Abstract
Mango (Mangifera indica) is one of the most popular tropical fruits around the world. It is also widely and commonly used in many cuisines and products in the food industry. However, the anaphylactic reaction caused by mango has long been reported as a major problem for consumption in recent years. To prevent allergens in mango, the best way is to avoid mango in the diet. In this study, a loop-mediated isothermal amplification (LAMP) assay was developed for the detection of mango in food. Four specifically designed LAMP primers targeting the internal transcribed sequence 1 (ITS1) of nuclear ribosomal DNA sequence regions were used to address the LAMP reaction for amplifying mango DNA. The results demonstrated that the detection of mango DNA was specifically validated by the LAMP primer. The sensitivity of LAMP for detecting mango DNA is equivalent to that of the traditional PCR method. The LAMP primer sets showed high specificity for detecting the DNA of mango and had no cross-reactions to other species. Moreover, when mango was mixed with other fruits at different ratios, no cross-reactivity for the detection of mango DNA was manifested during LAMP. Finally, genomic DNAs extracted from different heat-processed mangos were used as templates; the detection of mango DNA by LAMP was not significantly affected and was reproducible. As to this established LAMP herein, mango ingredients can be detected, and commercial foods containing mango can also be identified. This assay will be useful and have potential for the rapid detection of mango DNA in practical food markets.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Mango (Mangifera indica) belongs to the Anacardiaceae family and is one of the most popularly grown tropical fruits around the world. Mango contains high nutritional content, such as beta carotene and vitamin C, and possesses a mellow taste and unique flavour; thus, it is also called the “king of fruit” and is a very commonly and widely applied as a food constituent and is used in many cuisines [1]. In addition to its nutritive value for food production, mango also contains glycosides with biological activities such as antidiabetic, antiatherosclerotic, antimicrobial and antiviral properties [2]. Consequently, mango is considered an important economic fruit in the Southeast Asia region for applications in the food industry. Irrespective of its health benefits or nutritional value, the allergic characteristic of mango has long been reported as a major problem, causing anaphylaxis in recent years [3, 4]. To prevent mango-induced anaphylactic reactions, the best way for the patient is to not consume mango or mango-related food. Thus, correct management of food labelling, especially on information of allergens, is needed for most manufacturers in their products to lower consumers’ uncertainty and protect consumers' interests [5].
Currently, several hypersensitivity symptoms caused by mango have manifested, and these include angioedema, erythema, urticaria, wheezing dyspnoea, contact dermatitis and periorbital oedema [6]. Proteins in mango have been previously recognized as the main allergens causing anaphylaxis [7, 8]. Some researchers have also examined various peptides and proteins in mangos that might have allergic characteristics to humans [9, 10]. However, even though several potential allergens in mango have been reported, insufficient evidence has been identified regarding which kind of proteins in mango truly gives rise to the occurrence of allergies [11]. Moreover, some allergic proteins, such as the pan-allergen profilin, are found in certain vegetables, fruits, nuts and spices and are structurally similar to proteins in mangos. This has been shown that it becomes susceptible to cause a cross-reaction between mango and other species when protein-based allergen diagnosis is performed using specific IgE antibodies [12,13,14].
In addition to immunological assays, numerous well-developed methods for detecting mango in the diet have been reported based on DNA targets [15,16,17]. For DNA molecular assays, such as polymerase chain reaction (PCR)-based techniques, some specific DNA regions of mango were frequently chosen for amplification for the detection or authentication of the genomic DNA of mango. Due to the higher stability of DNA than that of the protein targets, it still displays good potential for the development of diagnostic methods for the detection of mango. In recent years, the principle of loop-mediated isothermal nucleic acid amplification (LAMP) has been well characterized and applied for the diagnosis of biomaterials in previous studies [18,19,20,21,22,23,24,25]. Using specific LAMP primers, DNA amplification is efficiently performed under isothermal conditions by catalysis of the Bst DNA polymerase. Thus, a thermal cycler is not strictly required. Currently, many bioresources, such as viruses, bacteria, and plant- and animal-derived samples, are employed for rapid diagnosis using the LAMP assay [19,20,21,22,23,24,25]. Accordingly, LAMP is a potential assay for developing a specific DNA-detection method.
In this study, isothermal DNA amplification was developed for the rapid and specific detection of mango DNA. The internal transcribed spacer (ITS) of ribosomal DNA was used to design the LAMP primer for the evaluation of the specificity and cross-reactivity. In addition, different imitated heat-processing methods to treat mango were also investigated to evaluate the feasibility of the LAMP method for detecting mango DNA and mango or mango-related products. To the best of our knowledge, this is the first report to detect mango DNA using the LAMP assay.
Materials and methods
Mango samples
All mango samples (Mangifera indica), two mango cultivars (Irwin and Jin-Huang) and commercial mango foods, including dried mango, mango iced tea, mango chocolate ball, mango pocky, mango hi-chew candy, mango cake, mango jelly, mango crispy crepes and mango jam, were collected from a local supermarket (Pingtung, Taiwan). The botanical origin of mango samples was verified by Professor Wen-Te Chang of the China Medical University (Taichung, Taiwan), and these samples were deposited at the Department of Food Science of National Pingtung University of Science and Technology.
DNA extraction
Total genomic DNA from mango samples and other fruits, including apple, guava, orange and pear, was extracted by a genomic extraction kit (AxyPrep Multisource Genomic DNA Miniprep Kit, Axygen Bioscience, CA, USA) according to a previous work [24]. The concentration of obtained mango genomic DNA was determined by spectrophotometry (NanoVue™, GE Healthcare, Piscataway, NJ, USA). Samples were stored at − 20 °C until needed.
LAMP primers
LAMP primers (outer primers, F3 and B3; inner primers, FIP and BIP) were used for detecting mango DNA and were designed based on the ITS1 consensus sequence of nuclear ribosomal DNA obtained from GenBank (https://www.ncbi.nlm.nih.gov) using the commercial software of Primer Explorer V4 (https://primerexplorer.jp; Eiken Chemical Co., Ltd., Tokyo, Japan). The accession numbers of ITS1 for the nuclear ribosomal DNA sequence of mango were used for consensus sequence alignment and included KF664199, KJ833758, KX347960, MF678502, AB071668, LN552225, AB598047 and AJ890466. The target position of LMAP primers is demonstrated and depicted in Fig. 1.
LAMP reaction
The LAMP reaction was performed in a reaction mixture containing 1 × Bst DNA polymerase buffer, 1 U of Bst DNA polymerase (New England Biolabs, Frankfurt, Germany), 0.5 μM outer primers (F3 and B3 primers each), 4 μM inner primers (FIP and BIP primers each), and 200 μM dNTPs each, as described previously [24]. For the different amount of sample’s genomic DNA used to each LAMP reaction, the mixtures were reacted at 55–64 °C for 60 min in a heating block (DNA engine, Biorad, CA, USA).
Detection of LAMP product
DNA electrophoresis with 2% agarose gel was used to detect the formation of LAMP product, and then the resulting gel was stained by ethidium bromide for observation of the presence of visible DNA bands, as described in a previous study [22].
Sensitivity of LAMP assay
Different amounts of mango genomic DNA (102, 10 ng, 1 ng, 10–1 ng and 10–2 ng, prepared by serial dilution) were used as template DNA to measure the sensitivity of LAMP.
DNA preparation of mango combined with other fruits at different ratios
The mango was weighted, combined with the assorted fruits (apple, guava, orange and pear were equivalently premixed) at different percentages of 50%, 10%, 5%, 1%, 0.1% and 0% mango and ground in a ceramic mortar and pestle after freezing using liquid nitrogen. All ground samples were taken and well mixed for the extraction of genomic DNA according to the above procedure.
PCR
The genomic DNA of mango was extracted and used as template in the polymerase chain reaction (PCR) mixture. PCR was carried out using outer primers (F3 and B3) of ITS1-based LAMP primers, as shown in Table 1. The PCR conditions were conducted in a 25 µL reaction mixture containing 1 × PCR buffer (75 mM Tris HCl [pH 9.0], 2 mM MgCl2, 50 mM KCl, and 20 mM (NH4)2SO4), 200 μM dNTP mix, 0.4 μM each of the primers, 1 U Taq DNA polymerase and 1 μL of various concentrations of template DNA. PCR mixtures were denatured at 94 °C for 90 s, followed by 35 cycles of 1 min at 94 °C, 1 min at 62 °C, and 1 min at 72 °C, with a final 10 min extension at 72 °C in a PCR Express Thermal Cycler (Hybaid, Ashford, UK). The PCR product was subjected to DNA electrophoresis with a 2% agarose gel and observed by the presence of visible DNA bands after staining with ethidium bromide.
Preparation of boiled and steamed mango
Mango samples were purchased from a local market, and 20 g of crushed samples were weighed for each sample for further boiling and steaming processes. For boiling mango, the mango samples were boiled in hot water at 95 °C for 20, 40, 60, 80, 100 and 120 min. After cooling in an ice bath, the genomic DNA of cooled mango samples was extracted according to a previously described method for LAMP and PCR. Mango samples were steamed using an autoclave. The mango samples were autoclaved at 121 °C for 20, 40, 60 and 80 min under a pressure of 15 psi. The steamed mango samples were subjected to an ice bath for cooling and then used for genomic DNA extraction, LAMP and PCR.
Results
Development of LAMP assay for the detection of mango
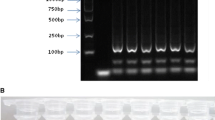
To establish a LAMP assay for the diagnosis of mango DNA, a set of specifically designed LAMP primers (including F3, B3, FIP and BIP illustrated in Table 1) was employed to amplify the genomic DNA of mango based on the consensus sequence of ITS1 in nuclear ribosomal DNA sequences available from GenBank (https://www.ncbi.nlm.nih.gov). When LAMP primers were used for the identification of mango DNA, LAMP products were amplified within 1 h if the amplicon of mango was present in the samples; a typical positive result demonstrated a pattern of ladder-like DNA fragments on the agarose gel (Fig. 2a, lane 1). In addition, DNAs from two different and popular cultivars, Irwin or Jin-Huang mango, were also detected using this LAMP primer set (Fig. 2b, lanes 2 and 3). In contrast, no LAMP products were shown on the agarose gel in the nontarget genomic DNA samples (Fig. 2a, lanes 2–5). Additionally, the LAMP reaction was not influenced by the genomic DNAs extracted from mango combined with DNA from other fruits at different percentages (Fig. 2c). At least 1% of mango genomic DNA was required in the total DNA of the sample for specific detection (Fig. 2c, lane 4). Taken together, these results demonstrated that this set of LAMP primers developed herein for the diagnosis of mango DNA is specific and validated for their use to amplify specific amplicons in mango samples.
Primer specificity and reactivity of the LAMP assay for the identification of mango DNA. The specificity of ITS1-based LAMP primers used for the detection of mango DNA was determined (a). Purified various fruits and mango genomic DNAs were used to perform LAMP. Lanes M and N represent 100 bp of the DNA ladder and the negative control, respectively. Lanes 1–5 represent different DNA: 1, mango; 2, apple; 3, guava; 4, orange; and 5, pear. Various DNA of mango cultivars were used to validate the primer specificities (b). Lanes M and N represent 100 bp of the DNA ladder and negative control, respectively. Lanes 1–3 represent different DNA: 1, positive control; 2, Irwin mango; and 3, Jin-Huang mango. The reactivity of LAMP primers for the identification of mango was analysed (c). The total genomic DNA extracted from assorted fruits (equal weights of premixed apple, guava, orange and pear) mixed with mango by different percentages verified the reactivity of the LAMP primers. Lane P: positive control (100% mango DNA), lanes 1–6: extracted DNA samples from 50%, 10%, 5%, 1%, 0.1% and 0% mango mixed with the assorted fruits by weight, respectively. Lanes M and N represent 100 bp of the DNA ladder and negative control, respectively
Sensitivity of LAMP for the detection of mango DNA
To evaluate the sensitivity of LAMP, the above LAMP primers were used to amplify the template DNAs of mango plants in various quantities. At least 1 ng of mango genomic DNA was required for its detection (Fig. 3a). Compared to the mango PCR primers (primers F3 and B3), 1 ng mango genomic DNA as the template was also needed for PCR (Fig. 3b). Specific DNA bands with a length of 178 bp were shown in the gel of the PCR results (Fig. 3b). Based on the above results, LAMP reactions were equally sensitive to PCR. Taken together, these results indicate that the sensitivities of LAMP methods developed in this work were compatible with PCR assays for the detection of mango DNA.
Effect of temperature range on the reactivity of LAMP assay
To assess the effect of temperature variation on the reactivity of the LAMP assay, several of the reaction temperatures for the initiation of LAMP were used. The temperatures ranging from 55 °C to 64 °C used for the reaction were all capable of producing LAMP products (Fig. 4). Compared to the intensity of DNA banding on the agarose gel, the LAMP product developed from the reaction temperature at 62.2 °C was higher than that of the others. In summary, broad reaction temperatures of LAMP were demonstrated herein for DNA amplification. This performance was applicable as a practical tool for the on-site detection of mango DNA without the strict requirement of temperatures.
Effect of the different heat-processing methods on mango authentication by LAMP assay
To investigate the effect of the heating process on mango authentication by LAMP assay, mango was taken to heat processing comprising a boiling bath and steam autoclave procedure. When LAMP primers were applied, Fig. 5a shows that the genomic DNA extracted from various mango samples with boiling processes for 20, 40, 60, 80, 100 and 120 min did not influence mango authentication by the LAMP assay (Fig. 5a). With regard to the DNA obtained from various steam-autoclaved mangos, heating times ranging from 20, 40, 60 to 80 min still revealed LAMP products on the agarose gel after performing the LAMP reaction (Fig. 6a). The PCR results for the detection of mango DNA were comparable to those of LAMPs, especially on the template DNA used in the reaction from boiled mango (Fig. 5b). These results specify that LAMP was well matched in strength to PCR for the authentication of mango DNA, as shown in Fig. 5a, b. It was worth noting that the specific PCR products were not detected in the mango samples subjected to a steam autoclave procedure for 20, 40 and 60 min (Fig. 6b). In contrast, LAMP patterns were detected in the obtained sample DNA from steam-autoclaved mango samples for 20, 40 and 60 min (Fig. 6a).
Analysis of LAMP (a) and PCR (b) products amplified from mango DNA after the boiling treatment. The LAMP product with a ladder-like pattern and a specific 178 bp PCR product was analysed by DNA electrophoresis. Lane M: 100 bp DNA ladder, lane N: negative control, and lane P: positive control. For the boiling procedure, mango samples were boiled for various times before DNA amplification. Different lanes represent mango samples boiled for 20, 40, 60, 80, 100 and 120 min
Analysis of LAMP (a) and PCR (b) products amplified from mango DNA after the steaming treatment. The LAMP product with a ladder-like pattern and a specific 178 bp PCR product was analysed by DNA electrophoresis. Lane M: 100 bp DNA ladder, lane N: negative control, and lane P: positive control. For the steaming procedure, mango samples were autoclaved for various times before DNA amplification. Different lanes represent that the mango sample was steamed for 20, 40, 60 and 80 min, respectively
Mango authentication in commercial mango foods by LAMP
To evaluate whether the LAMP assay established in this work was appropriate as an applied tool for identifying food ingredients containing mango, nine commercial mango foods were purchased from various local markets. Nine commercial mango foods, including dried mango, mango iced tea, mango chocolate ball, mango pocky, mango hi-chew candy, mango cake, mango jelly, mango crispy crepes, and mango jam, were extracted for total DNA for use in the LAMP reaction, and specific LAMP primers were used to authenticate the mango ingredients. As illustrated in Fig. 7a, five of nine commercial mango foods, including dried mango, mango pocky, hi-chew candy, mango cake and mango jelly, were identified and displayed typical gel patterns with positive results following the LAMP assay. For the PCR results, only three positive results, dried mango, mango pocky and hi-chew candy, were consistent with those from the LAMP detection (Fig. 7b). It was worth noting that the mango iced tea, mango chocolate ball, mango crispy crepes and mango jam were claimed and labelled as being artificial mango flavouring commercial foods. The above four commercial foods were not identified by LAMP and PCR as mango. The results obtained from the LAMP assay not only confirmed the PCR results, but LAMP can also be applied to more food authentication than that by PCR. Consequently, based on the established LAMP primers with specificity, sensitivity and validation, practical applications for mango authentication by LAMP can be used to provide more information on food allergens.
Electrophoretic analysis of LAMP (a) and PCR (b) products from commercial mango products. When mango authentication by LAMP and PCR was performed, the LAMP product with a ladder-like pattern and a specific 178 bp PCR product was analysed by DNA electrophoresis. Lanes M, N and P represent 100 bp of DNA ladder, negative control and positive control, respectively. Lanes 1–13 represent the sample DNA used for dried mango, mango iced tea, mango chocolate ball, mango pocky, mango hi-chew candy, mango cake, mango jelly, mango crispy crepes, and mango jam, respectively
Discussion
In recent years, LAMP has indicated its convenience and effectiveness in recognizing the identities of organisms in previously published studies [19,20,21,22,23,24,25]. Characteristically, LAMP belongs to an isothermal method for nucleic acid amplification since it was reported by Notomi et al. [18]. Thus, there is no need for a thermal cycler to amplify the DNA. Indeed, LAMP is quite easy to run for the on-site identification using simple heating with a water bath [18]. Herein, the specific aim was to develop a LAMP-based method with high specificity and rapid detection of mango DNA to avoid food allergens in the diet. Some molecular DNA techniques have been described to identify mango DNA [15,16,17]. However, to the best of our knowledge, this is the first report using LAMP to authenticate mango DNA in food. Mango has been reported as an allergen that causes anaphylaxis [11]. Thus, good food labelling might be a suitable way to advice customers on mango intake [26]. Before the bottleneck of protein-based diagnostic methods for mango are overcome, this developed LAMP technique for mango DNA detection can be applied in advance and practically to supervise and audit the correctness of food labelling, especially for food ingredients containing mango.
In this work, the consensus sequences of the internal transcribed spacer (ITS) of ribosomal DNA of mango were used. The ITS1 region is the most popular target chosen for mango identification using the DNA molecular technique. ITS has recently been recognized as marker DNA for species identification in previous studies [23, 24]. In our work, therefore, the ITS was used for designing LAMP primers for mango authentication. Thus, this paper positively established one set of LAMP primers that can be applied for the sensitive, specific, and rapid authentication of mango DNA. In fact, other DNA regions, such as ITS2 and profilin, were chosen to design LAMP primers. However, those ITS2- or profilin-based primer sets did not specifically allow the identification of mango DNA by LAMP (data not shown). Based on our previous preliminary data, ITS1 was chosen as a DNA marker for designing primers. After the validation of the LAMP primers, the specificity of LAMP was found to be compatible with the traditional PCR method for the identification of mango DNA. Moreover, the LAMP assay not only confirmed the PCR results but can also be applied to more food authentication than PCR (Fig. 7). Although the sensitivity of LAMP is not superior to PCR, LAMP can still be improved by involving loop primers to increase LAMP sensitivity [21]. It is worth noting that LAMP exhibited good effectiveness during the reaction within 1 h. Generally, conventional PCR requires 2–3 h for DNA amplification. Thus, LAMP is more suitable and flexible for on-site DNA detection in a short period than PCR using a simple heater instead of a precise thermal cycler. In addition, the LAMP assay did not strictly control the temperature of the reaction when LAMP was performed. In our case, the performing temperature of LAMP ranged from 55 to 64 °C. The performance of LAMP displayed its reproducibility at reaction temperatures between 55 and 64 °C (Fig. 4). Thus, LAMP might exhibit more tolerance to temperature variation during the initiation of DNA amplification. This advantage might reduce the appearance of false-negative results during authentication. In Taiwan, it is popular to purchase two other kinds of botanical varieties of mango, Irwin and Jin-Huang mangos, which are consumed depending on customer preference. Using the set of LAMP primers in this study, the amplicon DNA of the above mangos can be specifically detected by LAMP (Fig. 2b).
In this context, after boiling and steaming treatments, mango DNA was also detected when ITS1-based LAMP primers were used. Generally, processed commercial foods are composed of several ingredients in their formula. Thus, more processed food with complicated ingredients might become a problem and may interfere with their correctness during the identification of mango DNA. Mango DNA in commercial foods was verified by the LAMP method herein. It reflects the component of mango contaminated in the diet. This means that even though other DNA obtained from other ingredients were mixed with mango DNA, the LAMP specificity was not altered (Fig. 2c). This potential tool provides an alternative way for internal/external auditing to prevent allergen contamination by a food producer or government authorities [22, 24].
In summary, the LAMP-based method for mango DNA detection was developed in this study. This method can be applied as a potential tool to rapidly, specifically and sensitively authenticate mango DNA contained in mango-related foods to verify the presence of food allergens.
Conclusion
In this study, isothermal condition-based DNA amplification with high specificity and sensitivity was established for the rapid identification of mango DNA in food. This molecular method can be useful to set up a checkpoint of processed food for verifying the presence of mango.
References
Nutrient profile for mango from USDA SR-21, Available at https://nutritiondata.self.com/facts/fruits-and-fruit-juices/1952/2.
McGovern TW, LaWarre S (2001) Botanical briefs: the mango tree—Mangifera indica L. Cutis 67:365–366
Wu TC, Tsai TC, Huang CF, Chang FY, Lin CC, Huang IF, Chu CH, Lau BH, Wu L, Peng HJ, Tang RB (2012) Prevalence of food allergy in Taiwan: a questionnaire-based survey. Intern Med J 42:1310–1315
Hegde L, Venkatesh YP (2007) Anaphylaxis following ingestion of mango fruit. J Investig Allergol Clin Immunol 17:341–344
Food Standards Agency (2005) Draft of guidance on allergen management and consumer information. Available at http://www.reading.ac.uk/foodlaw/pdf/uk-05052-allergens.pdf
Shah A, Gera K (2014) Immediate hypersensitivity reaction with mango. Pneumonol Alergol Pol 82:445–453
Fasoli E, Righetti PG (2013) The peel and pulp of mango fruit: a proteomic samba. Biochim Biophys Acta 1834:2539–2545
Paschke A, Kinder ZK, Wigotzki M, Weßbecher R, Vieluf D, Steinhart H (2001) Characterization of allergens in mango fruit and ripening dependence of the allergenic potency. Food Agric Immunol 13:51–61
Andrade J, DeMagalhães T, Torres Toledo S, Beserra Nogueira BR, Cordenunsi FM, Lajolo ODN Jr (2012) 2D-DIGE analysis of mango (Mangifera indica L.) fruit reveals major proteomic changes associated with ripening. J Proteom 75:3331–3341
Renuse S, Harsha HC, Kumar P, Acharya PK, Sharma J, Goel, R, Kumar GS, Raju R, Prasad TS, Slotta T, Pandey A (2012) Proteomic analysis of an unsequenced plant—Mangifera indica. J Proteom 75:5793–5796
Besler M, Paschke A, Rodriguez J (2001) Allergen data collection: mango (Mangifera indica). Internet Symp Food Allerg 3:135–141
Vargas Correa JB, Sánchez Solís L, Farfán Ale JA, Noguchi H, Moguel Baños MT (1991) Vargas de la Peña, M.I. Allergological study of pollen of mango (Magnifera indica) and cross reactivity with pollen of piru (Schinus molle). Rev Alerg 38:134–138
Wellhausen A, Schöning B, Petersen A, Vieths S (1996) IgE binding to a new cross-reactive structure: a 35 kDa protein in birch pollen, exotic fruit and other plant foods. Z Ernahrungswiss 35:348–355
Song J, Zhang H, Liu Z, Ran P (2008) Mango profiling: cloning, expression and cross-reactivity with birch pollen profiling Bet v2. Mol Boil Rep 35:231–237
Srivastava N, Bajpai A, Chandra R, Rajan S, Muthukumar M, Srivastava MK (2012) Comparison of PCR based marker systems for genetic analysis in different cultivars of mango. J Environ Biol 33:159–166
Mansour H, Mekki LE, Hussein MA (2014) Assessment of genetic diversity and relationships among Egyptian mango (Mangifera indica L.) cultivers grown in Suez Canal and Sinai region using RAPD markers. Pak J Biol Sci 17:56–61
Souza IG, Valente SE, Britto FB, de Souza VA, Lima PS (2011) RAPD analysis of the genetic diversity of mango (Mangifera indica) germplasm in Brazil. Genet Mol Res 10:3080–3089
Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T (2000) Loop-mediated isothermal amplification of DNA. Nucleic Acids Res 28:e63
Huang CH, Lai GH, Lee MS, Lin WH, Lien YY, Hsueh SC, Kao JY, Chang WT, Lu TC, Lin WN, Chen HJ, Lee MS (2010) Development and evaluation of a loop-mediated isothermal amplification assay for rapid detection of chicken anaemia virus. J Appl Microbiol 108:917–924
Woźniakowski G, Tarasiuk K (2015) Visual detection of goose haemorrhagic polyomavirus in geese and ducks by loop-mediated isothermal amplification. Avian Pathol 44:311–318
Chen HJ, Lee MS, Lai JY, Lai GH (2015) Development of a loop-mediated isothermal amplification method for the rapid detection of the dioxin-degrading bacterium Ochrobactrum anthropi in soil. J Environ Manage 160:263–270
Lee MS, Huang JY, Lien YY, Sheu SC (2019) The rapid and sensitive detection of edible bird's nest (Aerodramus fuciphagus) in processed food by a loop-mediated isothermal amplification (LAMP) assay. J. Food Drug Anal 27:154–163
Lai GH, Chao J, Lin MK, Chang WT, Peng WH, Sun FC, Lee MS, Lee MS (2015) Rapid and sensitive identification of the herbal tea ingredient Taraxacum formosanum using loop-mediated isothermal amplification. Int J Mol Sci 16:1562–1575
Lee MS, Su TY, Lien YY, Sheu SC (2017) The development of loop-mediated isothermal amplification (LAMP) ssays for the rapid authentication of five forbidden vegetables in strict vegetarian diets. Sci Rep 7:44238
Lee MS, Hxiao HJ (2019) Rapid and sensitive authentication of Polygonum multiflorum (He-Shou-Wu) of Chinese medicinal crop using specific isothermal nucleic acid amplification. Ind Crop Prod 129:281–289
Vemula SR, Gavaravarapu SM, Mendu VV, Mathur P, Avula L (2014) Use of food label information by urban consumers in India– a study among supermarket shoppers. Public Health Nutr 17:2104–2114
Acknowledgements
This research was founded by Grants from the Ministry Science and Technology of Taiwan, ROC (Grant Number, MOST 103-2221-E-020-038-), and China Medical University (Grant Numbers CMU107-S-49, CMU 108-MF-91 and CMU 108-S-14).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declared that they have no conflict of interest.
Compliance with Ethics Requirements
This article does not contain any studies with human and animal subjects.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Sheu, SC., Tsou, PC., Lien, YY. et al. Rapid and specific detection of mango (Mangifera indica) in processed food using an isothermal nucleic acid amplification assay. Eur Food Res Technol 246, 759–766 (2020). https://doi.org/10.1007/s00217-020-03440-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00217-020-03440-z