Rosen ED, Walkey CJ, Puigserver P et al (2000) Transcriptional regulation of adipogenesis. Genes Dev 14:1293–1307
Zhu Y, Qi C, Korenberg JR et al (1995) Structural organization of mouse peroxisome proliferator-activated receptor γ (mPPAR γ) gene: alternative promoter use and different splicing yield two mPPAR γ isoforms. Proc Natl Acad Sci USA 92:7921–7925
Landschulz WH, Johnson PF, McKnight SL (1989) The DNA binding domain of the rat liver nuclear protein C/EBP is bipartite. Science 243:1681–1688
Yeh WC, Cao Z, Classon M et al (1995) Cascade regulation of terminal adipocyte differentiation by three members of the C/EBP family of leucine zipper proteins. Genes Dev 9:168–181
Cao Z, Umek RM, McKnight SL (1991) Regulated expression of three C/EBP isoforms during adipose conversion of 3T3-L1 cells. Genes Dev 5:1538–1552
Christy RJ, Kaestner KH, Geiman DE et al (1991) CCAAT/enhancer binding protein gene promoter: binding of nuclear factors during differentiation of 3T3-L1 preadipocytes. Proc Natl Acad Sci USA 88:2593–2597
Clarke SL, Robinson CE, Gimble JM (1997) CAAT/enhancer binding proteins directly modulate transcription from the peroxisome proliferator-activated receptor γ2 promoter. Biochem Biophys Res Commun 240:99–103
Lin FT, Lane MD (1994) CCAAT/enhancer binding protein α is sufficient to initiate the 3T3-L1 adipocyte differentiation program. Proc Natl Acad Sci USA 91:8757–8761
Tontonoz P, Hu E, Spiegelman BM (1994) Stimulation of adipogenesis in fibroblasts by PPARγ2, a lipid-activated transcription factor. Cell 79:1147–1156
Fajas L, Fruchart JC, Auwerx J (1998) Transcriptional control of adipogenesis. Curr Opin Cell Biol 10:165–173
Park SR, Oreffo RO, Triffitt JT (1999) Interconversion potential of cloned human marrow adipocytes in vitro. Bone 24:549–555
Justesen J, Pedersen SB, Stenderup K et al (2004) Subcutaneous adipocytes can differentiate into bone-forming cells in vitro and in vivo. Tissue Eng 10:381–391
Zhang X, Yang M, Lin L et al (2006) Runx2 overexpression enhances osteoblastic differentiation and mineralization in adipose-derived stem cells in vitro and in vivo. Calcif Tissue Int 79:169–178
Oki Y, Watanabe S, Endo T et al (2008) Mature adipocyte-derived dedifferentiated fat cells can trans-differentiate into osteoblasts in vitro and in vivo only by all-trans retinoic acid. Cell Struct Funct 33:211–222
Green H, Meuth M (1974) An established pre-adipose cell line and its differentiation in culture. Cell 3:127–133
Green H, Kehinde O (1979) Formation of normally differentiated subcutaneous fat pads by an established preadipose cell line. J Cell Physiol 101:169–171
Mikami Y, Takahashi T, Kato S et al (2008) Dexamethasone promotes DMP1 mRNA expression by inhibiting negative regulation of Runx2 in multipotential mesenchymal progenitor, ROB-C26. Cell Biol Int 32:239–246
Harada H, Tagashira S, Fujiwara M et al (1999) Cbfa1 isoforms exert functional differences in osteoblast differentiation. J Biol Chem 274:6972–6978
Tanabe N, Ito-Kato E, Suzuki N et al (2004) IL-1α affects mineralized nodule formation by rat osteoblasts. Life Sci 75:2317–2327
Sudhakar S, Katz MS, Elango N (2001) Analysis of type-I and type-II RUNX2 protein expression in osteoblasts. Biochem Biophys Res Commun 286:74–79
Lazarenko OP, Rzonca SO, Suva LJ et al (2006) Netoglitazone is a PPAR-gamma ligand with selective effects on bone and fat. Bone 38:74–84
Schilling T, Nöth U, Klein-Hitpass L et al (2007) Plasticity in adipogenesis and osteogenesis of human mesenchymal stem cells. Mol Cell Endocrinol 271:1–17
Kim SW, Her SJ, Kim SY et al (2005) Ectopic overexpression of adipogenic transcription factors induces transdifferentiation of MC3T3-E1 osteoblasts. Biochem Biophys Res Commun 327:811–819
Darlington GJ, Ross SE, MacDougald OA (1998) The role of C/EBP genes in adipocyte differentiation. J Biol Chem 273:30057–30060
Hong JH, Hwang ES, McManus MT et al (2005) TAZ, a transcriptional modulator of mesenchymal stem cell differentiation. Science 309:1074–1078
Yoshida CA, Furuichi T, Fujita T et al (2002) Core-binding factor β interacts with Runx2 and is required for skeletal development. Nat Genet 32:633–638
Miller J, Horner A, Stacy T et al (2002) The core-binding factor β subunit is required for bone formation and hematopoietic maturation. Nat Genet 32:645–649
Lee KS, Kim HJ, Li QL et al (2000) Runx2 is a common target of transforming growth factor β1 and bone morphogenetic protein 2, and cooperation between Runx2 and Smad5 induces osteoblast-specific gene expression in the pluripotent mesenchymal precursor cell line C2C12. Mol Cell Biol 20:8783–8792
Zhang YW, Yasui N, Ito K et al (2000) A RUNX2/PEBP2αA/CBFA1 mutation displaying impaired transactivation and Smad interaction in cleidocranial dysplasia. Proc Natl Acad Sci USA 97:10549–10554
Kim S, Koga T, Isobe M et al (2003) Stat1 functions as a cytoplasmic attenuator of Runx2 in the transcriptional program of osteoblast differentiation. Genes Dev 17:1979–1991
Zhou G, Zheng Q, Engin F et al (2006) Dominance of SOX9 function over RUNX2 during skeletogenesis. Proc Natl Acad Sci USA 103:19004–19009
Jheon AH, Ganss B, Cheifetz S et al (2001) Characterization of a novel KRAB/C2H2 zinc finger transcription factor involved in bone development. J Biol Chem 276:18282–18289
Hinoi E, Fujimori S, Wang L et al (2006) Nrf2 negatively regulates osteoblast differentiation via interfering with Runx2-dependent transcriptional activation. J Biol Chem 281:18015–18024
Dobreva G, Chahrour M, Dautzenberg M et al (2006) SATB2 is a multifunctional determinant of craniofacial patterning and osteoblast differentiation. Cell 125:971–986
Liu D, Scafidi J, Prada AE et al (2002) Nuclear phosphatases and the proteasome in suppression of STAT1 activity in hepatocytes. Biochem Biophys Res Commun 299:574–580
Zheng H, Guo Z, Ma Q et al (2004) Cbfa1/osf2 transduced bone marrow stromal cells facilitate bone formation in vitro and in vivo. Calcif Tissue Int 74:194–203
Skillington J, Choy L, Derynck R (2002) Bone morphogenetic protein and retinoic acid signaling cooperate to induce osteoblast differentiation of preadipocytes. J Cell Biol 159:135–146
Acknowledgments
We thank Prof. Gérard Karsenty (Department of Genetics and Development, Columbia University Medical Center) for Cbfa1-pCMV5 and 6 × OSE2/pGL3Basic, and Dr. Yoshikazu Mikami (Department of Anatomy, Nihon University School of Dentistry) for MKP-1/pcDNA3.1(+). This work was supported by grants from Dental Research Center, Nihon University School of Dentistry, Sato Fund, Nihon University School of Dentistry, and Graduate School of Dentistry, Nihon University.
Author information
Authors and Affiliations
Corresponding author
Additional information
The author has stated that there is no conflict of interest.
Rights and permissions
About this article
Cite this article
Takahashi, T. Overexpression of Runx2 and MKP-1 Stimulates Transdifferentiation of 3T3-L1 Preadipocytes into Bone-Forming Osteoblasts In Vitro. Calcif Tissue Int 88, 336–347 (2011). https://doi.org/10.1007/s00223-011-9461-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00223-011-9461-9