Abstract
Carrot is an important elicitor of food allergy. In this study, we developed a novel molecular detection assay based on loop-mediated isothermal amplification technology for the detection of carrot in food materials. The assay had sensitivity up to 4 pg of the purified carrot DNA and had no cross-reactions with other species, such as fennel, parsley, anise, caraway, cumin, walnut, pecan, peanut, cashew, almond, pistachio, sunflower seed, sesame seed, chestnut, soy, barley, wheat, oat, rye, rice, cabbage, green pepper, onion, celery, cucumber, beef, mutton, pork, chicken and shrimp. The validation study demonstrated high reproducibility and specificity. This assay was proved to be a potential tool for the detection and label management of carrot allergens in foods.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Naturally sweet, delicious and crunchy carrots (Daucus carota, belong to Umbelliferae family) are healthy additions that people can make to the vegetable list in the diet. Indeed, these root vegetables come with wholesome health benefiting compounds such as beta-carotenes, vitamin A, minerals and anti-oxidants in ample amounts. However, some people cannot consume carrots because of food allergies [1–4]. Although carrot is not listed as the “big eight” food allergies, which account for 90 % of all food-allergy reactions, it is also an important elicitor of food allergy [5]. Symptoms of a carrot allergy will vary with each person and can depend on how much carrot was consumed, whether the carrot was raw or cooked, and what other foods were eaten with the allergen [1, 2, 6]. Hypersensitivity to carrot is frequently associated with allergy to Apiaceae spices and sensitization to birch and mugwort pollens [7]. The most common signs and symptoms of carrot allergy include itching, swelling, and gastrointestinal discomfort. In most cases, these symptoms are bothersome but not life threatening and will dissipate within a few hours. Sometimes, a person’s reaction can be severe enough to constitute anaphylaxis and require emergency medical intervention [1, 8]. Complete and strict avoidance of the offending food is the only way to prevent these allergic reactions. To protect carrot-allergic consumers, carrot is required to be declared on food labels in some regulations [9, 10]. Even with the regulations, mistakes in food labeling and cross-contamination occur from time to time, though not always intentionally [11]. So, the development of a rapid, sensitive and specific method for the detection of carrot materials is in demand.
The routine methods for the detection of food allergens involve ELISA and PCR, the former is protein-based method, and the latter is a method operating on the DNA level. Due to changes of protein structure in food processing, and sometimes the case that special food matrix will inevitably affect the performance of the protein-based assays, the molecular detection assays are more desirable for processed products. Loop-mediated isothermal amplification (LAMP), a novel molecular amplification method, has been developed by Notomi [12]. Compared to PCR and real-time PCR, it amplifies the target sequence at a constant temperature without a significant influence of the co-presence of non-target DNA. Moreover, the set of specially designed primers can identify six distinct regions on the target genes, which adds highly to the specificity. The amount of DNA produced in LAMP is equivalent to the real-time PCR and higher than that of the PCR-based amplification. The cycling reaction continues with accumulation of 109 copies of target in less than an hour [10]. Since LAMP assay was developed, it has been widely used for diagnosis of disease, detection of pathogens and other fields [13–17]. In this study, we reported a LAMP-based assay by designing the specific primers to detect carrot materials in foods.
Materials and methods
Samples
Fifty-one carrot cultivars samples were used in our study, which were from different countries: 11 roots specimens were obtained from carrot planting base (Shouguang, China), and 40 seeds specimens were samples from the trading companies for exit-entry inspection and quarantine (14 imported from Japan, 11 from Korea, 6 from America, 5 from France, 4 from Australia). Other Umbelliferae plants such as fennel, parsley, anise, caraway, cumin and common allergenic food such as walnuts, pecans, peanuts, cashews, almonds, pistachios, sunflower seeds, sesame seeds, chestnuts, soy, barley, wheat, oats, rye, rice, beef, mutton, pork, chicken, shrimp, as well as common vegetables, such as cabbage, green peppers, onions, celery, cucumber were used for the analysis of assay specificity and were purchased in local supermarket in Qingdao. The processed foods were also purchased in local supermarket, too (Table 1).
DNA extraction
For DNA preparation, Plant DNA Mini-Prep Kit (Jiemen Biotech Co., Shanghai, China) was used according to the manufacturer’s instruction. Briefly, 100 mg ground sample was lysed with lysis buffer till no tissue was visible. Then, phenol/chloroform extraction was carried out. After centrifugation, the upper aqueous phase was transferred into a new tube, and then, the buffer that was provided with the kit was added to precipitate DNA. The resulting DNA pellet was dissolved and applied to the silica column. Following the column was washed twice with the washing buffer, the DNA bound to the column was eluted with elution buffer. The purity and concentration of DNA was determined by measuring the absorption at 260 and 280 nm. After UV quantification, the concentration of each DNA sample was adjusted to 10 ng/µL.
Primers design
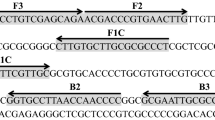
Primer design was performed using the online tool Primer Explorer version 4.0 (http://primerexplorer.jp/elamp4.0.0/index.html) on ITS1-5.8SRNA-ITS2 carrot gene (NCBI accession number: AY552527.1). According to the guide to LAMP primer designing, four set of six primers were chosen and synthesized by Sangon Biotech (Shanghai) Co., Ltd. These primers were purified by polyacrylamide gel electrophoresis. After experimental verification, one set of primers was used to develop LAMP assay. The sequences of primers are listed in Table 2.
Loop-mediated isothermal amplification (LAMP)
Optimized amplification system had a total volume of 25 μL: 1× ThermoPol reaction buffer (containing 20 mM Tris–HCl, 10 mM KCl, 10 mM (NH4)2SO4, 2 mM MgSO4, 0.1 % Triton X-100), 0.12 µmol/L forward outer primers (F3) and backward outer primer (B3), 0.96 µmol/L forward inner primer (FIP) and backward inner primer (BIP), 0.48 µmol/L forward loop primer (FLP) and backward loop primer (BLP), 1.3 mmol/L dNTP, 0.8 mol/L betaine, 6 mmol/L MgSO4, 0.32 U/L Bst DNA polymerase (New England Biolabs, USA), 4 μL of template DNA. Amplification was carried out in a general PCR machine (Authorized Thermal Cycler, Eppendorf, Germany), and the program was as follows: reaction at 63 °C for 40 min; termination at 80 °C for 5 min. Each sample was amplified in triplicate. After amplification, the 5 μL of LAMP amplified products was electrophoresed in 2 % agarose gel and stained with GelRed (Biotium, USA). Furthermore, the results of the reaction were detected with the naked eye under day light by addition of 1.0 μL of 1/10-diluted original SYBR Green I (Molecular Probes Inc.) to the mixture and observation of the solution color.
Inter-laboratory validation
The inter-laboratory validation study was conducted by five Chinese laboratories accredited by China National Accreditation Service for Conformity Assessment (referred to as CNAS). The validation study was designed to identify both the sensitivity and specificity of the testing system. Samples used in sensitivity test were prepared as follows: DNA was extracted from carrot and soybean, respectively. The concentration of each DNA sample was adjusted to 10 ng/µL. Then, the DNA solution of carrot was serially diluted with soybean DNA solution. Sensitivity was determined by evaluating five samples that contained 10, 1, 0.1, 0.01, 0.005 % carrot. Specific verification material included five species belong to Umbelliferae. DNA samples were delivered to each laboratory in ice box and preserved at −20 °C. Each sample was required to be tested three times with LAMP method.
Real-time PCR
DNA was amplified using Mastercycler (Eppendorf Realplex4, Germany). Amplification was carried out in a 96-well plate in a final volume of 25 μL: 1× Master Mix (containing HotStarTaq DNA polymerase, MgCL2, dNTP; Tiangen Biotech Co. Ltd., Beijing, China), 400 nmol/L each primer, 400 nmol/L probe, 40 ng template DNA. The forward primer was 5′-CGACAAGCAAGCTTTACTCCAA-3′, and the reverse primer was 5′-CGTCTGACACCCATGAGTCTGT-3′. The probe was 5′-FAM-TCAAAACAGCCTTGAAAAACCCCACCA-TAMRA-3′. PCR program was as follows: pre-denaturation at 95 °C for 2 min; 45 cycles of amplification (95 °C for 15 s, 60 °C for 40 s) [18]. Each sample was amplified in twice.
Results
Applicability of the assay
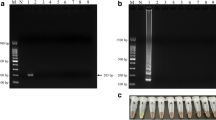
To determine the efficiency of the primers, a total number of 51 carrot samples were collected. All carrots samples were evident upon agarose gel electrophoresis as a ladder-like pattern on the gel, which is characteristic of the LAMP reaction and indicates the production of stem-loop DNA with inverted repeats of the target sequence, and the solution turned green in the presence of SYBR Green I, while the negative control remained orange with no amplification. Part of the results is shown in Fig. 1. Five carrot DNA samples were randomly chosen as LAMP template, and amplified in triplicate in one experiment and carried out three times. No difference in the results of intra-experimental and inter-experimental was observed. This implied the method had good repeatability and reproducibility.
The LAMP amplification result of partial carrots samples. a Electrophoresis of LAMP products in 2 % agarose gel stained with GelRed™ DNA staining solution. Lanes M marker (DL2000, Takara); N negative control; 1–6 from left to right, carrot samples from Japan, Korea, America, France, Australia and China, respectively. b Visual appearance of LAMP products after addition of 1 µL of 1/10 dilution of SYBR Green I to each reaction mixture under day light. N negative control; 1–6 from left to right, carrot samples from Japan, Korea, America, France, Australia and China, respectively
Ten commodities were purchased from local supermarket and detected with carrot specific primers to further validate the applicability of the assay. 8/10 were found to harbor carrot materials, including one labeled with “may contain traces of carrot” (Fig. 2). These commodities were detected with real-time PCR method too. Two samples labeled with “may contain traces of carrot” did not yield positive amplification (Table 3).
The LAMP amplification results of commodities. a Electrophoresis result. Lanes M marker (DL2000, Takara); N negative control; 1–10 from left to right, baby food in jar, infant cereal, cereal, noodles 1, noodles 2, vegetable juice 1, vegetable juice 2, fruit-flavored drink, dumpling and steamed stuffed bun. b Visual detection. N negative control; 1–10 from left to right, baby food in jar, infant cereal, cereal, noodles 1, noodles 2, vegetable juice 1, vegetable juice 2, fruit-flavored drink, dumpling and steamed stuffed bun
Sensitivity of the assay
To study the detection limit of the LAMP assay, serial tenfold dilutions of the carrot genomic DNA were tested. The initial amount of the purified DNA was 40 ng/25 μL. The results of the assay were analyzed through visual detection and accurately confirmed by gel electrophoresis. As can be seen from the results, the data were consistent between the two reading methods and as low as 4 pg of carrot genomic DNA, an equivalent of dilution 10−4, yielded reproducible positive results (Fig. 3), which implied the LAMP assay is highly sensitive.
The LAMP amplification results of serial dilutions of carrot DNA. a Electrophoresis result. Lane M marker (DL2000, Takara); N negative control; 1–6 various dilutions of carrot DNA: neat (10 ng), 10−1 (1 ng), 10−2 (100 pg), 10−3 (10 pg), 10−4 (1 pg), 10−5 (100 fg). b Visual detection. N negative control; 1–6 various dilutions of carrot DNA: neat (10 ng), 10−1 (1 ng), 10−2 (100 pg), 10−3 (10 pg), 10−4 (1 pg), 10−5 (100 fg)
Specificity of the assay
Specificity is always of primary concern. To determine the specificity of the assay, DNA were extracted from fennel, parsley, anise, caraway, cumin, cabbage, green peppers, onions, celery, cucumber, walnuts, pecans, peanuts, cashews, almonds, pistachios, sunflower seeds, sesame seeds, chestnuts, soy, barley, wheat, oats, rye, rice, beef, mutton, pork, chicken and shrimp and then amplified with carrot specific primers. No typical amplification bands and changes of color were observed, which demonstrated that primers had no cross-reactions with the species above.
Inter-laboratory reproducibility (reliability)
All the assay results showed high reproducibility of the parameters at the participating laboratories. Although there were differences in the carrot detection system used, i.e., instrument, the outputs obtained from each laboratory were consistent. 0.01 % DNA sample could be detected by all the participants. No cross-reactions with other closely related species were reported. Based on the AOAC worksheet (www.aoac.org/imis15_prod/AOAC_Docs/NEWS/09trad04_AOAC_binary-v2-3.xls), repeatability relative standard deviation (RSD(r)) and reproducibility relative standard deviation (RSD(R)) for the replicates in the inter-laboratory study were calculated. The full report is available in Table 4. These results clearly demonstrated the high reproducibility.
Discussion
Studies have showed that the allergic reactions to food are highly individual, in other words, enormous variation in the sensitivity and severity levels exists among allergen sensitive individuals. For some hypersensitive patients, even trace amount of an allergen can probably elicit life-threatening allergic reaction [1]. It is difficult to determine safe doses of allergens (i.e., thresholds), which has been a challenge to researchers. In view of the above-mentioned facts, the detection method for allergens should be as sensitive as possible. Internal transcribed spacer (ITS) refers to a piece of non-functional RNA situated between structural ribosomal RNAs (rRNA) on a common precursor transcript. Read from 5′ to 3′, this polycistronic rRNA precursor transcript contains the 5′ external transcribed sequence (5′ ETS), 18S rRNA, ITS1, 5.8S rRNA, ITS2, 28S rRNA and finally the 3′ ETS. Nowadays, sequence comparison of the ITS region is widely used in taxonomy and molecular phylogeny. One reason is that the ITS region is easy to amplify even from small quantities of DNA due to the high copy number of rRNA genes. Another is that it has a high degree of variation even between closely related species. In our study, even trace carrot (4 pg) can be detected with the developed LAMP method and no cross-reactions with other species occur. The sensitivity of LAMP method is much higher than that of the real-time PCR method we developed before [18]. The real-time PCR method targeted at antifreeze protein gene of carrot, and the limit of detection was only 100 pg carrot DNA. The LAMP assay actually showed higher sensitivity in detecting commodities compared to real-time PCR, where the PCR could not detect carrot in one of the samples. This suggests that it is feasible to use ITS1-5.8S-ITS2 gene segment as detection marker of carrot materials.
The LAMP method continues to attract the attention of researchers in many fields owing to its simplicity and rapidity of use. Research and development efforts on LAMP technology in recent years have focus on further simplifying the LAMP test process [19]. Hope is coming from integrating LAMP method into simple genetic tests in order to be used as point-of-care (POC) diagnostics [20]. Lateral flow devices has been explored as a means of detecting positive LAMP reactions [21, 22], which can be portable, and does not require instrumentation or electrical power. The developed LAMP method for carrot, with high sensitivity and specificity, is greatly helpful to the detection and label management of carrot allergen in foods. The further research consideration is to couple it with lateral flow. It will be a more rapid and easy-to-use method for timely diagnosis of carrot materials especially at POC.
References
Schiappoli M, Senna G, Dama A, Bonadonna P, Crivellaro M, Passalacqua G (2002) Anaphylaxis due to carrot as hidden food allergen. Allergol Immunopathol 30(4):243–244
Vieths S, Scheurer S, Ballmer-Weber B (2002) Current understanding of cross-reactivity of food allergens and pollen. Ann N Y Acad Sci 964:47–68
Roehr CC, Edenharter G, Reimann S, Ehlers I, Worm M, Zuberbier T, Niggemann B (2004) Food allergy and non-allergic food hypersensitivity in children and adolescents. Clin Exp Allergy 34(10):1534–1541
Moreno-Ancillo A, Gil-Adrados AC, Cosmes PM, Domínguez-Noche C, Pineda F (2006) Role of Dau c 1 in three different patterns of carrot-induced asthma. Allergol Immunopathol 34(3):116–120
Wangorsch A, Weigand D, Peters S, Mahler V, Fötisch K, Reuter A, Imani J, Dewitt AM, Kogel KH, Lidholm J, Vieths S, Scheurer S (2012) Identification of a Dau c PRPlike protein (Dau c 1.03) as a new allergenic isoform in carrots (cultivar Rodelika). Clin Exp Allergy 42(1):156–166
Bollen MA, Garcia A, Cordewener JH, Wichers HJ, Helsper JP, Savelkoul HF, van Boekel MA (2007) Purification and characterization of natural Bet v 1 from birch pollen and related allergens from carrot and celery. Mol Nutr Food Res 51(12):1527–1536
Moreno-Ancillo A, Gil-Adrados AC, Domínguez-Noche C, Cosmes PM, Pineda F (2005) Occupational asthma due to carrot in a cook. Allergol Immunopathol 33(5):288–290
Bohle B, Zwölfer B, Heratizadeh A, Jahn-Schmid B, Antonia YD, Alter M, Keller W, Zuidmeer L, van Ree R, Werfel T, Ebner C (2006) Cooking birch pollen-related food: divergent consequences for IgE- and T cell-mediated reactivity in vitro and in vivo. J Allergy Clin Immunol 118(1):242–249
Food Safety for Guangzhou Asian Games—Food Allergens Labeling, DBJ440100/T 28-2009
Food Safety for Universiade Shenzhen—Food Allergens Labeling
Añíbarro B, Seoane FJ, Múgica MV (2007) Involvement of hidden allergens in food allergic reactions. J Investig Allergol Clin 17(3):168–172
Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T (2000) Loop-mediated isothermal amplification of DNA. Nucleic Acids Res 28(12):E63
Wang Y, Wang Y, Xu H, Dai H, Meng S, Ye C (2014) Rapid and sensitive detection of Listeria ivanovii by loop-mediated isothermal amplification of the smcL gene. PLoS ONE 9(12):e115868
Duan YB, Ge CY, Zhang XK, Wang JX, Zhou MG (2014) Development and evaluation of a novel and rapid detection assay for Botrytis cinerea based on loop-mediated isothermal amplification. PLoS ONE 9(10):e111094
Xia Y, Guo XG, Zhou S (2014) Rapid detection of Streptococcus pneumoniae by real-time fluorescence loop-mediated isothermal amplification. J Thorac Dis 6(9):1193–1199
Ushijima H, Nishimura S, Thongprachum A, Shimizu-Onda Y, Tran DN, Pham NT, Takanashi S, Dey SK, Okitsu S, Yamazaki W, Mizuguchi M, Hayakawa S (2014) Sensitive and rapid detection of campylobacter species from stools of children with diarrhea in Japan by the loop-mediated isothermal amplification method. Jpn J Infect Dis 67(5):374–378
Kinoshita Y, Niwa H, Katayama Y (2014) Development of a loop-mediated isothermal amplification method for detecting Streptococcus equi subsp. zooepidemicus and analysis of its use with three simple methods of extracting DNA from equine respiratory tract specimens. J Vet Med Sci 76(9):1271–1275
Min S, Cheng-zhu L, Biao X, Hong-wei G, Chao L, Cai-xia L (2011) Detection of hazelnut allergens by real time PCR assay. Food Sci Technol 36(11):275–278
Mori Y, Kanda H, Notomi T (2013) Loop-mediated isothermal amplification (LAMP): recent progress in research and development. J Infect Chemother 19(3):404–411
Njiru ZK (2011) Rapid and sensitive detection of human African trypanosomiasis by loop-mediated isothermal amplification combined with a lateral-flow dipstick. Diagn Microbiol Infect Dis 69(2):205–209
Ge Y, Wu B, Qi X, Zhao K, Guo X, Zhu Y, Qi Y, Shi Z, Zhou M, Wang H, Cui L (2013) Rapid and sensitive detection of novel avian-origin influenza A (H7N9) virus by reverse transcription loop-mediated isothermal amplification combined with a lateral-flow device. PLoS ONE 8(8):e69941
Deng J, Pei J, Gou H, Ye Z, Liu C, Chen J (2014) Rapid and simple detection of Japanese encephalitis virus by reverse transcription loop-mediated isothermal amplification combined with a lateral flow dipstick. J Virol Methods 213C:98–105
Acknowledgments
This work was supported by Grants Nos. 2011IK011 and 2009IK254 from General Administration of Quality Supervision, Inspection and Quarantine of the People’s Republic of China.
Conflict of interest
None.
Compliance with Ethics Requirements
This article does not contain any studies with human or animal subjects.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sun, M., Gao, H., Xiao, X. et al. A novel loop-mediated isothermal amplification method for detection of the carrot materials in foods. Eur Food Res Technol 241, 295–302 (2015). https://doi.org/10.1007/s00217-015-2459-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00217-015-2459-1